Open Access Article
Open Access ArticleThe oxidation of (−)-epigallocatechin-3-gallate inhibits T-cell acute lymphoblastic leukemia cell line HPB-ALL via the regulation of Notch1 expression
Yu-Na Wang†
a,
Jing Wang†a,
Hao-Nan Yanga,
Bang-Lei Zhanga,
Pan Zhanga,
Pei-Yuan Suna,
Nin Zhanga,
Ya Wanga,
Jun Sheng*ac,
Xuan-Jun Wang*abc and
Cheng-Ting Zi *ab
*ab
aKey Laboratory of Pu-er Tea Science, Ministry of Education, Yunnan Agricultural University, Kunming, 650201, China. E-mail: shengj@ynau.edu.cn; wangxuanjun@gmail.com; zichengting@126.com
bCollege of Science, Yunnan Agricultural University, Kunming, 650201, China
cState Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan, Yunnan Agricultural University, Kunming, 650201, China
First published on 9th January 2020
Abstract
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematological malignancy, and commonly associated with activating mutations in the Notch1 pathway. (−)-Epigallocatechin-3-gallate (EGCG) is the most abundant and active catechin and has been shown to regulate Notch signaling. Taking into account the highly oxidizable and unstable of EGCG, we proposed that EGCG oxides may have greater potential to regulate Notch signaling than EGCG. In this study, we isolated and identified EGCG oxides (compound 2–4), using a chemical oxidation strategy, and evaluated for cytotoxicity against T-cell acute lymphoblastic leukemia cell line (HPB-ALL) by using the MTS assay. We found compound 3 significantly induced cell proliferation inhibition (38.3858 ± 1.67106 μM), cell apoptosis and cell cycle arrest in a dose-dependent manner. Remarkably, compound 3 inhibited expression of Notch1 compared with EGCG in HPB-ALL cells. Meanwhile, we found that compound 3 significantly inhibited c-Myc and Hes1, which are downstream target genes of Notch1. The findings demonstrate for the first time that an oxidation product of EGCG (compound 3) inhibits T-cell acute lymphoblastic leukemia cell line (HPB-ALL) and is a promising agent for cancer therapy deserving further research.
1. Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive blood malignancy, which is usually reflected as an invasive tumor and accounts for approximately 25% and 15% of adult acute lymphoblastic leukemia in adult and pediatric cohorts, respectively.1,2 Currently, T-ALL treatment schedules consist of high-dose chemotherapy, which is often associated with lack selectivity and displays significant toxic side effects.3–5 There is an urgent clinical need for optimized treatment stratification and more effective antileukemic drugs for the treatment of human T-ALL. Development of targeted drugs, increase of tumor selectivity and decrease of toxicity are the key topics which are currently being studied.Statistically, activating mutations in Notch1 occur in over 50% of T-ALLs,6–8 The deregulated Notch signaling affects diverse cellular processes involved in growth, proliferation, apoptosis and cellular metabolism by driving the expression of c-Myc, Hes1 and regulating other critical oncogenic signaling pathways.9–12 Many preclinical and clinical studies have explored the efficacy of targeting Notch in T-ALL, most commonly with γ-secretase inhibitors (GSIs).13
Natural products with various structures, unique biological activities and specific selectivity have served as important sources of lead compounds for antitumor agents which have been developed for clinical use.14 (−)-Epigallocatechin-3-gallate (EGCG, 1, Fig. 1) is the most abundant catechin (accounting for approximately 50% of total catechins) and has been reported to have stronger physiological activities.15,16 Previously, some studies have reported that several proteins have been identified as EGCG direct interactors including the trans-membrane receptor 67LR, metalloproteinases (MMPs), Pin1 and TGFR-II13.17 In addition, EGCG has the activity of regulating Notch signaling in the development of tumor angiogenesis and colorectal cancer.18–21 However, the use of EGCG is often hindered by problems such as easy oxidation, ready degradation in aqueous solutions and the poor intestinal absorbance.16,22
There have been many studies reporting that EGCG encapsulated into nanocarriers leads to its strengthened stability and higher therapeutic effects.23,24 In this study, we isolated and identified EGCG oxides by a chemical strategy, tested the ability of oxides with EGCG to inhibit T-cell acute lymphoblastic leukemia cell line (HPB-ALL), and further developed more effective antileukemic drugs for the treatment of human T-ALL.
