Open Access Article
Open Access ArticleA simple and rapid colorimetric detection of serum lncRNA biomarkers for diagnosis of pancreatic cancer†
Ut Kei Loua,
Chi Hin Wonga and
Yangchao Chen *ab
*ab
aSchool of Biomedical Sciences, Faculty of Medicine, The Chinese University of Hong Kong, Shatin, NT, Hong Kong. E-mail: yangchaochen@cuhk.edu.hk; Fax: +852 26035123; Tel: +852 39431100
bShenzhen Research Institute, The Chinese University of Hong Kong, Shenzhen 518087, China
First published on 25th February 2020
Abstract
A colorimetric assay is developed for detection of lncRNA HOTTIP by one-step reverse transcription-loop-mediated isothermal amplification (RT-LAMP) coupled with positively-charged gold nanoparticles ((+)AuNP) for diagnosis of pancreatic cancer. This assay allows simple, rapid, and sensitive quantification of lncRNA down to 50 copies.
1. Introduction
Pancreatic ductal adenocarcinoma (PDAC), accounting for 95% of pancreatic cancer cases, is one of the most common malignancies with a high mortality-to-incidence ratio of 98% in 2018.1,2 PDAC patients are often diagnosed at an advanced stage due to the lack of displayed symptoms, leading to poor prognosis with an overall survival of 6 months after diagnosis only.3 Available treatments remained ineffective with the majority of patients developing metastatic diseases eventually.4,5 Meanwhile, the most common circulating PDAC biomarker in blood, carbohydrate antigen 19-9 (CA-19-9), is not applicable for diagnosis due to limited specificity and sensitivity.6 This highlights an urgent demand for alternative biomarkers suitable for diagnosis of PDAC with high accuracy, simplicity and cost-effectiveness.Non-coding RNA (ncRNA) has emerged to be a new hallmark for cancer. Short and long ncRNAs play essential roles in regulating gene expression and cellular mechanisms associated with cancer initiation and progression. However, detection of short ncRNA like microRNA (miRNA) remains challenging due to the short sequence and high homology among family members.7 Long non-coding RNA (lncRNA) accounting for more than 80% of ncRNA has higher sequence specificity as well as tissue specificity.7 lncRNAs are capable of resisting RNase digestion and circulate in body fluids with enhanced stability.8 Therefore, these emphasize the significance of lncRNAs as promising biomarkers for cancer diagnosis and prognosis. lncRNA HOXA distal transcript antisense RNA (HOTTIP) plays an important role in PDAC by promoting cell proliferation and migration.9 Studies reported abnormal upregulation of HOTTIP in PDAC tumours and importantly in body fluids,10 suggesting HOTTIP as a competent circulating biomarker for diagnosis of PDAC.
Few approaches have been developed to detect lncRNA. Microarrays and RNA sequencing are well-established methods while requiring expensive instrumentation.11 Currently, quantitative reverse transcription PCR (RT-qPCR) remains the standard for lncRNA detection.7 However, RT and qPCR must be performed separately in two steps which are prone to contamination. Sample isolation and purification are often required for RT-qPCR, making the procedures complicated, costly and time-consuming. Such an approach is also less reliable for detection in body fluids due to a lack of standardized reference genes for normalization.7 Droplet digital PCR (ddPCR) has emerged as a new generation of PCR technology for quantification of miRNA and circular RNA (circRNA), with improved precision, sensitivity and reproducibility compared to RT-qPCR.12,13 Nevertheless, requirements of sample purity and quality, thermocycling conditions and relatively high cost of ddPCR limit its application for early detection. A novel cyclic enzymatic repairing-mediated signal amplification has been reported recently for fluorescent detection of lncRNA HOTAIR in cancer cells.14 Therefore, it is necessary to develop direct method for detection of lncRNAs in patient samples. Here, we applied a novel and simple approach for lncRNA detection using reverse transcription-coupled