Open Access Article
Open Access ArticleMass spectrometry-based metabolomics in health and medical science: a systematic review
Xi-wu Zhang,
Qiu-han Li,
Zuo-di Xu and
Jin-jin Dou*
Institute of Chinese Medicine, Heilongjiang University of Chinese Medicine, Heping Road 24, Harbin 150040, China. E-mail: doujinjin1980@126.com; Fax: +86-451-87266827; Tel: +86-451-87266827
First published on 17th January 2020
Abstract
Metabolomics is the study of the investigation of small molecules derived from cellular and organism metabolism, which reflects the outcomes of the complex network of biochemical reactions in living systems. As the most recent member of the omics family, there has been notable progress in metabolomics in the last decade, mainly driven by the improvement in mass spectrometry (MS). MS-based metabolomic strategies in modern health and medical science studies provide innovative tools for novel diagnostic and prognostic approaches, as well as an augmented role in drug development, nutrition science, toxicology, and forensic science. In the present review, we not only introduce the application of MS-based metabolomics in the above fields, but also discuss the MS analysis technologies commonly used in metabolomics and the application of metabolomics in precision medicine, and further explore the challenges and perspectives of metabolomics in the field of health and medical science, which are expected to make a little contribution to the better development of metabolomics.
1 Introduction
Metabolomics is a post-genomics discipline that combines high-throughput analysis techniques with bioinformatics, aimed at establishing high-speed, high-throughout, and comprehensive analysis of metabolites in biological samples. In the past two decades, great breakthroughs have been made in metabolomics that have propelled its evolution into an important tool in the field of medicine, especially in the study of disease-related biomarkers, toxicology, molecular mechanisms, or to provide more detailed information on human biochemistry.1,2 The science of metabolic phenotypes is beginning to show real value in explaining clinical and biological problems.3,4One main advantage of metabolomics is its ability to detect responses even when growth phenotypes are lacking, thus increasing functional readouts by orders of magnitude compared with traditional chemical or genetic growth screening. In any case, by revealing the composition, interaction or change of metabolites, the state of a given system can be described. Metabolomics can be a powerful tool for describing complex phenotypes and evaluating unique physiological responses.
The two main analytical techniques for metabolic analysis are 1H nuclear magnetic resonance (NMR) and MS. Both of these techniques can analyze a large number of small molecules coexisting in complex samples, including metabolite identification and quantification. NMR spectroscopy can directly identify and quantitatively analyze numerous analytes. Although the reproducibility of NMR is better than that of MS-based techniques, the sensitivity of NMR is lower and a larger sample size is required. In the field of medical science, clinical samples are particularly valuable; thus MS-based metabolomic technology is widely used in clinical research.5
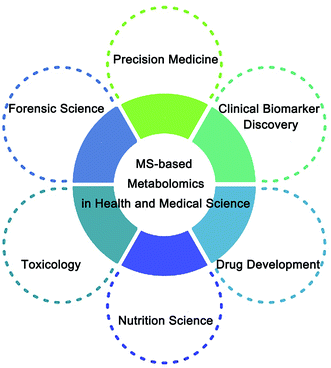
In this review, multiple MS-based analytical platforms for metabolomics are summarized, and the applications of metabolomics in health and medical fields are systematically introduced (Fig. 1). Furthermore, we discuss its limitations and provide a perspective for future developments in MS-based metabolomics.
2 The dilemma of health and medical science
The major health problems are no longer nutritional deficiencies, but how to promote health, how to effectively prevent diseases in advance, as well as the establishment and implementation of individualized therapies. Besides physical exercise to improve health, improvements in personal nutrition should also be strengthened. The focal point of disease prevention is to avoid the causes of the risk factors of disease, which mainly include three categories: primary prevention focuses on the various determinants around the entire population or high-risk groups. Secondary prevention comprises early detection and intervention. The third prevention objective is to improve recovery rates and reduce the risk of recurrence.As mentioned above, human health depends not only on genes, but also on the interaction of genes with different factors (lifestyle, environment, intestinal microflora, etc.). How to reflect the relationships and laws among them in scientific language is the key point that needs to be clarified at present, which will help us understand diseases, as well as implementing precision medical care.
3 Metabolomics in precision medicine
3.1 Pharmacometabolomics
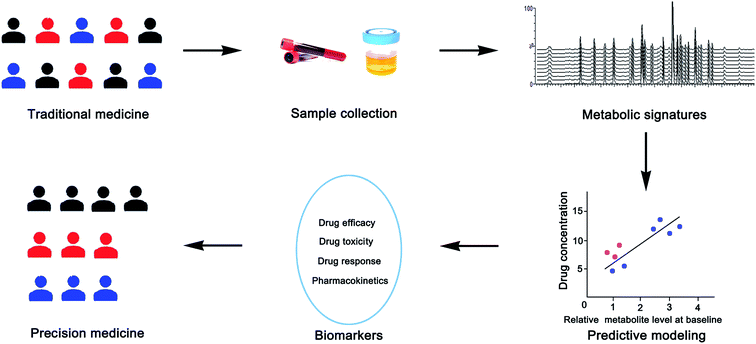
Pharmacometabolomics is considered to be the youngest and most promising branch of metabolomics.6 Pharmacometabolomics uses metabolomics to study the efficacy of drugs and helps us understand the mechanism of action and identify biomarkers associated with the organism's response to drug treatment, and further helps doctors predict the effectiveness and toxicity of therapeutic drugs to individuals.7–9 It can be used to monitor the body's response to drugs, and the response to a variety of external factors, which allows researchers to assess the drug's effectiveness and the patient's disease status. Therefore, it may be used to better define the mechanisms associated with different drug reactions in patients. Pharmacometabolomics is an effective complement to therapeutic drug monitoring (TDM) in clinical individualized drug therapy. TDM can be used to establish initial dosage regimens and to monitor drugs, while pharmacometabolomics can monitor a drug's levels and metabolites, and effectively predict the efficacy and toxicity of drugs, thus avoiding adverse reactions.10 A typical schematic diagram of pharmacometabolomic support for precision medicine is shown in Fig. 2.3.2 Precision medicine
With the continuous progress in medical treatment, the heterogeneity of disease is widely accepted; in other words, the same disease might have different effects on different individuals. One area in which metabolomics affects precision medicine is in drug response and monitoring. Since drug metabolism involves various enzymes, multiple organs, and varies with gender, age, diet, etc., it will be very difficult to predict the drug response of an individual based on genotype alone.10,11 Nevertheless, metabolomics can assess the sum of genotype, environmental and physiological effects, and can be applied to directly monitor drug reactions as well as to determine drug doses. The aim of metabolomics is not only to find novel diagnostic biomarkers to enable the early detection of disease, but also to find appropriate biomarkers for the selection of appropriate interventional therapies and the rational evaluation of current therapeutic responses.12,13 These characteristics of metabolomics provide a powerful tool for precision medicine. Metabolomics can reflect the interaction between the genetic and environmental effects of a downstream metabolic phenotype at a specific time.3.3 Clinical applications of metabolomics in precision medicine
Pharmacometabolomics has yielded certain exciting and medically useful results in drug dosage and response measurement. For instance, in the field of breast cancer treatment, an MS-based pharmacometabolomics for monitoring tamoxifen and its metabolites was used to predict the side effects and discontinuation rate.14 Side effects have been considered to be a significant predictor of tamoxifen discontinuation; thus, there is a need for reliable predictive factors of side effects of the anti-estrogen treatment.15 In the present study, over a 6 year period, longitudinal serum samples, patient-reported outcome measures and pharmacy records from 220 breast cancer patients were obtained. The serum concentrations of tamoxifen metabolites were measured by LC-MS/MS. After 2 years of follow up, a high concentration of tamoxifen, Z-4′-OHtam and tam-NoX had a profound association with vaginal dryness. This real-word data study suggested the pharmacometabolomic-based drug monitoring might be a helpful instrument for predicting side effects. A similar method also arose for drug monitoring in patients with Alzheimer's disease, whose dosage monitoring and drug efficacy are often difficult to estimate.16Spironolactone is the most effective drug for the treatment of resistant hypertension (RH), but there are differences between patients, as not all patients show a response, and the side effects are not negligible. M. Martin-Lorenzo et al. explored a metabolic prediction of treatment response.17 Urine was collected from 29 patients with RH and 13 patients with pseudo-RH. Samples were collected before and after administration of spironolactone, and RH patients were further classified as responders or non-responders. The contents of citrate and oxaloacetate were increased in RH patients, and together with α-ketoglutarate and malate, these 4 significantly changed metabolites showed an ability to discriminate between responders and non-responders with the area under curve (AUC) being 0.96. The results suggested that a disorder in the citric acid cycle and reactive oxygen species homeostasis control remained activated after the development of hypertension. A metabolomics study showed the changes before spironolactone treatment and predicted the future reactions of patients, and these markers will help to optimize the treatment with spironolactone in RH patients.