2. Results and discussion
2.1. Chemical oxidation and structural determination
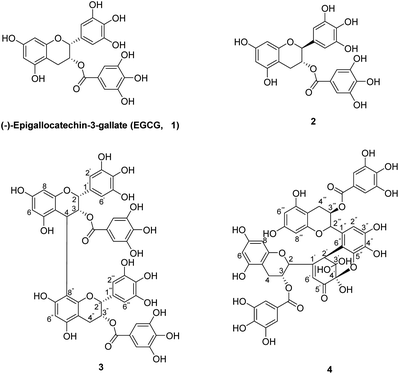
The chemical oxidation of EGCG has been reported under various oxidative conditions, including K3[Fe(CN)6],25 CuSO4,26 and CuCl2.25 In this research, the oxidation of EGCG was based on X. C. Wan and R. G. Bailey et al. method27,28 with some modification. EGCG was reacted with K3[Fe(CN)6]/NaHCO3 at 25 °C for 15 min to yield the EGCG oxides 2–4 in 1.3–6.3% yields (Fig. 1).Compound 2 was obtained as a white amorphous powder and shown to have the same molecular formula as EGCG by its HRESIMS. In the 1H-NMR spectra (Table 1), the proton at C-2 (5.52 ppm, d, J = 2.4 Hz) and C-3 (4.96 ppm, d, J = 2.0 Hz) appear as a double peak, indicating a cis-relationship between H-2 and H-3. Compound 3 was isolated as a yellow amorphous powder. The negative HRESIMS at m/z 913.7339 [M–H]− (calcd for C44H34O22, 913.7340) supporting a molecular formula of C44H34O22. The 1H-NMR and 13C-NMR spectra of compound 3 (Table 1) corresponded to the data reported in the literature.29 Compounds 4, a yellow amorphous powder, was assigned the molecular formula of C44H34O23 determined by the negative HRESIMS at m/z 929.1450 [M–H]− (calcd for C44H34O23, 929.1490), as well as from its NMR data (Table 1), which displayed a signal pattern similar to the data reported in the literatures.30,31
| C position | EGCG (δ) | Compound 2 (δ) | Compound 3 (δ) | Compound 4 (δ) | ||||
|---|---|---|---|---|---|---|---|---|
| δC | δH | δC | δH | δC | δH | δC | δH | |
| 2 | 78.6 | 5.56 (brs) | 78.6 | 5.52 (d, J = 2.4 Hz) | 76.3 | 5.23 (brs) | 94.0 | 4.47 (s) |
| 3 | 69.9 | 4.92 (s) | 69.9 | 4.96 (d, J = 2.0 Hz) | 69.0 | 4.90 (s) | 69.3 | 4.47 (s) |
| 4 | 26.8 | 3.03–2.92 (m, H) | 26.8 | 3.02–2.90 (m, H) | 26.9 | 4.72 (s) | 27.1 | 3.16–3.12 (m, H) |
| 5 (7) | 157.8 | 157.9 | 157.7 | 157.7 | ||||
| 6 | 96.5 | 5.94 (s) | 96.4 | 5.93 (s) | 96.6 | 5.88 (s) | 96.8 | 5.92 (d, J = 2.3 Hz) |
| 8 | 95.9 | 5.94 (s) | 95.8 | 5.93 (s) | 95.9 | 5.87 (s) | 95.8 | 5.90 (d, J = 2.3 Hz) |
| 9 | 157.8 | 157.9 | 157.7 | 157.0 | ||||