loop-mediated isothermal amplification (RT-LAMP), which functions at low temperature for rapid and highly sensitive detection.15 LAMP utilizes a set of multiple primers targeting 6 to 8 regions which enhances sensitivity and specificity for detection. Comparing to traditional RT-qPCR, one-step RT-LAMP takes 30–60 min at constant low temperature of 50–65 °C for reaction to proceed, without using sophisticated equipment.16 More importantly, LAMP provides another advantage of amplifying crude biological sample such as detergent-treated cerebrospinal fluid, heat-treated blood and stool samples while eliminating the need for nucleic acid extraction.17,18
Gold nanoparticles characterized by their optical properties and sensitivity are widely used for a range of biomedical applications, including genetic therapy, drug delivery and biosensors. Properties of the gold nanoparticles including sizes, shapes, charges and functionalities are dependent on the method of synthesis using various reducing and stabilizing agents.19 Gold nanoparticles are commonly applied for detection of nucleic acids, proteins and pathogens based on the red-shift effect upon reaction with aggregation of particles, leading to a color change from red to purple.20 However, such color shift is also induced by self-aggregation of nanoparticles in the absence of target molecules, which may affect reliability and clarity for colorimetric sensor detection.
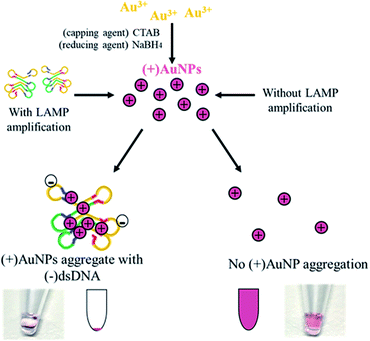
Herein, we proposed a colorimetric assay combining LAMP amplification with aggregation of positively-charged gold nanoparticles ((+)AuNPs) for detection of HOTTIP, as shown in Fig. 1. HOTTIP in serum is first amplified by LAMP assay. Negatively-charged double-stranded DNA (dsDNA) synthesised during LAMP precipitate (+)AuNPs, leading to a distinctive color change from red to colorless due to increased gravitational force of the AuNP-nucleic acid structure. Such decolorization process is triggered by increasing lncRNA concentration, so we hypothesized that upregulation of lncRNA biomarker in cancer samples would result in significant color change compared to normal samples.
2. Materials and methods
2.1 Materials and reagents
Bst3.0 polymerase, magnesium sulfate and 10× isothermal amplification buffer were purchased from New England Biolabs (Ipswich, MA, USA) while reverse transcriptase was ordered from Thermo Fisher Scientific (Vilnius, Lithuania). Six oligonucleotide primers for loop-mediated isothermal amplification of HOTTIP were designed through the PrimerExplorer V5 software available online (http://primerexplorer.jp/lampv5e/index.html), which includes forward inner primer (FIP), backward inner primer (BIP), forward outer primer (F3), backward outer primer (B3), loop forward primer (LF) and loop backward primer (LB) (Table S1†). Chloroauric acid (HAuCl4), cetyltrimethyl ammonium bromide (CTAB) and sodium borohydride (NaBH4) were obtained from Sigma-Aldrich (St. Louis, MO).2.2 Cell lines and mouse serum samples
PDAC cell lines PANC-1 and CFPAC-1 were obtained from American Type Culture Collection (Manassas, VA) and cultured in Dulbecco's Modified Eagle's Media (DMEM) containing 10% fetal bovine serum and 100 units per mL of penicillin and streptomycin. Total RNA from cell lines and genomic DNA were isolated using TRIzol-chloroform extraction and salt extraction respectively. Male BALB/c nude mice aged 4 to 6 weeks were obtained from Laboratory Animal Services Centre of the Chinese University of Hong Kong. All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Chinese University of Hong Kong and approved by the Animal Ethics Committee of Chinese University of Hong Kong.For in vivo mice xenograft model, CFPAC-1 cells were orthotopically injected into the pancreas of the mice. The tumor was allowed to grow for at least 8 weeks for tumor development before serum collection. For serum isolation, blood was allowed to clot for 30 minutes after collection at room temperature, followed by centrifugation at 2000g for 10 minutes.