4 Analytical techniques employed in MS-based metabolomics
MS is widely used in metabolomics due to its better sensitivity, high throughput, and its ability to detect more molecules in complex biological samples. The continuous development of MS technology is an important driving force for the development of metabolomics. Here we give a brief overview of three commonly used MS techniques in metabolomics, and Table 1 gives a brief comparison of these three MS-based metabolomic technologies.| Technology | Advantages | Disadvantages |
|---|---|---|
| GC-MS | • Mature technology | • Requires sample derivatization |
| • Comparatively cheap | • Relatively long time for sample analysis | |
| • Excellent separation reproducibility | • Unable to produce parent ions | |
| • Suitable for the detection of volatile metabolites | • Novel compound identification is difficult | |
| • Universal databases facilitate metabolite identification | ||
| LC-MS | • Excellent sensitivity | • Comparatively expensive |
| • Simple sample pretreatment process | • Lower reproducibility than GC-MS | |
| • Wide coverage of metabolite detection | • Not compatible with volatile metabolites | |
| • Relatively short time for sample analysis with sub-2 μm stationary phase particles | • Novel compound identification is difficult | |
| • Matching with multiple MS detectors | ||
| IMS | • In situ detection | • Ion suppression |
| • Providing location information of metabolites | • Imaging quality is affected by resolution | |
| • Relatively long time for sample analysis |
4.1 Gas chromatography-mass spectrometry (GC-MS)
GC-MS is the most mature chromatographic-MS technology.18 Compared with other technologies, GC-MS has the advantages of being relatively inexpensive and easy to operate, and better stability and repeatability. The principle of GC-MS determines that GC-MS can only analyze volatile substances or substances which are easily volatilized after derivatization. The most classical derivation method of GC-MS is the two-step method: oximation and silylation. The derivatization reaction contributes to the isolation and detection of metabolites.19 With the derivatization, GC-MS can also be used to analyze not only volatile and non-polar compounds, but also polar compounds such as fatty acids, amino acids, amines, sterols, and sugars.20,21 GC-MS has various commercial databases, such as NIST22 and Fiehn,23 which contain standardized EI source fragmentation information that can be considered to be its unique advantage over other MS-based technologies.24 A spectrum library search not only allows the direct identification of unknown compounds but also helps the characterization of chemical types or groups with similar structures to known constituents.4.2 Liquid chromatography-mass spectrometry (LC-MS)
Due to different ion sources and working modes of GC-MS and LC-MS, the data forms of the two platforms are also different. As to different ion scanning ranges, the data volume obtained by GC-MS is usually less than that for LC-MS. In addition, LC-MS has a higher tolerance to volatility, a wider metabolite coverage, and simpler approaches to sample preparation. Especially with the advent of UPLC, the separation degree, peak capacity and sensitivity have been greatly improved, and can withstand higher pressures in the range of 6000–19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 psi.25,26 At present, the UPLC system is the mainstream technology used in metabolomic research. Different chromatographic columns can be selected according to the polarity of the analytes. A reversed-phase chromatographic column can be used to analyze lipid-soluble metabolites, such as C18 or C8 stationary phases. Currently, most UPLC-MS-based metabolomic research has been carried out with a C18 column.27 However, some highly polar and charged metabolites are difficult to retain in a C18 column; thus, a hydrophilic chromatographic column is used to analyze hydrophilic metabolites. In addition, 2-dimensional/multi column approaches can be used to obtain more information about metabolites.28
000 psi.25,26 At present, the UPLC system is the mainstream technology used in metabolomic research. Different chromatographic columns can be selected according to the polarity of the analytes. A reversed-phase chromatographic column can be used to analyze lipid-soluble metabolites, such as C18 or C8 stationary phases. Currently, most UPLC-MS-based metabolomic research has been carried out with a C18 column.27 However, some highly polar and charged metabolites are difficult to retain in a C18 column; thus, a hydrophilic chromatographic column is used to analyze hydrophilic metabolites. In addition, 2-dimensional/multi column approaches can be used to obtain more information about metabolites.28
4.3 Imaging mass spectrometry (IMS)
Metabolite imaging involves the in vivo or in vitro detection and visualization of metabolites in tissues or cells. Whereas MS-based imaging techniques are usually used in in vitro detection. Imaging mass spectrometry (IMS) has been developed and applied to analyze the spatial distribution of chemical composition and molecular mass, and this is a relatively exciting and novel metabolomic technology. By using IMS, the spatial distribution of metabolites in tissues or cells can be visualized. Matrix assisted laser desorption ionization (MALDI) IMS is currently the most common commercial IMS mode, with the highest m/z range of 500 kDa and a spatial resolution of 20 μm. Continuing developments in MALDI-based imaging, such as matrix-free approaches, faster scan rates and smaller spot size, and improved software, have resulted in more explicit images with wider metabolite coverage.29 It seems that IMS might revolutionize the field of histology, just as immune-fluorescence staining did in the 1990s.The IMS technique may serve as a powerful tool to visualize the distribution of endogenous metabolites over time and space. MALDI imaging has been combined with MRI in the diagnosis of colon cancer, and can also be extended to whole-body imaging of vinblastine, such as in rats.30 IMS technology has become a high-throughput molecular histological research tool for molecular localization and function recognition. Through comprehensive and high-throughput characterization of metabolic changes in micro regions, IMS technology provides a new method and perspective for the study of metabolic mechanisms in disease. It is worth noting that the key challenges in MS imaging include: sample preparation (including optimization of cell fixation), statistical analysis of large datasets, and the balance between spatial resolution of tissue slice imaging and sensitivity of analyte detection.