| 10 | 99.4 | 99.4 | 99.0 | 99.7 | ||||
| 1′ | 130.8 | 130.7 | 129.5 | 156.5 | ||||
| 2′ | 106.9 | 6.49 (s) | 106.8 | 6.48 (s) | 108.3 | 6.74 (s) | 54.9 | 3.34 (s) |
| 3′ | 146.7 | 146.7 | 146.9 | 110.2 | ||||
| 4′ | 133.8 | 133.8 | 134.4 | 148.0 | ||||
| 5′ | 146.7 | 146.7 | 146.9 | 200.3 | ||||
| 6′ | 106.9 | 6.49 (s) | 106.8 | 6.48 (s) | 108.3 | 6.74 (s) | 124.0 | 5.96 (s) |
| 1′′ (1′′′) | 129.5 | 128.0 | ||||||
| 2′′ | 76.3 | 109.2 | 6.42 (s) | |||||
| 3′′ | 69.0 | 5.23–5.25 (m) | 140.1 | |||||
| 4′′ | 20.8 | 3.31–3.29 (m) | 130.9 | |||||
| 5′′ | 157.3 | 148.5 | ||||||
| 6′′ | 96.6 | 5.91 (d, J = 2.3 Hz) | 114.9 | |||||
| 7′′ (9′′) | 157.7 | |||||||
| 8′′ (8′′′) | 112.3 | 95.7 | 5.75 (d, J = 2.3 Hz) | |||||
| 10′′ (10′′′) | 99.0 | 99.2 | ||||||
| 2′′′ | 108.3 | 6.74 (s) | 78.5 | 5.45 (brs) | ||||
| 3′′′ | 145.4 | 66.4 | 4.57 (s) | |||||
| 4′′′ | 134.4 | 27.0 | 2.89–2.82 (m) | |||||
| 5′′′ (7′′′) | 145.4 | 157.7 | ||||||
| 6′′′ | 108.3 | 6.74 (s) | 96.6 | 5.80 (d, J = 2.3 Hz) | ||||
| 9′′′ | 157.0 | |||||||
| Galloyl 0 | 167.7 | 167.6 | 167.8 | 167.7 | ||||
| Galloyl 1 | 121.4 | 121.4 | 121.4 | 121.1 | ||||
| Galloyl 2 (6) | 110.2 | 6.94 (s, 2H) | 110.2 | 6.93 (2H, s) | 110.3 | 6.91 (s, 2H) | 110.2 | 6.95 (s, 2H) |
| Galloyl 3 (5) | 146.7 | 146.7 | 146.3 | 146.3 | ||||
| Galloyl 4 | 139.8 | 139.7 | 139.8 | 139.8 | ||||
2.2. Compound 3 is identified as the most potent cytotoxic compound against HPB-ALL cells
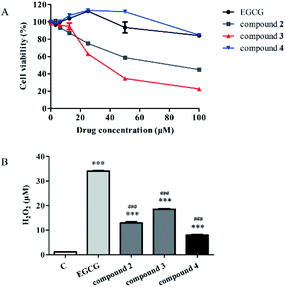
We tested the stability of compounds with H2O2 quantitative assay kit. As shown in Fig. 2A, EGCG was oxidized significantly. However, the stability of the compound 2, 3 and 4 was significantly improved compared with EGCG.Previous studies have identified several T-ALL cell lines that are sensitive to GSI treatment.6,32 To identify anticancer activity of EGCG oxides, we treated HPB-ALL cells with EGCG and its oxides in different concentrations and measured the cell viability using MTS assay (Fig. 2B). As shown in Fig. 2B, compound 3 appeared to be the most potent of these oxides. The IC50 value of compound 3 was 38.3858 ± 1.67106 μM in the HPB-ALL cells.