2.3 In vitro transcription of HOTTIP transcripts
HOTTIP expression plasmid was obtained from abm and was digested with XhoI to generate linearized DNA template. In vitro transcription of HOTTIP RNA was performed by MEGAscript T7 Transcription Kit according to the manufacturer's protocol.2.4 Loop-mediated isothermal amplification
Reactions were prepared on ice in Eppendorf PCR tubes. Standard LAMP was performed in incubator at 60 °C for one hour. Typical LAMP reaction volume was 5 μL, which contained 0.2 μM of F3 and B3 primers, 1.6 μM of FIP and BIP primers, 8 mM MgSO4, 12.5 U reverse transcriptase and 1.6 U Bst3.0 polymerase (NEB). 500 ng of PANC-1 DNA was used in positive reactions. For detection in serum sample, top-up water volume was replaced by diluted serum. After reaction, 1 μL of amplification products were analysed in 2% agarose gel electrophoresis stained in GelRed stain (IO Rodeo) under UV transilluminator.2.5 Preparation of positively-charged gold nanoparticles
(+)AuNPs were prepared according to Li's approach.21 Briefly, 1 mM of chloroauric acid (HAuCl4) and 10 mM of cetyltrimethyl ammonium bromide (CTAB) (Sigma-Aldrich) were mixed in a 17 mL solution to produce a yellowish solution, and the mixture was allowed to stirred in a shaker for 15 minutes. Then, 100 mM sodium borohydride (NaBH4) (Sigma-Aldrich) in 2 mL was added drop by drop with stirring. The mixture was stirred for another 8 min until there was a permanent color change to a reddish-purple solution. Subsequently, the (+)AuNP solution was filtered and stored at −4 °C before use.2.6 Characterization of gold nanoparticles
Size and shape of gold nanoparticles were characterized under transmission electron microscope (TEM, Hitachi, H-7700, Tokyo, Japan). Size distribution and zeta potential analysis were investigated using particle size analyser (Zetasizer Nano ZS90, Malvern Panalytical Ltd, England). Interaction between AuNP and LAMP was demonstrated by Fourier Transform Infrared Spectroscopy (FTIR) (Thermo Nicolet, Nexus). Data were presented as mean ± SD from three independent experiments.2.7 Optimization of absorbance wavelength for AuNP detection
Optimization of wavelength was conducted using PANC-1 gDNA in 1 μL reacting with 10 μL of (+)AuNP. Absorbance in the range of 400 nm to 700 nm were recorded after 1 hour using NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE).2.8 LAMP-(+)AuNP colorimetric detection of lncRNA HOTTIP in mice serum
Serum samples from 29 healthy mice and 13 mice xenografts were analysed. LAMP reaction in 5 μL was prepared in optimized conditions. Top-up water volume was replaced by diluted serum samples while 0.6 μL of serum was needed for each reaction. After LAMP, 1 μL of solution was added to 20 μL of (+)AuNP particles for decolorization at room temperature. Absorbance at 520 nm was measured after 30 min.2.9 Statistical analysis
Statistical analysis was performed by GraphPad Prism 6 and difference between two groups were analysed using one-way analysis of ANOVA. Statistical significance was considered when p-value (two-sided) smaller than 0.05.3. Results and discussion