In addition to the extensively used MS techniques mentioned above, some other platforms have also made significant progress in metabolomic applications, such as direct injection MS31 and capillary electrophoresis-mass spectrometry (CE-MS).32 Since there is no universal MS analytical technique for measuring each metabolite, users should choose the most suitable instrument for their research, and according to demand, give this comprehensive consideration, including resolution, sensitivity, throughput and cost.
The development of these MS-based metabolomic technologies has had a significant impact on several areas of biomedicine, with these advances not only making metabolomics more accessible and powerful, but also altering our understanding of disease, methods of drug discovery, and perspectives on the means of providing health care.
5 Application of MS-based metabolomics in medical science
Unlike other “omics” sciences, metabolomics can link both gene and environmental interactions. It represents not only the downstream output of the genome, but also the upstream input of the environment.33 In recent years, metabolomics has been applied in multiple fields, on account of its capability of detecting subtle changes in large datasets through comprehensive metabolic measurements. In general, MS-based metabolomics is a valuable implement in medical science, such as clinical biomarker discovery, drug development, nutrition science, toxicology, and forensic science.5.1 Metabolomic analysis for clinical biomarker discovery
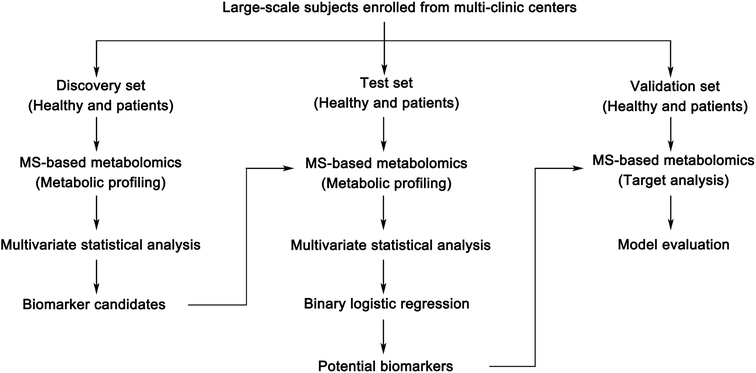
Success in treatment depends on the extent to which the disease is transmitted to the body, such as the stage of the disease; therefore, the early diagnosis of disease is particularly important. The life expectancy of patients can be improved by identifying specific biomarkers for disease diagnosis in the initial stage of a disease. Diagnosis of disease usually requires invasive examination, such as biopsies, or computed tomography scans, for which patients usually consider their radiation exposure. In 1846, the first tumor marker – the Bence Jones protein, confirmed by H. Bence-Jones – had a diagnostic value for multiple myeloma. But early diagnosis is not always feasible because the disease sometimes lacks typical clinical symptoms.34 One hundred years later, with the development of metabolomics, efficient screening of biomarkers has become a great possibility.35 Biomarkers have prospects for broad clinical application, including the following aspects: screening/diagnosis/detection, prognosis and prediction, and monitoring disease progress.35–37 Successful screening of biomarkers is very beneficial to clinics. It can provide prognostic information about disease outcomes, select and evaluate targeted therapies, and provide early intervention for patients, to help clinical decision-making and improve clinical outcomes. Herein, we briefly introduce the key developments in the discovery of clinical biomarkers in the past few years with MS-based metabolomic technologies that might pioneer clinical biomarkers in the future. Furthermore, the process of discovering clinical biomarkers based on MS-based metabolomics is summarized as Fig. 3.Metabolic phenotypes provide a new dimension in patient information, enabling patients to be stratified according to their metabolic profiles. Biomarkers for the diagnosis and prognosis of disease can play an important role in individualized treatment and precision medicine. Although the development prospects of biomarkers are tremendous, there are still many problems in the current stage. First, it is essential to select preclinical samples for biomarker detection. Large-scale, multi-center and comprehensive clinical information is a necessary factor. However, due to the cost and availability of samples, these classical large-scale cohort studies often use a single time point sample for metabolomic research. Samples of sequential points can increase the reliability of the results and contribute to research into disease progression. Moreover, although effective biomarkers are being used in clinical practice, such as carbohydrate antigen 125 for the early diagnosis of ovarian cancer and serum creatinine as an indicator of renal function,61 the development of these markers requires years and large-scale, multidisciplinary efforts. Among numerous potential biomarkers, less than 1% entered clinical practice.62 This not only indicates that it is very difficult to convert biomarkers into practical clinical applications, but their discovery and development are similar to drug discovery, and this is a great challenge, requiring a large amount of financial investment, rigorous science and technology, as well as networks of clinicians, biologists, specialists in bioinformatics, statisticians, clinical chemists and other experts.
5.2 Metabolomics analysis for drug development
Modern drug development involves many disciplines, including biology, chemistry and pharmacy. Traditional processes are needed to screen drugs in molecular libraries, to optimize hits, and search for molecular affinity, selectivity, metabolic stability, oral bioavailability, etc. Only after satisfying the above conditions can a small percentage of molecules enter the stage of being candidate drugs for clinical trials, with some of them eventually failing in phase III trials.63 Metabolomics, as a branch of systems biology, not only discovers exciting opportunities in diagnosis, but the application of toxicity/efficacy biomarkers in drug discovery has significantly accelerated the pace of drug discovery, and contributed to the formulation of appropriate clinical plans. By examining the changes in metabolic disturbances in vivo and in vitro, metabolomics can provide information on the efficacy and safety of drugs, which may not be available in other research processes. This function of metabolomics is usually used in preclinical studies of drug development, focusing on the safety of preclinical and translational target engagement biomarkers, as well as playing a vital role in the discovery of drug side effects. Furthermore, the results of a great number of metabolomic studies have broadened the understanding of the complexity and heterogeneity of diseases, thus contributing to the discovery of new targets in drug development.Metabolomics is often used in drug development to study the function of metabolic enzymes and to identify novel drug targets. Many drugs used clinically are enzyme inhibitors that regulate the activity of metabolic enzymes and maintain metabolic balance. A good example of the application of metabolomics in drug development is Enasidenib and Ivosidenib, two first-class drugs for the treatment of isocitrate dehydrogenase-1 (IDH-1) and IDH-2-mutated recurrence or refractory acute myeloid leukemia (AML). Dang et al.64 and Ward et al.65 with the aid of MS-based metabolomics demonstrated that the mutant enzymes of IDH-1 and IDH-2 can convert α-ketoglutarate into 2-hydroxyglutarate, which was considered to be an oncometabolite in glioma and AML. Subsequent studies confirmed that inhibitors of mutant IDH-1 inhibited the production of 2-hydroxyglutarate and the growth of tumors, giving prominence to the value of metabolomics in discovering new therapeutic targets.66 After a series of studies and clinical trails, Enasidenib, a new drug treatment for relapsed or refractory AML was approved. Ivosidenib, a sister drug to Enasidenib, an inhibitor for mutant IDH-1, was subsequently approved by the FDA.67 Metabolomics has been applied to almost every stage of the development of mutant IDH-1 and IDH-2 inhibitors, and has brought about new therapeutic options for relapsed or refractory AML patients.