2.3. Compound 3 blocks Notch1 processing in HPB-ALL cells
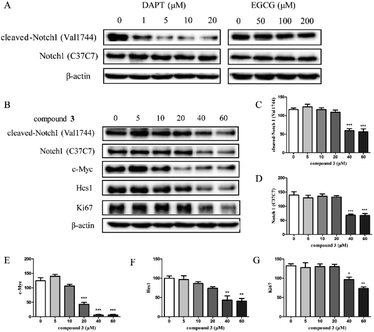
Cell growth and proliferation can be regulated by Notch1 in T-ALL.33 Previous studies have shown that EGCG regulates the inflammatory response by targeting Notch.34,35 In addition, compound 3 is a dimeric EGCG derivative, share a similar chemical structure with EGCG. Therefore, we speculated that the cytotoxicity of compound 3 may be partly due to the inhibition of Notch1 signaling pathway. To investigate the effects of compound 3 on Notch1 signaling pathway, we examined the expression of Notch1 and the downstream proteins in HPB-ALL cells (Fig. 3). As shown in Fig. 3B–F, HPB-ALL cells were treated with compound 3 at the concentration of 5, 10, 20, 40, 60 μM for 12 h and the expression level of Notch1, cleaved-Notch1, c-Myc and Hes1 were significantly decreased.7 Meanwhile, we also found that the expression of proliferation marker Ki67 was significantly reduced (Fig. 3B and G). These findings suggest that compound 3 inhibited the proliferation of HPB-ALL cells may be through down-regulating the expression of Notch1 and Ki67.2.4. Compound 3 induces cell cycle G2 arrest in HPB-ALL cells
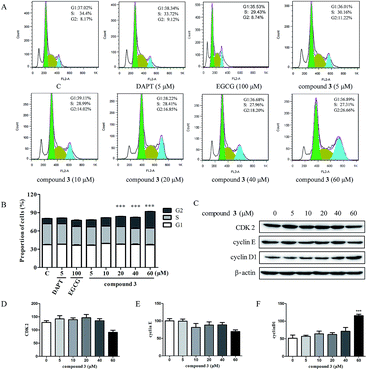
Given that compound 3 inhibited HPB-ALL cells proliferation, we wondered whether 3 causes cell cycle arrest. We treated the HPB-ALL cells with compound 3 (5, 10, 20, 40, 60 μM) for 48 h and analyzed their cell cycle profiles by propidium iodide (PI) staining (Fig. 4). As shown in Fig. 4A and B, in the HPB-ALL cells, compound 3 significantly increased the percentage of cells in G2 phase compared to control. Meanwhile, we examined the protein levels of several cell cycle (CDK2, cyclin E, cyclin D1). As shown in Fig. 4C–F, compound 3 reduced the expression levels of CDK2 and cyclin E, increased the expression levels of cyclin D1.2.5. Compound 3 induces apoptosis in HPB-ALL cells
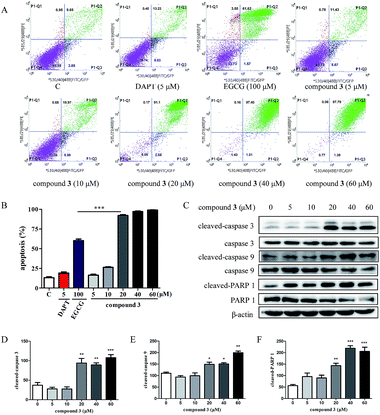
Previous studies have shown that EGCG exhibits the potential to promote apoptosis of tumor cells.36,37 Compound 3 has good cytotoxicity against HPB-ALL cell lines, suggesting that compound 3 mainly induces cell death in addition to causing cell cycle arrest. To test whether compound 3 induces apoptosis in HPB-ALL cells, we treated the cells with DAPT (5 μM), EGCG (100 μM) and compound 3 (5, 10, 20, 40, 60 μM) for 48 h, and measured the apoptosis of the HPB-ALL cells by FITC Annexin V staining and flow cytometry. Compound 3 significantly induced Annexin V positive apoptotic cells in HPB-ALL cells (Fig. 5A and B). Moreover, compound 3 induced the expression levels of caspase 3, caspase 9 and PARP 1, and increased the expression levels of cleaved-caspase 3, cleaved-caspase 9 and cleaved-PARP 1 (Fig. 5C–F).3. Experimental
3.1. Materials and methods