3.1 Optimization of LAMP conditions
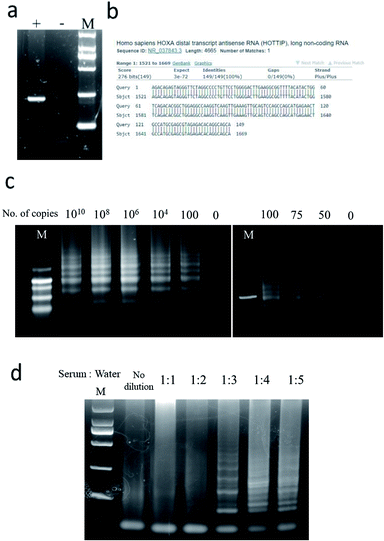
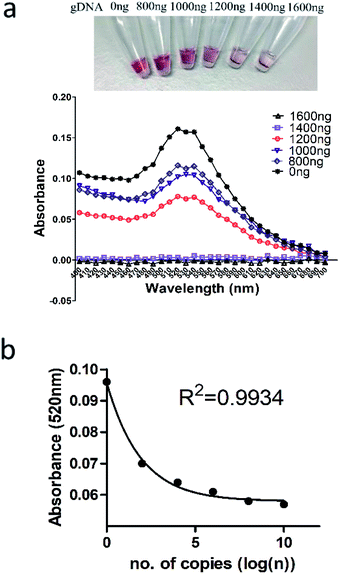
The combination and concentration of primers for LAMP was first determined (Fig. S1, ESI†). The optimal primer sets included 0.2 μM of outer F3 and B3 primers and 1.6 μM of inner FIP and BIP primers (Fig. S2, ESI†). With optimal primer conditions, experimental parameters including temperature, time, concentration of magnesium sulfate, reverse transcriptase and Bst3.0 polymerase were optimized systematically (Fig. S3, ESI†). Subsequent reactions were carried out using 8 mM MgSO4, 12.5 U reverse transcriptase, 16 U Bst3.0 polymerase at 60 °C for 60 min.Under the optimized conditions, specificity and sensitivity of LAMP for detection of HOTTIP were then evaluated. Since the outer primers (F3 and B3) create the basis of the loop structure for amplification in LAMP, we validated the specificity of these primers. PCR was carried out using outer F3 and B3 primers. Analysis of PCR product by agarose gel electrophoresis showed a single band with expected size (Fig. 2a). Sanger sequencing of the PCR product also confirmed the specificity of primers for HOTTIP (Fig. 2b). We then evaluated the sensitivity of the LAMP assay over a range from 1010 to 0 copies of HOTTIP. As low as 50 copies of HOTTIP could be detected in our assay (Fig. 2c). Since components in serum such as amino acids or salts may inhibit LAMP assay by binding to essential magnesium ions and polymerase enzymes,22 we next determined the optimal dilution of serum for HOTTIP detection. Four-fold dilution of serum could achieve the highest efficiency of HOTTIP detection (Fig. 2d).
3.2 Characterization of (+)AuNPs and interaction with LAMP products
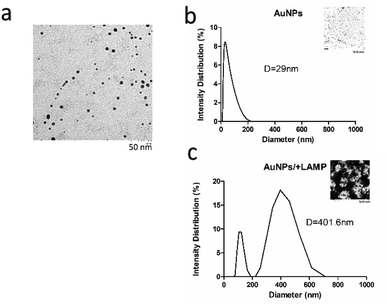
Size and shape of (+)AuNPs after synthesis were characterized under transmission electron microscopy (TEM), showing spherical gold nanoparticles with estimated average size of 10 ± 1.5 nm (Fig. 3a). Dynamic light scattering (DLS) revealed size distribution of nanoparticles with hydrodynamic diameter of 29 ± 3.4 nm (Fig. 3b), while TEM would provide a more accurate measurement of the particle hard core size.23 Polydispersity index (PDI) of 0.33 ± 0.04 under DLS analysis demonstrated uniformity and monodispersity of the gold nanoparticles.After incubation of gold nanoparticles with LAMP reaction products, there was a rightward shift of increase in average particles size distribution from 29 nm to 401.6 ± 52.1 nm estimated by DLS, with large aggregates observed under TEM (Fig. 3c). Two size distribution patterns were shown, with a major peak indicating large aggregation between LAMP products and nanoparticles, while unreacted particles or smaller aggregates contributed to the lower peak. This suggested that aggregation of gold nanoparticles occurred as a result of interaction between AuNP and LAMP products.