5.3 Metabolomics analysis for nutrition science
Epidemiological and clinical studies have provided clear evidence since the 1970s that many diseases with high morbidity and mortality are related to diet, including diabetes, cardiovascular diseases and a range of cancers.68 It is known that selected foods, nutrients, and dietary patterns interact with various metabolic processes to reduce or increase the risk of disease: for instance, high salt intake increases blood pressure,69 and high red meat intake increases the incidence of type 2 diabetes mellitus,70 cardiovascular disease,71 and cancers.72Diet, dietary patterns and other environmental factors play a vital role in the prevention and development of many diseases. Food intake is an important environmental factor, because food ingredients may alter gene expression and structural modification, thereby altering the composition of proteins and metabolites. Thus, it is an important part of health-related research. Metabolomics, as an experimental method in nutrition science, is gradually maturing.73 It is a useful method of analysis to reveal dietary changes and related systemic biological results. Nutrition metabolomics has become a high-throughput and sensitive method for identifying and characterizing biochemical pathways.74 It is also the basis for revealing the complex relationship between dietary exposure and chronic diseases with metabolic phenotypic changes.75
The emergence of nutritional metabolomics has two main objectives: (1) to determine the effects of dietary compounds on host metabolism within a certain period of time; (2) to determine the biomarkers of dietary intake or phytochemical dose-dependent metabolites.76,77 The application of metabolomics in nutrition research can generally be divided into four main fields: identification of dietary biomarkers, identification of dietary-related diseases and disease biomarkers, application of nutrition as a tool to identify molecular mechanisms in dietary intervention research, and precision nutrition.78,79 These areas are mainly focused on determining the effects of dietary compounds on the host metabolism after consumption for a certain period of time, and determining biomarkers of metabolites related to dietary intake or phytochemical dose dependence, which have a significant impact on a study of the diet–health relationship.80
Compared with traditional dietary assessment methods, the application of metabolomics provides a more objective dietary intake measurement method for the identification of dietary intake biomarkers, which greatly improves our ability to assess dietary intake.81
At present, the discovery of dietary biomarkers is mainly concentrated on single biomarkers. To give impetus to this area, combinations or panels of biomarkers might provide more accurate measurements of dietary exposure.82 The application of metabolomics in dietary intervention research enables researchers to study the mechanism of intervention and thus ascertain the effects of diet/food on metabolic pathways. Precise nutrition can be defined as customized dietary advice for individuals. A recent study showed that personalized dietary recommendations improved individual eating habits in a randomized controlled trial compared with general healthy dietary guidelines,83 showing a vital role in the development and provision of precise nutrition.
Metabolomics has broad prospects for application in nutritional epidemiology, and it is expected that it will play a leading role in explaining the interaction between diet and health. It is necessary to validate the new biomarkers found in a study, and long-term studies are needed to identify dietary biomarkers that reflect habitual dietary intake. Nutrition science will benefit greatly from the appropriate application of metabolomics.
5.4 Metabolomic analysis for toxicology
Metabolomics has gradually expanded from the initial screening of biomarkers for diseases to the field of toxicology research.84,85 In toxicology, metabolomics is the most relevant discipline to classical knowledge about disturbed biochemical pathways. It can quickly identify potential targets of dangerous compounds and provide information about target organs, evaluate toxicity changes in organisms from a holistic perspective, predict toxicity and discover toxicity-related biomarkers, which are helpful to improving our understanding of the toxicity mechanisms of given compounds.Incidents with Yunnan Baiyao and other safety problems with traditional Chinese medicines (TCM) occur frequently, which seriously hinders the development and clinical application of TCMs. Toxicity and adverse reactions of TCM have become a research hotspot in academia. In virtue of the complex composition of TCM, its multi-component and multi-target mode of action coincides with the ideas of metabolomics, and the use of metabolomics to study the toxicity of TCM has become a prevalent technical method. Yunnan Baiyao is a traditional Chinese prescription with the magical effect of promoting blood clotting and hemostasis. But its clinical application has been widely questioned because it contains the poisonous TCM of Aconiti kusnezoffii radix. Correct assessment of the toxicity of Yunnan Baiyao will not only help to eliminate misunderstanding about Yunnan Baiyao, but also promote the development of TCM. Ren et al. performed MS-based metabolomics to study the toxic effects of Yunnan Baiyao on normal rats.86 According to the results of the study, the toxicity of Aconiti kusnezoffii radix was observed earlier than in histopathology. Moreover, by comparing the changes in metabolite callback under the action of Aconiti kusnezoffii radix, processed Aconiti kusnezoffii radix, Yunnan Baiyao and Yunnan Baiyao lacking Aconiti kusnezoffii radix, with a secondary focus on metabolic pathways, the key metabolic pathways that may play a role in attenuation were determined. Furthermore, the compatibility and detoxification effects of Yunnan Baiyao were clarified. However, this study only clarifies one aspect of the mechanism of compatibility and detoxification of Yunnan Baiyao, and corresponding animal models should be established to evaluate the toxicity of Yunnan Baiyao under the effective state of treatment, which will make the study more comprehensive.
A series of clinical and experimental poisoning cases with cinnabar and realgar are related to overdose or long-term use of drugs.87 An-gong Niuhuang Wan just contains these two poisonous TCMs; moreover, they are the active ingredients in An-gong Niuhuang Wan for neuroprotection.88 Nevertheless, the toxicity of cinnabar and realgar can be reduced by compatibility with other TCMs. Xia et al. established an ultra-high performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UPLC/Q-TOFMS)-based metabolomics approach to reveal the protective role of other herbs in An-gong Niuhuang Wan against the hepatorenal toxicity of cinnabar and realgar.89 The results showed that the toxicity of the prescription was lower than that of cinnabar and realgar alone. In addition, other TCMs have anti-inflammatory and attenuation effects by regulating the metabolism of glycerophospholipid, arachidonic acid, linoleic acid, ether lipid and sphingolipid. These studies provide more scientific evidence for the safety of clinical applications of TCM.
5.5 Metabolomics analysis for forensic science
The overall objective of forensic science is to provide tools for all actors involved in criminal investigations and procedures in order to achieve fairer tracking, effective investigation of criminal science, and evidence for prosecution for or elimination from a suspected crime.90 The characteristics of most forensic samples are complex and they are available in trace amounts. Sample types include blood, urine, saliva, faeces, tissues, etc., similar to the samples in metabolomics. Furthermore, MS is undoubtedly a powerful analytical technology better suited to the exigencies of forensic samples and analyte identification.The identification of endogenous and exogenous chemicals embodied in latent fingerprints is vital for forensic science to obtain evidence of criminal identities and contact with specific chemicals. Groeneveld et al. performed a “basics” and the potential of MALDI IMS for the detection of drugs and their metabolites in fingermarks.91 In this study, 17 compounds from five different categories of drugs, amphetamine, alkaloids, opioids, cannabinoids and specialty drugs, were selected. The number of species included in MALDI-MS profiling and imaging analyses represents the importance of detecting forensic-related substances, such as drug abuse and its metabolites, in pre-developed marks. The overall results showed that, although the drug signal intensity is much lower in many cases, as a pre-development effect, their intensity was still enough to provide an image of the ridge detail. The data presented by this research confirmed the powerful potential of implementing MALDI IMS in forensic casework.
Opiates are derivatives of opium alkaloids, the most representative of which are morphine, codeine and heroin. Heroin is the second most popular recreational drug, more abusers die each year from heroin abuse than from any other illicit drug, and it causes a great many people to commit crimes. With the aid of MS-based metabolomics, heroin abuse and related mechanisms were gradually revealed.92 The biomarkers of 6-acetylmorphine, morphine, codeine, codeine-6-glucuronide, 6-acetylcodeine, noscapine, papaverine, and thebaine have been utilized and recommended for obtaining the most reliable forensic results. Furthermore, profound alterations in L-tryptophan, 5-hydroxytryptamine and 5-hydroxyindoleacetate were suggested as potential non-specific biomarkers of long-term heroin addiction.93 The above-mentioned exogenous components and endogenous metabolites will undoubtedly help to reveal a link between metabolic disorders and addiction, with relevant clinical and forensic implications. In light of drug dependence being a highly complex disease, combining reward effects, tolerance, and withdrawal effects, longitudinal studies need to be carried out to approve the aforementioned exogenous components and endogenous metabolites. In addition, with the emergence of new drugs in the clandestine market, and new degradation pathways of existing compounds, it is a great challenge for MS-based metabolomics and a never-ending task for analytical chemists to develop the most appropriate method.
6 Limitations and future perspective
Nowadays, in the medical science field, people have gradually become interested in describing the characteristics of health status at the molecular level in order to develop personalized treatments, early diagnosis, nutritional adjustments and rapid and sensitive detection in forensic science, etc. MS-based metabolomics, as a vital part of systems biology, has made remarkable progress in health and medical science. However, limitations and challenges still exist in its continuing exploration.In clinical research, it is necessary to further explore the effects of race, gender, age, dietary intake, physical activity, intestinal microflora, and stress, so as to promote an understanding of human metabolome, realize the real integration of metabolomics and big data, and thus improve data elucidation. The acquisition speed, accuracy, sensitivity and coverage of MS still require further improvement. New processes for the accurate identification of metabolites, rapid acquisition methods, and novel detection methods with high derivative efficiency should be developed to achieve large-scale coverage of metabolites and to facilitate data processing. Although metabolomics can identify a great quantity of metabolites, researchers usually do not identify metabolites that are not found in databases or websites. The exploration of unknown metabolites is a real challenge and research direction in metabolomics. In addition, some standards are difficult to obtain, and the existence of cis- and trans-isomers can also lead to inaccurate identification of metabolites. Thus, it is necessary to speed up the construction of standard product databases and information sharing.
Current MS-based metabolomic analysis mostly adopts relative quantification or qualitative analysis with fewer examples of quantitative analysis. The accurate concentration of metabolites in vivo has an important influence on the interpretation of metabolite function. How to simultaneously quantify multiple metabolites is the short board of current metabolomics? The ideal state is to use stable isotope-assisted metabolomics to track reaction substrates and determine the role of metabolites in metabolic pathways.94 Furthermore, the diversity of many enzyme substrates, in addition to the complexity of metabolic networks, make it difficult to influence the content of a single metabolite without affecting others. This in turn increases the difficulty in assessment of metabolite function. Moreover, we should also pay attention to the analysis of precious/scarce available samples. The information contained in these samples is of great scientific value, and might be key to solving some difficult problems.
It must be admitted that metabolomics is not a separate discipline, and the future direction of development must be a combination of multiple omics, so as to better understand the biological system. In order to better implement and simplify multi-omics technology, more vigorous computing power and new bioinformatic algorithms need to be developed. For metabolomics, especially for metabolomic technology applied in health and the medical science field, a series of common criteria should be established for metabolomic research design, method optimization and data processing etc., so as to ensure the authenticity and validity of the research and data sharing in different laboratories.
Metabolomics can undoubtedly help accelerate drug development, reducing costs and time. Introducing new biomarkers into existing biomarkers will help to assess disease-related risks, as well as being used in precision medicine, selecting appropriate treatment methods for patients, and monitoring their response to treatment. Metabolomics will also have a great impact on the development of other medical fields, paving the way for new medical strategies. With the development of the medical health industry entering a new era, the emergence and development of new technologies, such as big data, artificial intelligence and other new technologies, will surely release a huge space for the improvement and optimization of medical services. In the future, automation and artificial intelligence will also enter into the sample preparation process and the complex and massive data analysis process.
7 Conclusion
MS-based metabolomics technology has provided exciting opportunities in the field of health and medical science. As mentioned above, opportunities and challenges coexist. It is believed that, with the continuous progress in technologies, there will be more and more effective biomarkers for the diagnosis of clinical diseases, the widespread implementation of personalized treatment, improvement in national nutrition and physical fitness, the scientific assessment of toxic substances and breakthroughs in forensic medicine. There are grounds to expect that MS-based metabolomics technology will be gradually applied in clinical practice.Abbreviations
| AFP | α-Fetoprotein |
| AML | Acute myeloid leukemia |
| AUC | Area under curve |
| AD | Alzheimer's disease |
| CA19-9 | Carbohydrate antigen 19-9 |
| CAD | Coronary artery disease |
| CE | Capillary electrophoresis |
| CEA | Carcinoembryonic antigen |
| CFS | Cerebrospinal fluid |
| CKD | Chronic kidney disease |
| HCC | Hepatocellular carcinoma |
| ICC | Intrahepatic cholangiocarcinoma |
| IHD | Isocitrate dehydrogenase |
| IMS | Imaging mass spectrometry |
| GC | Gas chromatography |
| GSC | Gastric cancer |
| LCA | Lung cancer |
| LC | Liquid chromatography |
| MALDI | Matrix assisted laser desorption ionization |
| MS | Mass spectrometry |
| NMR | Nuclear magnetic resonance |
| OA | Osteoarthritis |
| RH | Resistant hypertension |
| TCM | Traditional Chinese medicines |
| TDM | Therapeutic drug monitoring |
| TFA | Nonesterified and esterified fatty acids |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by grants from the Key Program of Natural Science Foundation of State (Grant No. 81873161), the Chinese Postdoctoral Science Foundation (2018W631), and the Natural Science Foundation of Heilongjiang Province (H2018050, LH2019H102).References
- B. Hocher and J. Adamski, Metabolomics for clinical use and research in chronic kidney disease, Nat. Rev. Nephrol., 2017, 13(5), 269–284 CrossRef PubMed.
- C. H. Johnson, J. Ivanisevic and G. Siuzdak, Metabolomics: beyond biomarkers and towards mechanisms, Nat. Rev. Mol. Cell Biol., 2016, 17(7), 451–459 CrossRef PubMed.
- P. D. Rainville, G. Theodoridis and R. S. Plumb, et al., Advances in liquid chromatography coupled to mass spectrometry for metabolic phenotyping, TrAC, Trends Anal. Chem., 2014, 61, 181–191 CrossRef.
- M. Fessenden, Metabolomics: small molecules, single cells, Nature, 2016, 540(7631), 153–155 CrossRef PubMed.
- A. D. Kennedy, B. M. Wittmann and A. M. Evans, et al., Metabolomics in the clinic: a review of the shared and unique features of untargeted metabolomics for clinical research and clinical testing, J. Mass Spectrom., 2018, 53(11), 1143–1154 CrossRef.
- T. A. Clayton, J. C. Lindon and O. Cloarec, et al., Pharmaco-metabonomic phenotyping and personalized drug treatment, Nature, 2006, 440(7087), 1073–1077 CrossRef.
- I. Du Preez and D. T. Loots, Novel insights into the pharmacometabonomics of first-line tuberculosis drugs relating to metabolism, mechanism of action and drug-resistance, Drug Metab. Rev., 2018, 50(4), 466–481 CrossRef.
- Q. Huang, J. Aa and H. Jia, et al., A Pharmacometabonomic Approach To Predicting Metabolic Phenotypes and Pharmacokinetic Parameters of Atorvastatin in Healthy Volunteers, J. Proteome Res., 2015, 14(9), 3970–3981 CrossRef.
- A. Backshall, R. Sharma and S. J. Clarke, et al., Pharmacometabonomic profiling as a predictor of toxicity in patients with inoperable colorectal cancer treated with capecitabine, Clin. Cancer Res., 2011, 17(9), 3019–3028 CrossRef.
- R. Kaddurah-Daouk, R. Weinshilboum and Pharmacometabolomics Research N, Metabolomic Signatures for Drug Response Phenotypes: Pharmacometabolomics Enables Precision Medicine, Clin. Pharmacol. Ther., 2015, 98(1), 71–75 CrossRef.
- E. E. Balashova, D. L. Maslov and P. G. Lokhov, A Metabolomics Approach to Pharmacotherapy Personalization, J. Pers. Med., 2018, 8(3), 28 CrossRef.
- M. Jacob, A. L. Lopata and M. Dasouki, et al., Metabolomics toward personalized medicine, Mass Spectrom. Rev., 2019, 38(3), 221–238 CrossRef.
- L. Puchades-Carrasco and A. Pineda-Lucena, Metabolomics Applications in Precision Medicine: An Oncological Perspective, Curr. Top. Med. Chem., 2017, 17(24), 2740–2751 CrossRef.
- T. Helland, K. B. Hagen and M. E. Haugstoyl, et al., Drug monitoring of tamoxifen metabolites predicts vaginal dryness and verifies a low discontinuation rate from the Norwegian Prescription Database, Breast Cancer Res. Treat., 2019, 177(1), 185–195 CrossRef.
- E. A. Bowles, D. M. Boudreau and J. Chubak, et al., Patient-reported discontinuation of endocrine therapy and related adverse effects among women with early-stage breast cancer, J. Oncol. Pract., 2012, 8(6), e149–e157 CrossRef.
- S. Ponnayyan Sulochana, K. Sharma and R. Mullangi, et al., Review of the validated HPLC and LC-MS/MS methods for determination of drugs used in clinical practice for Alzheimer's disease, Biomed. Chromatogr., 2014, 28(11), 1431–1490 CrossRef CAS.
- M. Martin-Lorenzo, P. J. Martinez and M. Baldan-Martin, et al., Citric Acid Metabolism in Resistant Hypertension: Underlying Mechanisms and Metabolic Prediction of Treatment Response, Hypertension, 2017, 70(5), 1049–1056 CrossRef CAS.
- D. J. Beale, F. R. Pinu and K. A. Kouremenos, et al., Review of recent developments in GC-MS approaches to metabolomics-based research, Metabolomics, 2018, 14(11), 152 CrossRef.
- J. M. Halket, D. Waterman and A. M. Przyborowska, et al., Chemical derivatization and mass spectral libraries in metabolic profiling by GC/MS and LC/MS/MS, J. Exp. Bot., 2005, 56(410), 219–243 CrossRef CAS.
- S. C. Yan, Z. F. Chen and H. Zhang, et al., Evaluation and optimization of sample pretreatment for GC/MS-based metabolomics in embryonic zebrafish, Talanta, 2020, 207, 120260 CrossRef CAS PubMed.
- Y. Yang, Y. Yin and X. Chen, et al., Evaluating different extraction solvents for GC-MS based metabolomic analysis of the fecal metabolome of adult and baby giant pandas, Sci. Rep., 2019, 9(1), 12017 CrossRef PubMed.
- Y. Simon-Manso, M. S. Lowenthal and L. E. Kilpatrick, et al., Metabolite profiling of a NIST Standard Reference Material for human plasma (SRM 1950): GC-MS, LC-MS, NMR, and clinical laboratory analyses, libraries, and web-based resources, Anal. Chem., 2013, 85(24), 11725–11731 CrossRef CAS.
- T. Kind, G. Wohlgemuth and D. Y. Lee, et al., FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry, Anal. Chem., 2009, 81(24), 10038–10048 CrossRef CAS.
- M. Vinaixa, E. L. Schymanski and S. Neumann, et al., Mass spectral databases for LC/MS and GC/MS-based metabolomics: state of the field and future prospects, TrAC, Trends Anal. Chem., 2016, 78, 23–35 CrossRef CAS.
- J. R. Mazzeo, U. D. Neue and M. Kele, et al., Advancing LC Performance with Smaller Particles and Higher Pressure, Anal. Chem., 2005, 77(23), 460–467 CrossRef.
- A. F. Nassar, T. Wu and S. F. Nassar, et al., UPLC-MS for metabolomics: a giant step forward in support of pharmaceutical research, Drug Discovery Today, 2017, 22(2), 463–470 CrossRef CAS.
- N. L. Kuehnbaum and P. Britz-McKibbin, New advances in separation science for metabolomics: resolving chemical diversity in a post-genomic era, Chem. Rev., 2013, 113(4), 2437–2468 CrossRef CAS.
- H. Gika, C. Virgiliou and G. Theodoridis, et al., Untargeted LC/MS-based metabolic phenotyping (metabonomics/metabolomics): the state of the art, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2019, 1117, 136–147 CrossRef CAS.
- J. Sekula, J. Niziol and W. Rode, et al., Gold nanoparticle-enhanced target (AuNPET) as universal solution for laser desorption/ionization mass spectrometry analysis and imaging of low molecular weight compounds, Anal. Chim. Acta, 2015, 875, 61–72 CrossRef CAS PubMed.
- P. H. Pevsner, J. Melamed and T. Remsen, et al., Mass spectrometry MALDI imaging of colon cancer biomarkers: a new diagnostic paradigm, Biomark. Med., 2009, 3(1), 55–69 CrossRef CAS PubMed.
- Y. Zhang, L. Qiu and Y. Wang, et al., High-throughput and high-sensitivity quantitative analysis of serum unsaturated fatty acids by chip-based nanoelectrospray ionization-Fourier transform ion cyclotron resonance mass spectrometry: early stage diagnostic biomarkers of pancreatic cancer, Analyst, 2014, 139(7), 1697–1706 RSC.
- K. Sasaki, H. Sagawa and M. Suzuki, et al., Metabolomics Platform with Capillary Electrophoresis Coupled with High-Resolution Mass Spectrometry for Plasma Analysis, Anal. Chem., 2019, 91(2), 1295–1301 CrossRef CAS.
- D. S. Wishart, Emerging applications of metabolomics in drug discovery and precision medicine, Nat. Rev. Drug Discovery, 2016, 15(7), 473–484 CrossRef CAS.
- G. Lubes and M. Goodarzi, GC-MS based metabolomics used for the identification of cancer volatile organic compounds as biomarkers, J. Pharm. Biomed. Anal., 2018, 147, 313–322 CrossRef CAS.
- M. M. Khamis, D. J. Adamko and A. El-Aneed, Mass spectrometric based approaches in urine metabolomics and biomarker discovery, Mass Spectrom. Rev., 2017, 36(2), 115–134 CrossRef CAS.
- L. C. Martial, R. E. Aarnoutse and M. Mulder, et al., Dried blood spot sampling in psychiatry: perspectives for improving therapeutic drug monitoring, Eur. Neuropsychopharmacol., 2017, 27(3), 205–216 CrossRef CAS.
- M. Paulzen, T. W. Goecke and E. Stickeler, et al., Sertraline in pregnancy – therapeutic drug monitoring in maternal blood, amniotic fluid and cord blood, J. Affective Disord., 2017, 212, 1–6 CrossRef CAS PubMed.
- M. Shipkova, S. Rapp and R. Rigo-Bonnin, et al., Therapeutic Drug Monitoring of Everolimus: Comparability of Concentrations Determined by 2 Immunoassays and a Liquid Chromatography Tandem Mass Spectrometry Method, Ther. Drug Monit., 2017, 39(2), 102–108 CrossRef CAS.
- Y. Fan, Y. Li and Y. Chen, et al., Comprehensive Metabolomic Characterization of Coronary Artery Diseases, J. Am. Coll. Cardiol., 2016, 68(12), 1281–1293 CrossRef.
- Z. D. Liu, A. H. Zhang and H. Sun, et al., Two decades of new drug discovery and development for Alzheimer's disease, RSC Adv., 2017, 7, 6046–6058 RSC.
- S. A. Love, J. C. Seegmiller and J. Kloss, et al., Urine Creatinine Concentrations in Drug Monitoring Participants and Hospitalized Patients, J. Anal. Toxicol., 2016, 40(8), 659–662 CrossRef CAS.
- M. Prince, A. Comas-Herrera, M. Knapp, et al., World Alzheimer report 2016: improving healthcare for people living with dementia: coverage, quality and costs now and in the future, Alzheimer's Disease International, 2016 Search PubMed.
- M. Kim, S. Snowden and T. Suvitaival, et al., Primary fatty amides in plasma associated with brain amyloid burden, hippocampal volume, and memory in the European Medical Information Framework for Alzheimer's Disease biomarker discovery cohort, Alzheimer's Dementia, 2019, 15(6), 817–827 CrossRef.
- J. Afonso, S. Lopes and R. Goncalves, et al., Proactive therapeutic drug monitoring of infliximab: a comparative study of a new point-of-care quantitative test with two established ELISA assays, Aliment. Pharmacol. Ther., 2016, 44(7), 684–692 CrossRef CAS.
- Z. K. Ye, Y. L. Chen and K. Chen, et al., Therapeutic Drug Monitoring of Vancomycin: A Guideline of the Division of Therapeutic Drug Monitoring, Chinese Pharmacological Society, J. Antimicrob. Chemother., 2016, 71(11), 3020–3025 CrossRef.
- D. Q. Chen, G. Cao and H. Chen, et al., Identification of serum metabolites associating with chronic kidney disease progression and anti-fibrotic effect of 5-methoxytryptophan, Nat. Commun., 2019, 10(1), 1476 CrossRef.
- F. Bray, J. Ferlay and I. Soerjomataram, et al., Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, Ca-Cancer J. Clin., 2018, 68(6), 394–424 CrossRef.
- H. B. El-Serag and K. L. Rudolph, Hepatocellular carcinoma: epidemiology and molecular carcinogenesis, Gastroenterology, 2007, 132(7), 2557–2576 CrossRef CAS.
- P. Luo, P. Yin and R. Hua, et al., A Large-scale, multicenter serum metabolite biomarker identification study for the early detection of hepatocellular carcinoma, Hepatology, 2017, 67(2), 662–675 CrossRef.
- D. DiPrinzio and R. Sethi, Will Adding Methadone to Controlled Substance Monitoring Programs Help Psychiatrists Prevent Prescription Drug Overdoses?, Primary Care Companion for CNS Disorders, 2016, 18(2) DOI:10.4088/PCC.15l01871.
- J. Lock, R. K. Patel and C. Brookes, et al., Clinical Experience of Prescribing Dabigatran Etexilate With Dronedarone: The Role of Therapeutic Drug Monitoring, Ther. Drug Monit., 2016, 38(6), 813–814 CrossRef.
- J. L. Ren, D. Zhang and Y. J. Liu, et al., Simultaneous Quantification of Serum Nonesterified and Esterified Fatty Acids as Potential Biomarkers to Differentiate Benign Lung Diseases from Lung Cancer, Sci Rep., 2016, 6, 34201 CrossRef CAS.
- P. Petryszyn and A. Wiela-Hojenska, Economic Issues in Therapeutic Drug Monitoring, Acta Pol. Pharm., 2016, 73(3), 599–604 CAS.
- Y. El Samad, J. P. Lanoix and Y. Bennis, et al., Tolerability and Plasma Drug Level Monitoring of Prolonged Subcutaneous Teicoplanin Treatment for Bone and Joint Infections, Antimicrob. Agents Chemother., 2016, 60(10), 6365–6368 CrossRef.
- J. Bai, J. T. Wang and K. C. Mei, et al., Real-time monitoring of magnetic drug targeting using fibered confocal fluorescence microscopy, J. Controlled Release, 2016, 244(Pt B), 240–246 CrossRef CAS.
- P. Herviou, E. Thivat and D. Richard, et al., Therapeutic drug monitoring and tyrosine kinase inhibitors, Oncol. Lett., 2016, 12(2), 1223–1232 CrossRef CAS.
- J. M. Garcia Ruiz de Morales, D. Pascual-Salcedo and F. Llinares Tello, et al., Anti-tumor necrosis factor drug therapy: the usefulness of monitoring drug levels and anti-drug antibodies in clinical practice, Med. Clin., 2016, 147(9), 410–416 CrossRef.
- Y. Mukai, K. Wada and M. Fujimoto, et al., Long-term impact of therapeutic drug monitoring on the risk of hypoglycemia in HOCM patients on cibenzoline therapy, Int. J. Clin. Pharmacol. Ther., 2016, 54(10), 795–803 CrossRef CAS.
- X. Li, S. B. Yang and Y. P. Qiu, et al., Urinary Metabolomics as a Potentially Novel Diagnostic and Stratification Tool for Knee Osteoarthritis, Metabolomics, 2010, 6(1), 109–118 CrossRef CAS.
- C. M. Rebholz, B. Yu and Z. Zheng, et al., Serum metabolomic profile of incident diabetes, Diabetologia, 2018, 61(5), 1046–1054 CrossRef CAS.
- E. P. Diamandis, Cancer biomarkers: can we turn recent failures into success, JNCI, J. Natl. Cancer Inst., 2010, 102(19), 1462–1467 CrossRef CAS.
- S. E. Kern, Why your new cancer biomarker may never work: recurrent pattern and remarkable diversity in biomarker failure, Cancer Res., 2012, 72(23), 6097–6101 CrossRef CAS.
- A. Mullard, New drug costs US $2.6 billion to develop, Nat. Rev. Drug Discovery, 2014, 13, 877 Search PubMed.
- L. Dang, D. W. White and S. Gross, et al., Cancer-associated IDH1 mutations produce 2-hydroxyglutarate, Nature, 2009, 462, 739–744 CrossRef CAS.
- P. S. Ward, J. Patel and D. R. Wise, et al., The common feature of leukemia associated IDH1 and IDH2 mutations is a neomorphicenzyme activity converting α-ketoglutarate to 2-hydroxyglutarate, Cancer Cell, 2010, 17, 225–234 CrossRef CAS.
- G. Kontostathi, J. Zoidakis and N. P. Anagnou, et al., Proteomics approaches in cervical cancer: focus on the discovery of biomarkers for diagnosis and drug treatment monitoring, Expert Rev. Proteomics, 2016, 13(8), 731–745 CrossRef CAS.
- A. Hilgers and M. Schaefer, Systematic Adverse Drug Reaction Monitoring of Patients Under Newer Antiepileptic Drugs Using Routine Clinical Data of Inpatients, Drugs - Real World Outcomes, 2016, 3(2), 209–221 CrossRef.
- E. G. Armitag, F. J. Rupérez and C. Barbas, Metabolomics of diet-related diseases using mass spectrometry, TrAC, Trends Anal. Chem., 2013, 52, 61–73 CrossRef.
- T. L. Archibald, D. E. Murrell and S. D. Brown, Chromatographic methods in HIV medicine: application to therapeutic drug monitoring, Biomed. Chromatogr., 2018, 32(2), e4170 CrossRef.
- M. Khoubnasabjafari, E. Rahimpour and A. Jouyban, Exhaled breath condensate as an alternative sample for drug monitoring, Bioanalysis, 2018, 10(2), 61–64 CrossRef CAS.
- D. Ternant, C. Passot and A. Aubourg, et al., Model-Based Therapeutic Drug Monitoring of Infliximab Using a Single Serum Trough Concentration, Clin. Pharmacokinet., 2018, 57(9), 1173–1184 CrossRef CAS.
- A. R. Reinink, A Pharmacokinetic Rationale for Proactive Therapeutic Drug Monitoring of Anti-TNF Drugs, Am. J. Gastroenterol., 2017, 112(12), 1892–1893 CrossRef CAS.
- M. Guasch-Ferre, S. N. Bhupathiraju and F. B. Hu, Use of Metabolomics in Improving Assessment of Dietary Intake, Clin. Chem., 2018, 64(1), 82–98 CrossRef CAS.
- E. E. McGee, R. Kiblawi and M. C. Playdon, et al., Nutritional Metabolomics in Cancer Epidemiology: Current Trends, Challenges, and Future Directions, Curr. Nutr. Rep., 2019, 8, 187–201 CrossRef PubMed.
- S. Biswas, R. Mengji and S. Barman, et al., ‘AIE + ESIPT’ assisted photorelease: fluorescent organic nanoparticles for dual anticancer drug delivery with real-time monitoring ability, Chem. Commun., 2017, 54(2), 168–171 RSC.
- P. Marquet, L. Albano and J. B. Woillard, et al., Comparative clinical trial of the variability factors of the exposure indices used for the drug monitoring of two tacrolimus formulations in kidney transplant recipients, Pharmacol. Res., 2018, 129, 84–94 CrossRef CAS.
- A. O'Gorman and L. Brennan, The role of metabolomics in determination of new dietary biomarkers, Proc. Nutr. Soc., 2017, 76(3), 295–302 CrossRef.
- M. F. Silva, C. Ribeiro and V. M. F. Goncalves, et al., Liquid chromatographic methods for the therapeutic drug monitoring of methotrexate as clinical decision support for personalized medicine: a brief review, Biomed. Chromatogr., 2018, 32(5), e4159 CrossRef.
- Y. Sun, H. Jensen and N. J. Petersen, et al., Concomitant monitoring of implant formation and drug release of in situ forming poly(lactide-co-glycolide acid) implants in a hydrogel matrix mimicking the subcutis using UV-vis imaging, J. Pharm. Biomed. Anal., 2018, 150, 95–106 CrossRef CAS.
- A. Tebani and S. Bekri, Paving the Way to Precision Nutrition Through Metabolomics, Front. Nutr., 2019, 6, 41 CrossRef.
- C. Lanvers-Kaminsky, A. Ruffer and G. Wurthwein, et al., Therapeutic Drug Monitoring of Asparaginase Activity-Method Comparison of MAAT and AHA Test Used in the International AIEOP-BFM ALL 2009 Trial, Ther. Drug Monit., 2018, 40(1), 93–102 CAS.
- C. E. Argoff, D. P. Alford and J. Fudin, et al., Rational Urine Drug Monitoring in Patients Receiving Opioids for Chronic Pain: Consensus Recommendations, Pain Med., 2018, 19(1), 97–117 CrossRef PubMed.
- J. Theobald, X. Cheng and A. Ghanem, et al., Monitoring cytochrome P450 activity in living hepatocytes by chromogenic substrates in response to drug treatment or during cell maturation, Arch. Toxicol., 2018, 92(3), 1133–1149 CrossRef CAS.
- S. A. Ranamukhaarachchi, C. Padeste and M. Dubner, et al., Integrated hollow microneedle-optofluidic biosensor for therapeutic drug monitoring in sub-nanoliter volumes, Sci. Rep., 2016, 6, 29075 CrossRef CAS PubMed.
- Z. K. Ye, K. Chen and Y. L. Chen, et al., A protocol for developing a clinical practice guideline for therapeutic drug monitoring of vancomycin, J. Huazhong Univ. Sci. Technol., Med. Sci., 2016, 36(3), 469–472 CrossRef CAS PubMed.
- J. L. Ren, H. Sun and H. Dong, et al., A UPLC-MS-based metabolomics approach to reveal the attenuation mechanism of Caowu compatibility with Yunnan Baiyao, RSC Adv., 2019, 9, 8926–8933 RSC.
- B. G. J. Dekkers, M. Bakker and K. C. M. van der Elst, et al., Therapeutic Drug Monitoring of Posaconazole: An Update, Curr. Fungal Infect. Rep., 2016, 10, 51–61 CrossRef.
- L. Dalfino, F. Bruno and N. Brienza, Does the journey toward efficacy and safety of high dose beta-lactams pass through therapeutic drug monitoring?, Minerva Anestesiol., 2016, 82(9), 923–925 Search PubMed.
- F. B. Xia, A. Li and Y. S. Chai, et al., UPLC/Q-TOFMS-Based Metabolomics Approach to Reveal the Protective Role of Other Herbs in An-Gong-Niu-Huang Wan Against the Hepatorenal Toxicity of Cinnabar and Realgar, Front. Pharmacol., 2018, 9, 618 CrossRef PubMed.
- P. C. Liang, Y. C. Chen and C. F. Chiang, et al., Doxorubicin-modified magnetic nanoparticles as a drug delivery system for magnetic resonance imaging-monitoring magnet-enhancing tumor chemotherapy, Int. J. Nanomed., 2016, 11, 2021–2037 CAS.
- G. Groeneveld, M. de Puit and S. Bleay, et al., Detection and mapping of illicit drugs and their metabolites in fingermarks by MALDI MS and compatibility with forensic techniques, Sci. Rep., 2015, 5, 11716 CrossRef CAS.
- E. Germovsek, A. Kent and T. Metsvaht, et al., Development and Evaluation of a Gentamicin Pharmacokinetic Model That Facilitates Opportunistic Gentamicin Therapeutic Drug Monitoring in Neonates and Infants, Antimicrob. Agents Chemother., 2016, 60(8), 4869–4877 CrossRef CAS.
- M. Selwet, M. Galbas and R. Slomski, et al., Monitoring of Virulence Genes, Drug-Resistance in Campylobacter coli Isolated from Golden Retrievers, Pol. J. Microbiol., 2016, 65(2), 237–240 Search PubMed.
- C. Jang, L. Chen and J. D. Rabinowitz, Metabolomics and Isotope Tracing, Cell, 2018, 173(4), 822–837 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2020 |