(−)-Epigallocatechin-3-gallate was purchased from Chengdu Proifa Technology Development Co., Ltd (Chengdu, China); 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) purchased from Promega (Madison, WI, USA). H2O2 quantitative assay kit was provided by sangon biotech (Shanghai, China). HPB-ALL cell line were purchased from the American Type Culture Collection (ATCC, Manassas, Virginia, USA); antibodies against cleaved-Notch1, Notch1, Hes1, cleaved-caspase 3, caspase 3, cleaved-caspase 9, caspase 9, CDK2 and β-actin were obtained from Cell Signaling Technology (Beverly, MA, USA). Antibodies against c-Myc, cleaved-PARP1, PARP1 and Ki67 were obtained from Abcam (Lake Placid, NY, USA). Antibodies against cyclin E and cyclin D1 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). All reagents were commercially available and used without further purification unless indicated otherwise. The melting points were measured by an X-4 melting point apparatus and were uncorrected. Mass spectra (MS) and high-resolution mass spectra (HRMS) data were recorded on an Agilent 6540 Q-Tof (ESIMS) (Agilent, California, USA). Proton nuclear magnetic resonance (1H-NMR) and carbon-13 nuclear magnetic resonance (13C-NMR) spectra were recorded on Bruker AVANCE III 500 MHz (Bruker BioSpin GmbH, Rheinstetten, Germany) instruments, using tetramethylsilane (TMS) as an internal standard: chemical shifts (δ) are given in ppm, coupling constants (J) in Hz, the solvent signals were used as references (CD3OD: δC = 49.0 ppm; residual CH3OH in CD3OD: δH = 4.78 ppm). Silica gel (200–300 mesh) for column chromatography and silica GF254 for thin-layer chromatography (TLC) were produced by Qingdao Marine Chemical Company (China).3.2. Oxidation of EGCG and characteristic data of compounds 2–4
To a solution of EGCG (200 mg, 0.45 mmol) in 10 mL of distilled water was added dropwise a solution of potassium hexacyanoferrate (III) (300 mg, 0.90 mmol) and solium hydrogen carbonate (100 mg, 1.2 mmol) in 10 mL of distilled water at 25 °C, pH 7.0 for 15 min. Then, the pH of resulting mixture was adjusted to 2.0 by addition of saturated aqueous citric acid (2 mL). The reaction mixture was extracted with ethyl acetate (10 mL × 3), and the combined organic layer was dried over sodium sulfate (s). The solvent was evaporated under vacuum and the residue was purified by column chromatography (silica gel, chloroform![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol = 4
methanol = 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 → 1
1 → 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, in 0.2% acetic acid) to afford compound 2 (4.9 mg, 2.5%), compound 3 (12.6 mg, 6.3%), and compound 4 (2.5 mg, 1.3%).
1, in 0.2% acetic acid) to afford compound 2 (4.9 mg, 2.5%), compound 3 (12.6 mg, 6.3%), and compound 4 (2.5 mg, 1.3%).
Compound 2: white amorphous powder, 1H-NMR (CD3OD, 500 MHz) and 13C-NMR (CD3OD, 125 MHz) data: see Table 1; ESIMS: m/z 457 [M–H]−, HRESIMS was calculated for C22H17O11 [M–H]− 457.0849 and was found to be 457.0850. Compound 3: yellow amorphous powder, 1H-NMR (CD3OD, 400 MHz) and 13C-NMR (CD3OD, 100 MHz) data: see Table 1; ESIMS: m/z 457 [M–H]−, HRESIMS was calculated for C44H33O22 [M–H]− 913.7340 and was found to be 913.7339. Compound 4: yellow amorphous powder, 1H-NMR (CD3OD, 500 MHz) and 13C-NMR (CD3OD, 125 MHz) data: see Table 1; ESIMS: m/z 457 [M–H]−, HRESIMS was calculated for C44H33O23 [M–H]− 929.1490 and was found to be 929.1450.