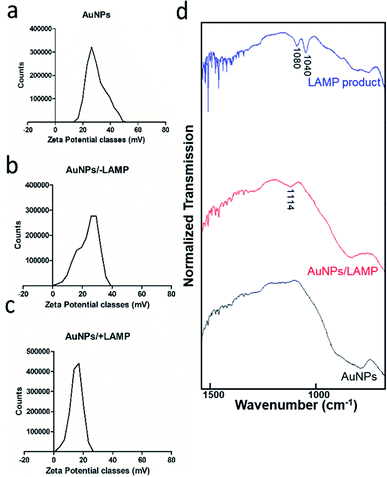
Zeta potential analysis of the gold nanoparticles confirmed positive nature (+34.5 ± 2.5 mV) of particles (Fig. 4a). There was a reduction in zeta potential to +22.9 ± 4.6 mV after incubation with LAMP components due to neutralization of positive charges of gold nanoparticles by negatively-charged primers and dNTPs (Fig. 4b). Moreover, the zeta potential further reduced to +15.8 ± 1.6 mV in the presence of LAMP reaction products due to absorption of negatively-charged nucleic acid polymers formed in LAMP to positive-charged gold nanoparticles (Fig. 4c).
To further validate the interaction between LAMP and AuNP, FTIR spectra of purified LAMP products, AuNP, and LAMP/AuNP reaction complexes were collected (Fig. 4d). From the LAMP product spectrum, two distinct peaks observed at approximately 1040 cm−1 and 1080 cm−1 were representative of C–C bond vibration in DNA ribose sugar and phosphate group (P–O) respectively.24,25 Aggregates formed after incubation of AuNP with LAMP products demonstrated a broader peak with a shift to 1114 cm−1, which was combination of the two DNA bands. The bands indicative for DNA were absent in AuNP alone, confirming the interaction between LAMP products and nanoparticles.
3.3 Optimization and sensitivity of the colorimetric (+)AuNPs detection
The analytical performance of positively-charged gold nanoparticles on detection of LAMP products were evaluated. Using genomic DNA ranging from 0 to 1600 ng reacting with 10 μL of (+)AuNP, the optimal wavelength at 520 nm was determined for DNA detection (Fig. 5a). Since the (+)AuNP would precipitate with negatively-charged LAMP components including dNTP and primers, aggregation with LAMP product and decolorization of the (+)AuNP reaction mixture would be interfered. This may potentially lead to false positive results (Fig. S4a in the ESI†). Therefore, LAMP components to (+)AuNPs ratio was optimized to minimize background decolorization. 20 μL of (+)AuNPs per μL of LAMP reaction mixture could overcome the interference brought by the negatively-charged dNTP and primers in LAMP (Fig. S4b in the ESI†). We then coupled (+)AuNPs with LAMP over a range from 1010 to 100 copies of HOTTIP as input for detection. As expected, there was a negative correlation between HOTTIP copy number and absorbance measured (Fig. 5b).3.4 LAMP-AuNP colorimetric detection of lncRNA HOTTIP in normal and cancer mice serum
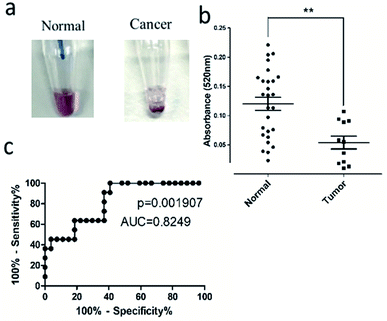
To investigate the capability of the LAMP-(+)AuNP assay in detecting HOTTIP in PDAC serum, the assay was applied for detection in mice serum with PDAC. CFPAC-1 cells were orthotopically injected to the pancreas in BALB/c nude. Mice serum was collected when tumour was developed. LAMP was carried out using diluted normal and PDAC mice serum samples with gold nanoparticles added subsequently. Absorbance was measured after 30 min. In general, reaction mixture turned colorless for tumor samples while remained red for normal samples (Fig. 6a). In addition, color