3.3. Measurement of the stability of compounds
The medium containing the different compounds (50 μM) was incubated at 37 °C for 30 min. According to the instructions, the medium and operating fluid was transferred into a 96-well plate. The plate was shaken for 30 s and left at room temperature for 20 min. The optical density (OD) values were measured at 595 nm with a Flex Station 3 Multi-Mode Microplate Reader (Molecular Devices).3.4. Cell toxicity assay
HPB-All cells were placed into 96-well plates at a density of 2.0 × 105 cells per well and treated with medium free of phenol red and different concentrations of EGCG and EGCG oxides. Cells was incubated at 37 °C for 48 h. 20 μL of MTS solution was transferred to each well, and then the plate was incubated at 37 °C for 4 h. The OD values were measured at 490 nm with Flex Station 3 Multi-Mode Microplate Reader (Molecular Devices).3.5. Western blot analysis
HPB-ALL cells were seeded in the 100 mm plates at a density of 6.0 × 106 cells per well and treated with compound 3 at different concentrations in cell culture flasks, and then the cells were harvested and lysed in lysis buffer to assess the expression of Notch1, c-Myc, Hes1, caspase 3, caspase 9, PARP 1, Ki67, cyclin E, cyclin D1 and CDK2. Extracted proteins samples were quantified using bicinchoninic acid (BCA) assays; equal amounts were loaded 8% SDS-polyacrylamide gel and then transferred onto polyvinylidene fluoride (PVDF) membranes (EMD Millipore Corporation, Merck Life Sciences, KGaA, Darmstadt, Germany). Membranes containing protein blots were blocked with 5% non-fat milk in TBST at room temperature for 1 h and then incubated with the primary antibodies at 4 °C overnight. Following a wash with TBST, membranes were incubated with secondary antibodies for 1 h. Detection was performed using a FluorChem E System (ProteinSimple, Santa Clara, CA, United States).3.6. Cell cycle analysis
HPB-ALL cells were seeded in the 6-well plates at a density of 1.0 × 106 cells per well, harvested, and washed twice with PBS. After The cells were synchronized for 48 h in medium containing 3% FBS in the presence or absence of DAPT, EGCG and compound 3, and then washed in ice-cold PBS and fixed in 70% ethanol at 4 °C for 24 h. Then the cells were washed twice with cold PBS and stained in buffer containing PI (10 μg mL−1) and RNase. After 30 min incubation in the dark on ice, cell cycle distributions were detected and analyzed by flow cytometry (BD FACSCalibur, USA).3.7. Cell apoptosis assay
HPB-ALL cells were seeded in the 6-well plates at a density of 1.0 × 106 cells per well. After the cells were pretreated with DAPT, EGCG and compound 3 and incubated for 48 h in medium containing 3% FBS. Following collection and suspension in binding buffer, the cells were incubated with 5 μL of Annexin V and 5 μL of PI (BD Biosciences) for 15 minutes in darkness. Detection was performed using a flow cytometry (BD FACSJazz TM Cell Sorter, 655487, USA).4. Conclusions
In summary, we isolated and identified EGCG oxides (compound 2–4), using a chemical oxidation strategy, and evaluated for cytotoxicity against T-cell acute lymphoblastic leukemia cell line (HPB-ALL) by using MTS assay. Among them, compound 3 showed the highest anticancer activity with its IC50 values at 38.3858 ± 1.67106 μM. We founding that compound 3 inhibited the proliferation of HPB-ALL cells may be through down-regulating the expression of Notch1 and Ki67. Compound 3 induced cell cycle G2 arrest in HPB-ALL cells, and also induced the expression levels of the apoptotic maker protein (caspase 3, caspase 9 and PARP 1). We demonstrated for the first time that an oxidation product of EGCG (compound 3) inhibits T-cell acute lymphoblastic leukemia cell line HPB-ALL via the regulation of Notch1 expression.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
We are grateful to the National Nature Science Foundation of China for financial support (No. 21602196); the Yunnan Provincial Science and Technology Department (No. 2017ZF003, 2017FD084, and 2017FG001-046).Notes and references
- A. A. Ferrando, D. S. Neuberg, J. Staunton, M. L. Loh, C. Huard and S. C. Raimondi, Cancer Cell, 2002, 1, 75–87 CrossRef CAS.
- E. Baghbani, B. Baradaran, F. Pak, L. Mohammadnejad, D. Shanehbandi, B. Mansoori, V. Khaze, N. Montazami, A. Montazami and P. Kokhaei, Biomed. Pharmacother., 2017, 86, 41–47 CrossRef CAS PubMed.
- A. J. Barrett, M. M. Horowitz, B. H. Pollock, M. J. Zhang, M. M. Bortin and G. R. Buchanan, N. Engl. J. Med., 1994, 331, 1253–1258 CrossRef CAS PubMed.
- J. M. Goldberg, L. B. Silverman, D. E. Levy, V. K. Dalton, R. D. Gelber and L. Lehmann, J. Clin. Oncol., 2003, 21, 3616–3622 CrossRef PubMed.
- P. V. Vlierberghe and A. Ferrando, J. Clin. Invest., 2012, 122, 3398–3406 CrossRef PubMed.
- A. P. Weng, A. A. Ferrando, L. Woojoong, J. P. Morris, L. B. Silverman and S. I. Cheryll, Science, 2004, 306, 269–271 CrossRef CAS PubMed.
- C. Grabher, H. Von Boehmer and A. T. Look, Nat. Rev. Cancer, 2006, 6, 347–359 CrossRef CAS PubMed.
- I. Pinto, M. Duque, J. Gonçalves, P. Akkapeddi, M. L. Oliveira, R. Cabrita, J. A. Yunes, S. K. Durum, J. T. Barata and R. Fragoso, Oncogene, 2019, 1–12 Search PubMed.
- V. Giambra, C. R. Jenkins, H. Wang, S. H. Lam, O. O. Shevchuk and O. Nemirovsky, Nat. Med., 2012, 18, 1693–1698 CrossRef CAS PubMed.
- S. Jarriault, B. O. Le, E. Hirsinger, O. Pourquié, F. Logeat and C. F. Strong, Mol. Cell. Biol., 1998, 18, 7423–7431 CrossRef CAS PubMed.
- S. Weijzen, P. Rizzo, M. Braid, R. Vaishnav, S. M. Jonkheer, A. Zlobin, B. A. Osborne, S. Gottipati, J. C. Aster, W. C. Hahn, M. Rudolf, K. Siziopikou, W. M. Kast and L. Miele, Nat. Med., 2002, 8, 979–986 CrossRef CAS PubMed.
- A. P. Wen, Genes Dev., 2006, 20, 2096–2109 CrossRef PubMed.
- N. Takebe, D. Nguyen and S. X. Yang, Pharmacol. Ther., 2014, 141, 140–149 CrossRef CAS PubMed.
- C. T. Zi, Y. S. Gao, L. Yang, S. Y. Feng, Y. Huang, L. Sun, Y. Jin, F. Q. Xu, F. W. Dong, Y. Li, Z. T. Ding, J. Zhou, Z. H. Jiang, S. T. Yuan and J. M. Hu, Front. Chem., 2019, 7, 434 CrossRef PubMed.
- C. Cabrera, R. Gimenez and M. C. Lopez, J. Agric. Food Chem., 2003, 51, 4427–4435 CrossRef CAS PubMed.
- X. Zhang, J. Wang, J. M. Hu, Y. W. Huang, X. Y. Wu, C. T. Zi, X. J. Wang and J. Sheng, Molecules, 2016, 21, 620 CrossRef PubMed.
- A. Negri, V. Naponelli, F. Rizzi and S. Bettuzzi, Nutrients, 2018, 10, 1–24 CrossRef PubMed.
- L. T. Gu, J. Yang, S. Z. Su, W. W. Liu, Z. G. Shi and Q. R. Wang, Neurochem. Res., 2015, 40, 1211–1219 CrossRef CAS PubMed.