changes were quantified by measuring the absorbance at 520 nm. The LAMP-(+)AuNP assay for HOTTIP detection in PDAC had sensitivity of 69% and specificity of 83% at a cut-off value of 0.06 (Fig. 6b). Area under the receiver operating characteristic (ROC) curve confirmed the good performance of this diagnostic assay (Fig. 6c). Collectively, our results demonstrated the utility of LAMP-(+)AuNP assay for diagnosis of PDAC by detection of lncRNA HOTTIP in diluted serum.4. Conclusions
In summary, our study proposed a rapid colorimetric approach for detection of lncRNA HOTTIP in diluted serum for diagnosis of PDAC using the LAMP-(+)AuNP assay. A major advantage of this assay is its feasibility in diluted serum without the need of RNA extraction and purification. The reaction includes two steps carried out in single tube under isothermal condition for one hour with high simplicity and cost-effectiveness. LAMP detection has high sensitivity down to 50 copies of HOTTIP with enhanced specificity using multiple primers without complicated probe design. Decolorization induced by aggregation of nanoparticles is distinctive between normal and cancer samples and which can be further validated by quantitative measurement, confirming reliability of the assay. Therefore, the LAMP-(+)AuNP assay has great potential for clinical screening of PDAC based on lncRNA detection. Multiple biomarkers may be utilized to improve accuracy of detection. Regarding to identification of urine-based and saliva-based biomarkers in cancer diagnosis,26–28 we aim at developing a completely non-invasive diagnostic approach using alternative biological fluids which would be investigated in the future.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is supported by grant support: General Research Fund, Research Grants Council of Hong Kong [CUHK462713, 14102714, 4171217 to Y. C.]; National Natural Science Foundation of China [81672323 to Y. C.]; Direct Grant from CUHK to YC.Notes and references
- M. Ilic and I. Ilic, World J. Gastroenterol., 2016, 22, 9694–9705 CrossRef PubMed.
- E. B. Andrew, G. H. Yasmin, F. Harold and L. L. Aimee, World J. Gastroenterol., 2014, 20, 11182–11183 CrossRef PubMed.
- D. F. Megan, A. A. Melissa, I. L. Christopher, J. D. R. Anneclaire and J. B. Deborah, Cancer Epidemiol., Biomarkers Prev., 2005, 14, 1766–1773 CrossRef PubMed.
- P. E. Oberstein and K. P. Olive, Ther. Adv. Gastrointest. Endosc., 2013, 6, 321–337 CrossRef PubMed.
- C. L. Wolfgang, J. M. Herman, D. A. Laheru, A. P. Klein, M. A. Erdek, E. K. Fishman and R. H. Hruban, Ca-Cancer J. Clin., 2013, 63, 318–348 CrossRef PubMed.
- K. E. Poruk, D. Z. Gay, K. Brown, J. D. Mulvihill, K. M. Boucher, C. L. Scaife and S. J. Mulvihill, Curr. Mol. Med., 2013, 13, 340–351 CAS.
- T. Shi, G. Gao and Y. Cao, Dis. Markers, 2016, 2016, 1–10 CrossRef PubMed.
- L. Bolha, M. Ravnik-Glavač and D. Glavač, Dis. Markers, 2017, 2017, 1–14 CrossRef PubMed.
- Y. Cheng, I. Jutooru, G. Chadalapaka, J. C. Corton and S. Safe, Oncotarget, 2015, 6, 10840–10852 Search PubMed.
- Y. Wang, Z. Li, S. Zheng, Y. Zhou, L. Zhao, H. Ye, X. Zhao, W. Gao, Z. Fu, Q. Zhou, Y. Liu and R. Chen, Oncotarget, 2015, 6, 35684–35698 Search PubMed.
- S. Uchida, High-Throughput, 2017, 12, DOI:10.3390/ht6030012.
- F. E. Ahmed, N. C. Ahmed and M. M. Gouda, Integr. Cancer Sci. Ther., 2018, 5, 1–7 Search PubMed.
- C. Wang, Q. Ding, P. Plant, M. Basheer, C. Yang, E. Tawedrous, A. Krizova, C. Boulos, M. Farag, Y. Cheng and G. M. Yousef, Clin. Biochem., 2019, 67, 54–59 CrossRef CAS PubMed.
- Y. Zhang, X. Wang, X. Su and C. Zhang, Chem. Commun., 2019, 55, 6805–6952 RSC.
- D. J. Grab, O. V. Nikolskaia, N. Inoue, O. M. Thekisoe, L. J. Morrison, W. Gibson and J. S. Dumler, PLoS Neglected Trop. Dis., 2011, 5, e1249 CrossRef CAS PubMed.
- D. G. Wang, J. D. Brewster, M. Paul and M. Peggy, Molecules, 2015, 20, 6048–6059 CrossRef CAS PubMed.
- D. J. Grab, O. V. Nikolskaia, N. Inoue, O. M. Thekisoe, L. J. Morrison, W. Gibson and J. S. Dumler, PLoS Neglected Trop. Dis., 2011, 5, e1249 CrossRef CAS PubMed.
- Z. K. Njiru, PLoS Neglected Trop. Dis., 2012, 6, e1572 CrossRef PubMed.
- Y. C. Yeh, B. Creran and V. M. Rotello, Nanoscale, 2012, 4, 1871–1880 RSC.
- D. Vilela, M. C. González and A. Escarpa, Anal. Chim. Acta, 2012, 751, 24–43 CrossRef CAS PubMed.
- Z. Li, X. Miao, K. Xing, A. Zhu and L. Ling, Biosens. Bioelectron., 2015, 74, 687–690 CrossRef CAS PubMed.
- H. Kaneko, T. Kawana, E. Fukushima and T. Suzutani, J. Biochem. Biophys. Methods, 2007, 70, 499–501 CrossRef CAS PubMed.
- F. D. Sikkema, M. Comellas-Aragones, R. G. Fokkink, B. J. M. Verduin, J. J. L. M. Cornelissen and R. J. M. Nolte, Org. Biomol. Chem., 2007, 5, 54–57 RSC.
- T. Rolim, J. Cancino and V. Zucolotto, Biomed. Microdevices, 2015, 17, 1–9 CrossRef CAS PubMed.
- B. R. Wood, Chem. Soc. Rev., 2016, 45, 1980–1998 RSC.
- A. Øverbye, T. Skotland, C. J. Koehler, B. Thiede, T. Seierstad, V. Berge, K. Sandvig and A. Llorente, Oncotarget, 2015, 6, 30357–30376 CrossRef PubMed.
- O. E. Bryzgunova, M. M. Zaripov, T. E. Skvortsova, E. A. Lekchnov, A. E. Grigor'eva, I. A. Zaporozhchenko, E. S. Morozkin, E. I. Ryabchikova, Y. B. Yurchenko, V. E. Voitsitskiy and P. P. Laktionov, PLoS One, 2016, 11, e0157566 CrossRef PubMed.
- T. Machida, T. Tomofuji, T. Maruyama, T. Yoneda, D. Ekuni, T. Azuma, H. Miyai, H. Mizuno, H. Kato, K. Tsutsumi, D. Uchida, A. Takaki, H. Okada and M. Morita, Oncol. Rep., 2016, 36, 2375–2381 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Details of Experimental section, tables and figures. See DOI: 10.1039/c9ra07858d |
| This journal is © The Royal Society of Chemistry 2020 |