- H. Xie, J. Q. Sun, Y. Q. Chen, M. Zong, S. J. Li and Y. Wang, Oxid. Med. Cell. Longevity, 2015, 2015, 1–10 CrossRef PubMed.
- Y. Gao, G. O. Rankin, Y. Tu and Y. C. Chen, Int. J. Oncol., 2016, 48, 281–292 CrossRef CAS PubMed.
- H. Jin, W. Gong, C. Zhang and S. Wang, OncoTargets Ther., 2013, 6, 145–153 CrossRef CAS PubMed.
- S. Kitao, T. Matsudo, M. Sasaki, T. Horiuchi and H. Sekine, Biosci. Biotechnol. Biochem., 1995, 59, 2167–2169 CrossRef CAS.
- O. Krupkova, S. J. Ferguson and K. Wuertz-Kozak, J. Nutr. Biochem., 2016, 37, 1–12 CrossRef CAS PubMed.
- G. Andreia, I. Frias, N. A. Neves, M. Pinheiro and S. Reis, BioMed Res. Int., 2017, 2017, 1–15 Search PubMed.
- Y. Takino, H. Imagawa, Y. Aoki and T. ocirc ko Ozawa, Agric. Biol. Chem., 1963, 27, 562–568 CrossRef CAS.
- T. Shii, M. Miyamoto, Y. Matsuo, T. Tanaka and I. Kouno, Chem. Pharm. Bull., 2011, 59, 1183–1185 CrossRef CAS PubMed.
- X. C. Wan, H. E. Nursten, C. Ya, A. L. Davis, J. P. G. Wilkins and A. P. Davies, J. Sci. Food Agric., 1997, 74, 401–408 CrossRef CAS.
- R. G. Bailey, H. E. Nursten and I. Mcdowell, J. Sci. Food Agric., 1993, 63, 455–464 CrossRef CAS.
- J. P. de Mello, F. Petereit and A. Nahrstedt, Phytochemistry, 1996, 41, 807–813 CrossRef CAS.
- S. Valcic, A. Muders, N. E. Jacobsen, D. C. Liebler and B. N. Timmermann, Chem. Res. Toxicol., 1999, 12, 382–386 Search PubMed.
- T. Tanaka, S. Watarumi, Y. Matsuo, M. Kamei and I. Kouno, Tetrahedron, 2003, 59, 39–47 Search PubMed.
- T. Palomero, M. L. Sulis, M. Cortina, P. J. Real, K. Barnes, M. Ciofani, E. Caparros, J. Buteau, K. Brown, S. L. Perkins, G. Bhagat, A. M. Agarwal, G. Basso, M. Castillo, S. Nagase, C. Cordon-Cardo, R. Parsons, J. C. Zúñiga-Pflücker, M. Dominguez and A. Ferrando, Nat. Med., 2007, 13, 1203–1210 CrossRef CAS PubMed.
- M. Paganin and A. Ferrando, Blood Rev., 2011, 25, 83–90 CrossRef CAS PubMed.
- T. F. Wang, Z. M. Xiang, Y. Wang, X. Li, C. Y. Fang, S. Song, C. L. Li, H. S. Yu, H. Wang, L. Yan, S. M. Hao, X. J. Wang and J. Sheng, Front. Immunol., 2017, 8, 433 Search PubMed.
- Y. W. Huang, Q. Q. Zhu, X. Y. Yang, H. H. Xu, B. Sun, X. J. Wang and J. Sheng, FASEB J., 2018, 33, 1–12 Search PubMed.
- W. H. Yang, Y. C. Fong, C. Y. Lee, T. R. Jin, J. T. Tzen, T. M. Li and C. H. Tang, J. Cell. Biochem., 2011, 112, 1601–1611 CrossRef CAS PubMed.
- Y. Q. Wang, J. L. Lu, Y. R. Liang and Q. S. Li, Molecules, 2018, 23, 23–34 Search PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |