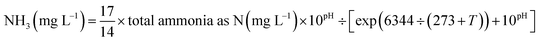Open Access Article
Open Access ArticleEnhanced nitrite accumulation under mainstream conditions by a combination of free ammonia-based sludge treatment and low dissolved oxygen: reactor performance and microbiome analysis†
Heng Yu a,
Zhiyong Tianb,
Jiane Zuo*a and
Yonghui Song*b
a,
Zhiyong Tianb,
Jiane Zuo*a and
Yonghui Song*b
aState Key Joint Laboratory of Environment Simulation and Pollution Control, School of Environment, Tsinghua University, Beijing 100084, China. E-mail: h-yu@mail.tsinghua.edu.cn; jiane.zuo@tsinghua.edu.cn; Fax: +86-10-84917906; Fax: +86-10-62784521; Tel: +86-10-84914787 Tel: +86-10-62772455
bState Key Laboratory of Environmental Criteria and Risk Assessment, Chinese Research Academy of Environmental Sciences, Department of Urban Water Environmental Research, Beijing 100012, China. E-mail: Hkytzy2008@163.com; songyh@craes.org.cn; Fax: +86-10-84917906; Fax: +86-10-84915194; Tel: +86-10-84914787 Tel: +86-010-84915308
First published on 10th January 2020
Abstract
Partial nitritation under mainstream conditions is one of the major bottlenecks for the application of deammonification processes to municipal wastewater treatment plants. This study aimed at evaluating the combination effect of a side-stream free ammonia (FA) treatment and low dissolved oxygen (0.2 ± 0.1 mg L−1) on inhibiting nitrite oxidizing bacteria (NOB) from enhancing nitrite accumulation in long-term lab-scale experiments. Two continuous floccular sludge reactors treating low-strength synthetic wastewater (60 mg N–NH4+ L−1 without COD) with a fixed nitrogen loading rate of 0.22 ± 0.03 g N per L per day were operated in a varied temperature range of 7–31 °C, with one acting as the experimental reactor and the other as the control. Side-stream sludge treatment with a stepwise elevation of FA concentration (65.2–261.1 mg NH3 L−1) was carried out every day in the experimental reactor; the nitrite accumulation ratio (NAR, (NO2–N/(NO2−–N + NO3−–N) × 100%)) in the experimental reactor was always about twice that in the control one. Quantitative PCR (q-PCR) and high-throughput sequencing analyses showed the dominant NOB was mostly Nitrobacter, while there was an alternating trend between Nitrobacter and Nitrospira. Even though the whole microbial communities of each experimental stage between the two reactors were relatively clustered due to an incomplete NOB washout, three abundant metabolisms (amino acid metabolism, pyruvate metabolism and nitrogen metabolism) and key functional genes of nitrification predicted by PICRUSt in the experimental reactor were enriched, providing a better understanding of nitrite accumulation. These results have demonstrated that the positive hybrid effects of FA side-stream sludge treatment and a low DO could enhance nitrite accumulation. It is expected that a complete washout of NOB would be achieved after further process optimization.
1. Introduction
In order to reduce the cost of nitrogen removal from wastewater, the biological nitrogen removal (BNR) process in wastewater treatment plants (WWTPs) has progressively shifted from conventional nitrification-denitrification to an anammox-based process.1 The implementation of the anammox-based process to mainstream nitrogen removal could decrease aeration cost by 63%, save nearly 100% of organic matter addition, and also reduce sludge production and nitrous oxide emission.2 However, when it comes to treating low-strength ammonium wastewater, partial nitritation, as the rate-limiting step for the anammox-based process, has been considered as one of the major challenges.3The achievement of a stable partial nitritation relies on the enrichment of ammonia-oxidizing bacteria (AOB) and the repression or washout of nitrite-oxidizing bacteria (NOB). In previous studies on the treatment of ammonium-rich wastewater, several critical operational parameters were identified for a successful partial nitritation, such as nitrogen loading rate (NLR), free ammonia (FA), free nitrous acid (FNA), temperature and low dissolved oxygen (DO).4,5 Among these, only maintaining a low DO was available to typical municipal wastewater. Low DO (<1.0 mg L−1) with intermittent aeration has been used to repress or eliminate NOB under mainstream conditions.6–8 However, compared with Nitrobacter, Nitrospira with higher affinities for nitrite and DO was enriched under long-term low DO, and NOB would outcompete AOB, which brought about the failure of mainstream partial nitritation.9–11 These contradicting results in published research indicate that the regime of NOB guilds under long-term low DO needed to be further checked and clarified.
Recently, FNA and FA have been proposed as the chemical inhibitors of NOB during side-stream sludge treatment to achieve a stable nitrite accumulation under mainstream conditions.12–14 The objective of the above research aimed to operate a shortcut nitrogen removal process to save a carbon source. Synthetic wastewater was used to simulate a real mixture of municipal wastewater and an anaerobic digestion suspension with a COD concentration of 300 mg L−1. To remove simultaneously nitrogen and carbon in a sequence batch reactor, DO ranged from 1.5 to 2 mg L−1 in the aeration period. However, the specific research background was completely inconsistent with a mainstream anammox-based nitrogen removal system. Thus, the effectiveness of the strategies for eliminating NOB in a mainstream autotrophic nitrogen removal process needs to be checked further. Additionally, between FA and FNA, due to the availability of FA from reject water with a low C/N and high ammonium concentration,15,16 the cost-effective FA strategy could be well-incorporated to most of existing WWTPs.13 Thus, it is significant to examine partial nitritation performance under a long-term low DO in a continuous process when a side-stream sludge treatment using FA is implemented.
In addition, two common genera of NOB in wastewater treatment are Nitrobacter and Nitrospira,17 which exhibit opposing metabolic characteristics due to their affinities for nitrite and DO: while Nitrobacter is an r-strategist with a faster growth rate under a high nitrite concentration and DO, Nitrospira is a K-strategist with a higher population density under nitrite and DO limitation.18 It was speculated that the presence of Nitrobacter could repress Nitrospira, enhancing the stability of partial nitritation.19,20 According to the complex ecological niches within NOB, it would be extremely difficult for the repression of versatile NOB to be validated using a single strategy. It is advisable that the development of different strategies is required to cope with a specific thriving NOB. Thus, sustainable nitritation could be attained under mainstream conditions if different strategies are combined to repress NOB activity.
In order to overcome NOB adaption to low DO and get a more stable NOB repression, the objective of this study was to investigate the feasibility of the repression of NOB and achieve mainstream nitrite buildup through the combination strategy of a side-stream sludge process with FA and low DO, which was carried out in a lab-scale continuous floccular sludge reactor. Meanwhile, an in-depth understanding of the community evolution of functional microbial guilds under low-term DO was performed in another reactor serving as the control.
2. Materials and methods
2.1 Reactor set-up and operation
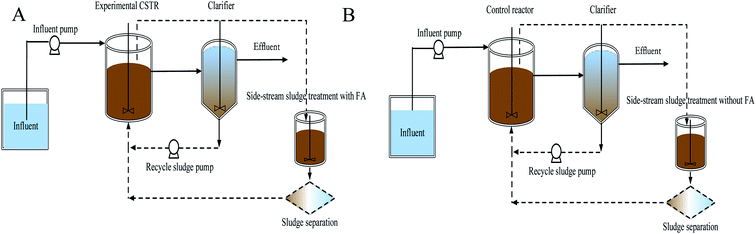
Two lab-scale continuous stirred tank reactors (CSTR) were operated in parallel, as shown in Fig. 1. Each reactor with a working volume of 1.7 L was followed by a clarifier. Compressed air was aerated through a diffuser stone located at the bottom of the reactor and manually manipulated to maintain dissolved oxygen (DO) in the liquid bulk throughout the experiments. However, one reactor with a side-stream sludge using FA was the experimental reactor and the other served as the control reactor without FA treatment. Settled nitrifying sludge in the clarifier was returned to the reactor with a peristaltic pump at a 100% recycle rate. Each reactor was equipped with a mechanical mixer (180 rpm) to maintain complete mixing. The operational temperature and pH were monitored but not controlled. pH in the reactors was about 7.9 ± 0.2 during the experiments. No excessive sludge was discharged during the experiments, but a small amount of biomass was washed out in the daily reactor operations at a MLVSS of 6 ± 1.2 mg L−1.Based on the FA concentration applied to the side-stream sludge treatment, reactor operation could be divided into 4 phases (Table 1). Initially, about 300 mL of nitrifying sludge was taken out from the CSTR every day and allowed to settle for 30 min in a 500 mL cylinder. To prevent the possible NOB adaption to FA, a series of (NH4)2SO4 (1.4, 2.8, 4.2 and 5.7 g) amounts were dissolved in 100 mM Tris–HCl buffer (pH = 8) to prepare the target FA (65.2, 130.6, 195.9 and 261.1 mg NH3 L−1) in different phases. Extra KHCO3 was also added at times to strengthen the buffer capacity. Subsequently, the settled nitrifying sludge from the experimental and the control reactor was recovered to the initial volume using FA-involved and FA-free Tris–HCl buffer, respectively, transferred to 500 mL serum flasks and incubated in an orbital shaker (150 rpm) in a dark climate room at a temperature of 25 °C for 24 h. Finally, FA-treated and -untreated nitrifying sludges were washed with the effluent of the reactors to remove FA residue and returned to the respective reactors. The FA concentration was calculated as a function of total ammonium, operational temperature and pH by equilibrium as follows:15
| Phase | Days | Temperature (°C) | Exposition to FA (mg NH3 L−1) of side-stream sludge treatment | |
|---|---|---|---|---|
| Experimental reactor | Control reactor | |||
| a NA: not applicable. | ||||
| I | 1–74 | 26.6 ± 3.8 | 65.2 | NA |
| II | 75–97 | 25.6 ± 2.2 | 130.6 | NA |
| III | 98–152 | 18.0 ± 3.5 | 195.9 | NA |
| IV | 153–209 | 12.2 ± 2.3 | 261.1 | NA |
2.2 Wastewater and the characteristics of inoculums
Two reactors were continuously fed with synthetic wastewater with an average ammonium concentration of 60 mg N L−1 and a hydraulic retention time (HRT) of 6 h, corresponding to an approximate NLR of 0.24 g N per L per day. The feed, a simulated pretreated municipal wastewater from anammox-based WWTP, also contained (g L−1): 1.0 KHCO3, 0.0272 KH2PO4, 0.3 MgSO4·7H2O, and 0.18 CaCl2·2H2O and 1 ml L−1 of both trace element solutions I and II.21 The reactors were inoculated with nitrifying sludge from a sequencing batch reactor (SBR), which had been operated for more than 150 days and treated with synthetic high-strength ammonium wastewater (800–1000 mg N L−1). The dominant AOB and NOB were Nitrosomonas spp. and Nitrobacter spp., respectively, whereas Nitrospira spp. were not detected by qPCR and high throughout sequencing before the start of the experiments.2.3 Analytical method
The concentrations of nitrite and nitrate were measured using an ion exchange chromatograph (ICS-2100, Thermo, USA) equipped with a Thermo Scientific IonPac AS11-HC anion column. The ammonium concentration was determined colorimetrically using commercial reagents (Hach, USA). pH and DO were determined by a pH meter (Orion star A214, Thermo scientific, USA) and a portable DO sensor (Oxi 330i, WTW, Germany), respectively.2.4 Microbial community analysis
3. Results and discussion
3.1 Reactor performance
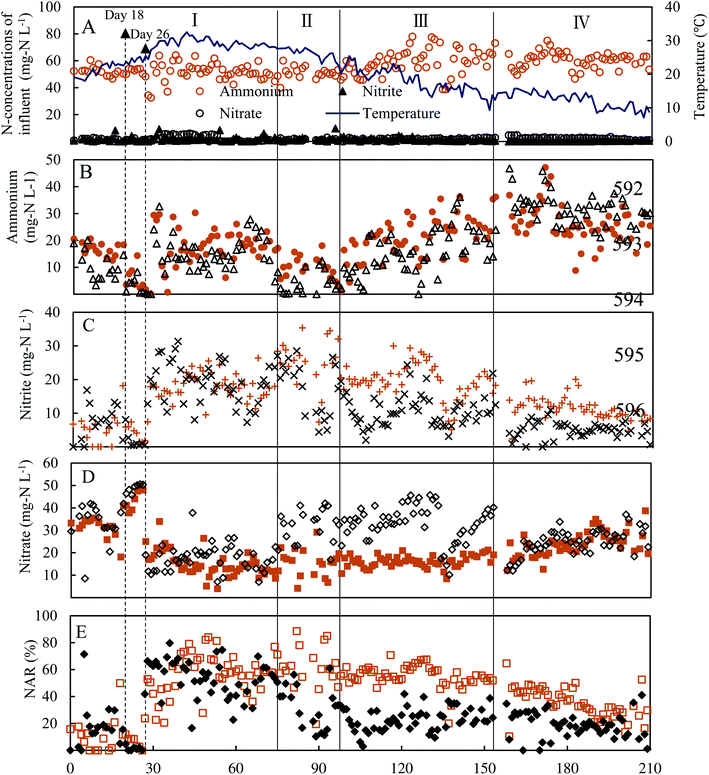
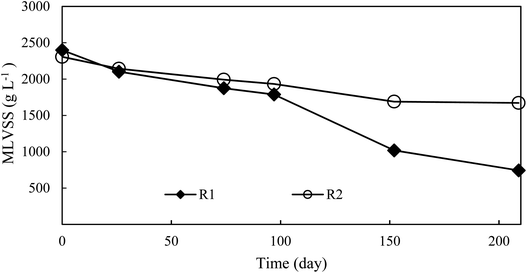
At the onset of the experiment, nitrite buildup was observed in the effluent of two reactors, especially in the control reactor, in which a higher nitrite was detected (Fig. 2C). However, after adaptation to the operational parameters for 18 days, the undesirable complete nitrification occurred in low DO (0.5 ± 0.1 mg L−1) at an aeration rate of 60 mL min−1. Thus, on day 26, the aeration rate was decreased and fixed at 6 mL min−1. An immediate increase of the effluent nitrite concentration of the two reactors emerged from day 27 on. During days 27–74 of phase I, the average concentrations of effluent ammonium, nitrite and nitrate in the experimental reactor were 17.6 ± 7.3 mg N L−1, 17.9 ± 4.0 mg N L−1 and 15.4 ± 5.7 mg N L−1, respectively, similar to the control reactor (Fig. 2C), indicating FA treatment in phase I did not take effect in repressing NOB and nitrite accumulation was associated with a lower DO (0.2 ± 0.1 mg L−1). Meanwhile, similar nitrite accumulation ratios (NAR) (NO2–N/(NO2−–N + NO3−–N) × 100%) in the two reactors were of 55.8 ± 15.6% and 53.8 ± 13.9%, respectively, indicating that NOB repression through FA treatment was inefficient (Fig. 2E). A possible reason was that the effect of FA treatment was counteracted due to an activity recovery of NOB in the bulk liquid. In phase II, as the FA concentration was elevated to 130.6 mg NH3 L−1 in the side-stream sludge treatment, distinct profiles of effluent ammonium, nitrite and nitrate in two reactors were established. Effluent nitrite concentration and NAR in the experimental reactor increased to 24.8 ± 7.0 mg N L−1 and 62 ± 16.2%, with maximum values of 35.4 mg N L−1 and 88%, respectively. Although effluent nitrite was relatively stable in the control, NAR decreased to 33 ± 13% due to a high residual nitrate of 31.9 ± 7.3 mg N L−1. During 55 days of phase III, in spite of increasing FA to 195.9 mg NH3 L−1, nitrite accumulation in two reactors was not enhanced further. The effluent nitrite concentration and NAR in the experimental reactor were 20.0 ± 5.4 mg N L−1 and 55.4 ± 8.4%. The corresponding values for the control were 10.6 ± 4.7 mg N L−1 and 23.1 ± 8.6%. Compared with the control reactor, a better nitrite accumulation performance was still maintained in the experimental reactor. From day 103 onwards, the two reactors were operated below 20 °C. To some extent, the lower operational temperature from 25.6 ± 2.2 °C in phase II to 18.0 ± 3.5 °C in phase III possibly attenuated the effect of the combined strategies, due to a more sensitive temperature dependence of AOB than of NOB.24 However, even though the operational temperature fluctuated between 15.9 and 7.1 °C in phase IV, the NAR of 36.4 ± 11.5% in the experimental reactor was still twice that in the control, indicating that FA treatment still played an important role in NOB repression under low temperature conditions. During the entire period of the experiments, a significant difference in NAR between two reactors was established. Meanwhile, nitrite was always markedly detected in the control reactor, indicating that low DO acted as the inhibitor of NOB and favored nitrite accumulation. These results clearly demonstrated that the combination of FA treatment and low DO was more effective in repressing NOB activity than single DO limitation for achieving a stable nitrite buildup. In fact, complete washout of NOB was not attained by FA treatment, inconsistent with the latest study, where FA treatment with a concentration of 255 mg NH3 L−1 was carried out at the outset.13 NOB partial adaptation to FA treatment and low DO was one plausible explanation for the failure of the complete elimination of NOB in this study.11,13 In addition, unlike the operational mode of this study, a stable partial nitritation for low-strength wastewater could be attributed to a lasting repression of NOB activity in other studies.25,26 However, the average NLR of 0.22 ± 0.03 kg N per m per day in this study was much lower than that in similar studies, where NLR was in the range 0.63–0.80 kg N per m per day.25–28 Thus, a higher NAR could be anticipated with the elevation of NLR. Besides, effluent ammonium concentration was lower in the control than that of the experimental reactor most of the time (Fig. 2B), indicating that AOB activity was also inhibited by FA treatment. Previous studies have found that FA concentration in the range of 10–150 mg NH3 L−1 could significantly inhibit AOB activity.15,29 FA concentration (65.2–261.1 mg NH3 L−1) used in the side-stream sludge treatment was within or beyond the range. However, compared with AOB, NOB was literally subject to a more serious FA inhibition with the higher NAR in the experimental reactor.The variation pattern of biomass concentration between two reactors was similar (Fig. 3). As expected, the slight decrease in the biomass concentration of the control reactor from the initial value of 2.3 g VSS L−1 to 1.7 g VSS L−1 was related to a low NLR. The biomass concentration of the experimental reactor decreased from the initial value to a final value of 0.7 g VSS L−1. The significant biomass loss was the result of FA treatment, which quickened the sludge hydrolysis rate.30
3.2 Microbial community compositions
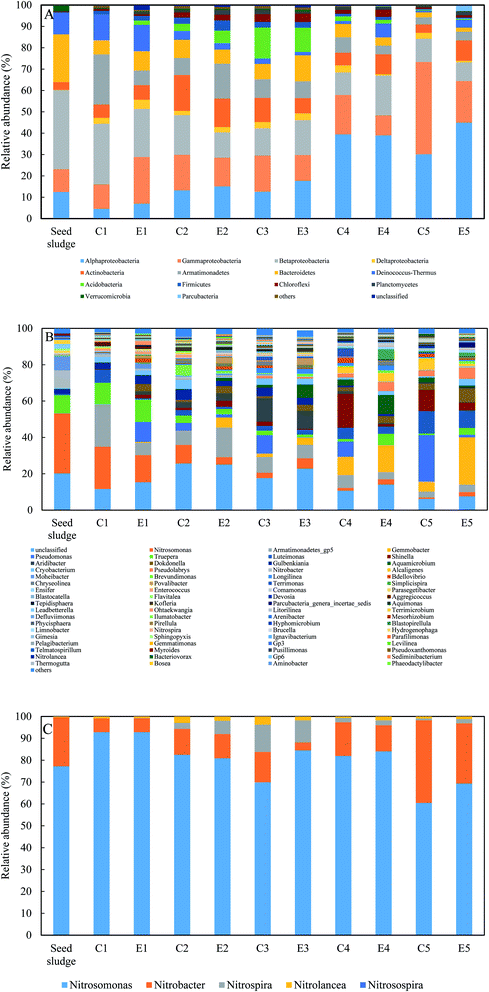
The microbial communities of two reactors were characterized by the IIIumina Miseq paired-end sequencing of 16S rRNA gene (Fig. 4). With the exception of the phylum Armatimonadetes, the phyla that dominated in these samples were in line with previous studies.31,32 The five most abundant bacterial phlya were Proteobacteria, Actinobacteria, Armatimonadetes, Bacteroidetes, and Deinococcus–Thermus in sequence (Fig. 4A). The relative abundance of Proteobacteria excluding AOB and NOB continuously increased from 17.5% in the seed sludge to 70.5% in E5 and 85.8% in C5. Within Proteobacteria, the same trends were observed in the four Alpha-, Beta-, Gamma- and Delta-subclasses of proteobacteria. The sum of Alpha- and Gammaproteobacteria excluding AOB accounted for the largest proportion of proteobacteria. The possible reason was that these subclasses had a preference for inorganic environments and thrived on mineralizing soluble microbial products (SMP).33 Conversely, a radical decrease in the AOB and NOB guilds from 42.7% in the seed sludge to 3.4% in E5 and 1.2% in C5 could be observed. The phylum Armatimonadetes increased steeply after start-up with a maximum of 23.5% in C1 and 16.4% in E2, respectively, and was stable eventually. The reason for this phylum being enriched in this study is not unclear. However, Armatimonadetes has been regularly detected in the autotrophic anaerobic ammonium oxidation process through high-throughput sequencing.34 The relative abundance of the other three phyla was in the range of the aforementioned publications.At genus level, similar dynamics of the functional bacterial guilds in the two reactors are shown in Fig. 4B and C. During the experiments, only AOB guild was Nitrosomonas with an initial 33.0% relative abundance. As the operational temperature decreased, Nitrosomonas reduced the population to lower than 1% in C5 and 2.34% in E5. In the seed sludge, the NOB guild consisted of Nitrobacter and Nitrolancea with relative abundances of 9.6% and 0.2%, respectively, and the genus Nitrospira was not detected. Subsequently, the genus Nitrobacter quickly decreased to lower than 1.6% in 26 days and finally fluctuated between 0.2 and 1.6% in the two reactors. If the fraction of Nitrobacter was dominant within the NOB guild, it could repress the growth of Nitrospira and enhance the stability of partial nitritation.19,27 The dominated NOB population in the two reactors was almost Nitrobacter for except E3 in phase II (Fig. 3C). This fact was supported since Nitrobacter as an r-strategist microorganism is favored at a high nitrite concentration in bulk liquid.35 The emergence of Nitrospira in C2 and E2 was related to the long-term low DO and the dominant NOB shift from Nitrobacter to Nitrospira could be regarded as a strategy for reducing the negative effect of a low DO on complete nitrification.11 However, the shift was not reached for the nitrite accumulation always detected in the control reactor, demonstrating that the role of low DO in repressing NOB was not neglected. Meanwhile, a higher nitrite accumulation in the experimental reactor indicated that FA treatment in phase II (130.6 mg NH3 L−1) could inhibit the activity of Nitrospira, not contradictory with the previous study, where an FA concentration of 12.1 mg NH3 L−1 was ineffective in the inhibition of Nitrospira.36 Because the FA concentration in the side-stream sludge treatment was nearly eleven times higher than that in the literature, Nitrolancea was not favored at the operational conditions of this study for its thermophilic ecological niche. The relative abundance of Nitrospira increased to 0.5% in C3 and 0.7% in E3. However, when the temperature dropped to 7.1 °C, the Nitrospira abundance decreased to 0.01% in C5 and 0.06% in E5. The changing pattern of Nitrospira with temperature is in line with a positive correlation.37 Pseudomonas as the heterotrophic nitrifier has shown a potential capacity for ammonium oxidation in low-DO nitrifying reactors.38 The relative abundance of Pseudomonas in the control gradually increased to 25.6% in C5 with a lower temperature and even outcompeted the AOB guild. The presence of Pseudomonas in the experimental reactor was very unstable, indicating that the actual ammonium oxidation activity of Pseudomonas was possibly influenced by the FA treatment and needs further investigation.
Although no organic carbon was spiked into synthetic wastewater, most of OTUs detected in these samples did not represent nitrifiers and were regarded as potential heterotrophic bacteria growing on SMP of the functional bacteria or preying on NOB.33,39 Numerous heterotrophic species affiliated to the dominant phyla were identified in this study. The mutual reaction between nitrifiers and heterotrophs was profitable for them. Surprisingly, although DO was maintained at about 0.2 mg L−1, many dominant heterotrophs during the experiments were identified as obligatory aerobic or facultative anaerobic bacteria, such as Armatimonadetes_gp5, Gemmobacter, Pseudomonas, Truepera, Luteimona and Shinella. It is unclear why aerobic microbial groups were enriched at such a low DO.
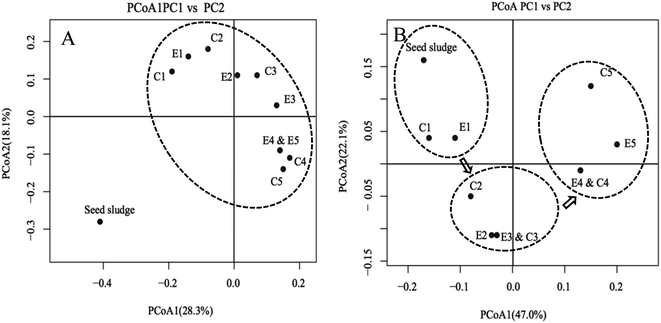
In order to discern the population shift of the overall microbial community among the samples, PCoAs of unweighted and weighted Unifrac metrics were performed (Fig. 5). After the start of the experiment, a significant separation between seed sludge and other samples was most likely due to the low-strength ammonia wastewater allowing the survival of more bacterial species (Fig. 5A). This prediction was supported by the observation that the number of OTUs of the seed sludge was substantially lower than those of others (ESI Table S2†). Based on the PCoA of weighted Unifrac metrics, eleven samples could be roughly divided into three clusters (Fig. 5B), indicating that a significant microbial community shift in the two reactors occurred. The microbial community in the experimental reactor remained relatively consistent with that in the control reactor and the samples from the two reactors in the same phase tended to cluster together. The introduction of FA treatment with a low frequency to the experiment reactor had a minor influence on shaping the microbial community structure. A specific microbial community would be established when the complete washout of NOB was achieved.40,41 Thus, it was reasonable that a relatively consistent microbial community between two reactors was observed before NOB washout. Temperature could significantly influence the nitrifying community.17 Some studies have shown that decreasing the temperature not only reduced the diversity and structure of AOB and NOB guilds, but also changed the species richness of the microbial community.42 When the temperature dropped from 10 to 5 °C, the OTU amount of the nitrifying sludge reduced to half of the initial level.42 In this study, after adaption of the seed sludge to operational parameters, OTUs of the microbial community showed a positive correlation between OTUs and temperature (Table S2†). Therefore, it was proposed that the evolution of the microbial community was more associated with temperature.
3.3 Microbial functional prediction
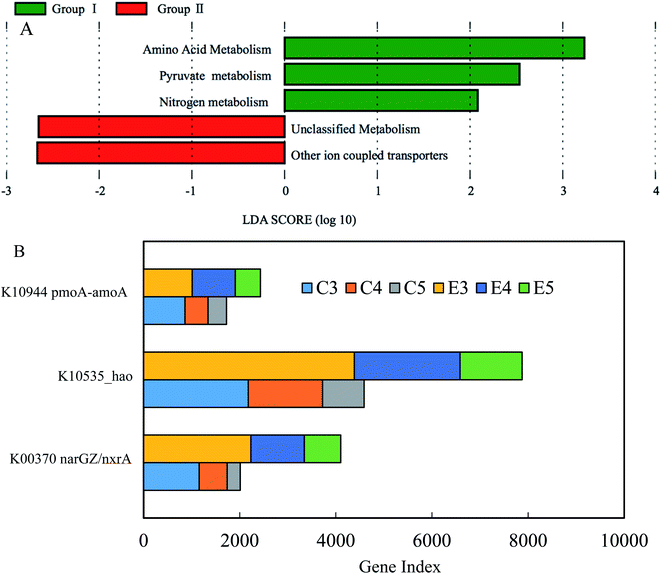
Compared to microbial taxonomy using the taxonomic marker of 16S rRNA gene, microbial functional profiles would be more important than taxonomic profiles, especially under consistent microbial community compositions between two reactors in this study. In order to clarify the distinct NAR between the reactors, PICRUSt was applied to predict the metagenomics functional content (KEGG pathways) (Fig. 6).23 Due to differential nitrite accumulation between two reactors during phases II–IV, group I from the experimental reactor consisted of E3, E4 and E5, and group II from the control reactor consisted of C3, C4 and C5. The predicted results through the differential abundance analysis with LEfSe43 revealed that amino acid metabolism, pyruvate metabolism and nitrogen metabolism were significantly enriched in group I, while unclassified metabolism and unclassified other ion coupled transporters dominated in group II (Fig. 6A). In recent studies, it has been demonstrated that FA pretreatment at the optimized value of 303 mg NH3 L−1 could enhance efficiently the solubilization rate of waste sludge.30 The FA level used in the side-stream FA treatment during phases II–IV was higher than the lowest value of 103 mg NH3 L−1 initially testified in the literature, which released various soluble biodegradable substances containing all kinds of proteins and pyruvates. Moreover, the amino acid metabolism and pyruvate metabolism have been characterized in the mineralization process of cyanobacteria-derived particulate organic matter and anaerobic sludge processes.44,45 Thus, these side-products of nitrifying sludge via FA treatment possibly elicited the enrichment of the amino acid metabolism and the pyruvate metabolism. The prevailing nitrogen metabolism was mainly composed of nitrification under the current operational parameters. As discussed above, NAR over phases II–IV was markedly higher in the experimental reactor than that in the control, while a higher effluent nitrate in the control occurred, indicating that the more active nitritation and nitratation processes prevailed in the two reactors, respectively. Previous studies indicated that NOB was more sensitive to a low DO than AOB, and nitrite accumulation could be achieved under limited DO.46,47 However, it is known that nitritation is the rate-limiting step of full nitrification in long-term low DO.48 Literally, the potential capacity for full nitrification was higher in the experimental reactor. Additionally, although nitrogen loss was insignificant in the two reactors, the denitrification process was probably favored for the derivatives of FA treatment. This was further confirmed by more abundant functional denitrifying genes (see ESI Fig. S1†). According to these results, the enriched metabolisms in the experimental reactor were reasonable.Furthermore, the predicted pattern of functional genes associated with nitrification is illustrated in Fig. 6B. The ammonia monooxygenase subunit A (amoA) gene and hydroxylamine oxidoreductase (hao) had a higher predicated gene index in the experimental reactor, compared with the control. The same pattern of multifunctional nitrate reductase subunit α (nxrA) was unexpected between the two reactors, inconsistent with the respective reactor performances. A plausible explanation was that nitrite accumulation depended on the extent of the deviation from the ratio among the three genes detected during nitrification. The gene ratio for amoA to hao to nxrA in the experimental reactor was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.92
1.92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.59, and the ratio in the control was 1
0.59, and the ratio in the control was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.28
2.28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.86. Notably, as the three functional genes were designated for critical gene annotation of the nitrification process in the KEGG database, other similar functional genes related to the nitrification process should be taken into consideration to obtain a complete and accurate prediction of nitrogen conversion. Fig. S2† presents these similar functional genes of nitrification process. After the addition of the similar functional genes, the ratios for amoA/B/C to hao to nxrA/B in two reactors were 1
0.86. Notably, as the three functional genes were designated for critical gene annotation of the nitrification process in the KEGG database, other similar functional genes related to the nitrification process should be taken into consideration to obtain a complete and accurate prediction of nitrogen conversion. Fig. S2† presents these similar functional genes of nitrification process. After the addition of the similar functional genes, the ratios for amoA/B/C to hao to nxrA/B in two reactors were 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.16
1.16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.55 and 1
0.55 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.31
1.31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.80, respectively. In spite of the lack of the reference ratio of three genes under complete nitrification, the portion of nxrA/B was significantly lower in the experimental reactor. The result complied with the higher nitrite accumulation in the experimental reactor.
0.80, respectively. In spite of the lack of the reference ratio of three genes under complete nitrification, the portion of nxrA/B was significantly lower in the experimental reactor. The result complied with the higher nitrite accumulation in the experimental reactor.
3.4 AOB and NOB quantification
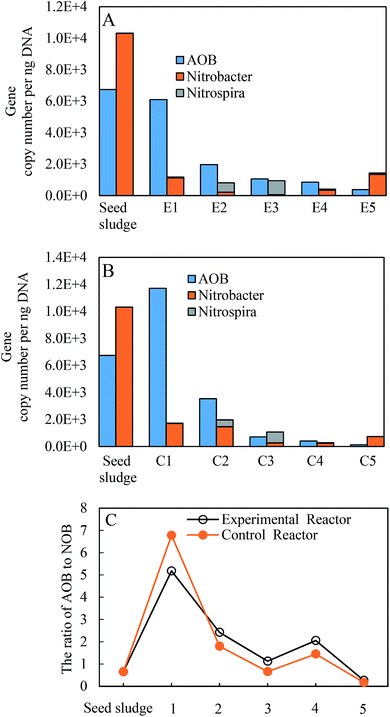
Functional AOB and NOB guilds in the two reactors were further checked (Fig. 7). After the inoculation, both AOB and NOB of the two reactors decreased from 6.7 × 103 to 1.2 × 102 gene copies per ng-DNA and 1.0 × 104 to 7.3 × 102 gene copies per ng-DNA at the end of the experiments, respectively, and were approximately 1–2 orders of magnitude lower than the initial values. Correspondingly, the fractions of AOB and NOB went sharply down from 8.8 to 1.7% and from 13.4 to 0.2% until phase II, respectively, and finally fluctuated within a narrow range of 0.05–1% over time. This was understandable given that seed sludge was collected from the full nitrification reactor fed with high-strength ammonia wastewater. The feature of a low abundance of nitrifying species has been identified in a nitrification reactor under long-term low DO.49,50 Within NOB, Nitrospira began to proliferate in phase II, and exceeded the native-dominated Nitrobacter in phase III. However, as the temperature decreased, Nitrospira almost vanished in the following phases. Nitrobacter as the dominant NOB genus was always detected in successful partial nitritations for low-strength ammonia wastewater in the biofilm system, even at low temperatures.26,27,51 However, even though the ratio of Nitrospira to Nitrosomonas was below 0.07 in the continuous floc sludge reactor, the dominant Nitrospira with a considerably low relative abundance of 1.2% resulted in the failure of nitrite accumulation.10 In this study, the final outcompeting of Nitrobacter could be attributed to the operational conditions, among which selective seed sludge excluding Nitrospira was one of key factors. Furthermore, a major difference was the higher ratios of AOB to NOB in the experimental reactor (Fig. 7C), which was still below the range of 4.4–172.5 in similar literature.26,27,51,52 Significant nitrite accumulation under such a low ratio was almost never reported.4. Conclusions
The present study has demonstrated that the introduction of the combination of a side-stream sludge treatment using FA and low DO could more effectively enhance nitrite accumulation than the single DO limitation. Even though NAR fluctuated in a range of 85–13% with operational temperature decreasing from 30.6 to 7.1 °C, a higher nitrite accumulation was established in the experimental reactor than that in the control reactor. Microbial communities between the two reactors showed relative consistency. Nitrosomonas was always the dominant AOB, but the dominated NOB was variable. The proliferation of Nitrospira was not supported due to nitrite accumulation, and Nitrobacter was the dominant NOB for most of the experimental duration. The microbial function and key functional genes of nitrification predicted by PICRUSt in the partial nitritation reactor could be explained well by process performance and operation. For the robustness of partial nitritation under mainstream conditions, future studies should put an emphasis on optimizing the process operation parameters to prevent the adaption of NOB to FA and low DO.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank financial support from the National Major Program of Science and Technology for Water Pollution Control and Governance (Fund number, 2013ZX07202-010, 2012ZX07202-005, PR China).References
- B. Kartal, J. G. Kuenen and M. C. M. Van Loosdrecht, Science, 2010, 328, 702–703 CrossRef CAS PubMed.
- M. S. M. Jetten, S. J. Horn and M. C. M. Van Loosdrecht, Water Sci. Technol., 1997, 35, 171–180 CrossRef CAS.
- Y. Cao, M. C. M. van Loosdrecht and G. T. Daigger, Appl. Microbiol. Biotechnol., 2017, 101, 1365–1383 CrossRef CAS PubMed.
- Y. Ma, Y. Peng, S. Wang, Z. Yuan and X. Wang, Water Res., 2009, 43, 563–572 CrossRef CAS.
- C. Hellinga, A. A. J. C. Schellen, J. W. Mulder, M. C. M. Van Loosdrecht and J. J. Heijnen, Water Sci. Technol., 1998, 37, 135–142 CrossRef CAS.
- Z. Zheng, S. Huang, W. Bian, D. Liang, X. Wang, K. Zhang, X. Ma and J. Li, Bioresour. Technol., 2019, 283, 213–220 CrossRef CAS PubMed.
- Y. Miao, L. Zhang, Y. Yang, Y. Peng, B. Li, S. Wang and Q. Zhang, Bioresour. Technol., 2016, 218, 771–779 CrossRef CAS PubMed.
- A. Malovanyy, J. Trela and E. Plaza, Bioresour. Technol., 2015, 198, 478–487 CrossRef CAS.
- W. Liu, W. Chen, D. Yang and Y. Shen, Bioresour. Technol., 2019, 275, 272–279 CrossRef CAS PubMed.
- W. Liu and D. Yang, Bioresour. Technol., 2017, 224, 94–100 CrossRef CAS PubMed.
- G. Liu and J. Wang, Environ. Sci. Technol., 2013, 47, 5109–5117 CrossRef CAS PubMed.
- Q. Wang, L. Ye, G. Jiang, S. Hu and Z. Yuan, Water Res., 2014, 55, 245–255 CrossRef CAS PubMed.
- Q. Wang, H. Duan, W. Wei, B.-J. Ni, A. Laloo and Z. Yuan, Environ. Sci. Technol., 2017, 51, 9800–9807 CrossRef CAS PubMed.
- H. Duan, L. Ye, X. Lu and Z. Yuan, Environ. Sci. Technol., 2019, 53, 1937–1946 CrossRef CAS PubMed.
- A. C. Anthonisen, R. C. Loehr, T. B. Prakasam and E. G. Srinath, J. - Water Pollut. Control Fed., 1976, 48, 835–852 CAS.
- H. Yun and D. Kim, J. Chem. Technol. Biotechnol., 2003, 78, 377–383 CrossRef CAS.
- S. Siripong and B. E. Rittmann, Water Res., 2007, 41, 1110–1120 CrossRef CAS PubMed.
- H. Daims, S. Lucker and M. Wagner, Trends Microbiol., 2016, 24, 699–712 CrossRef CAS PubMed.
- J. H. Ahn, T. Kwan and K. Chandran, Environ. Sci. Technol., 2011, 45, 2734–2740 CrossRef CAS PubMed.
- R. Nogueira and L. F. Melo, Biotechnol. Bioeng., 2006, 95, 169–175 CrossRef CAS PubMed.
- A. A. V. De Graaf, P. De Bruijn, L. A. Robertson, M. S. M. Jetten and J. G. Kuenen, Microbiology, 1996, 142, 2187–2196 CrossRef.
- Y. IchYu, C. Lee, J. Kim and S. Hwang, Biotechnol. Bioeng., 2005, 89, 670–679 CrossRef PubMed.
- M. G. I. Langille, J. Zaneveld, J. G. Caporaso, D. Mcdonald, D. Knights, J. A. Reyes, J. C. Clemente, D. E. Burkepile, R. L. V. Thurber and R. Knight, Nat. Biotechnol., 2013, 31, 814–821 CrossRef CAS PubMed.
- S. W. H. Van Hulle, H. J. P. Vandeweyer, B. D. Meesschaert, P. A. Vanrolleghem, P. Dejans and A. Dumoulin, Chem. Eng. J., 2010, 162, 1–20 CrossRef CAS.
- V. Poot, M. Hoekstra, M. Geleijnse, M. C. M. Van Loosdrecht and J. Perez, Water Res., 2016, 106, 518–530 CrossRef CAS PubMed.
- C. Reino, M. E. Suarezojeda, J. Perez and J. Carrera, Water Res., 2016, 101, 147–156 CrossRef CAS PubMed.
- E. Isanta, C. Reino, J. Carrera and J. Perez, Water Res., 2015, 80, 149–158 CrossRef CAS PubMed.
- M. Piculell, M. Christensson, K. Jonsson and T. Welander, Water Sci. Technol., 2015, 73, 1253–1260 Search PubMed.
- S. Villaverde, P. A. Garciaencina and F. Fdzpolanco, Water Res., 1997, 31, 1180–1186 CrossRef CAS.
- W. Wei, X. Zhou, D. Wang, J. Sun and Q. Wang, Water Res., 2017, 118, 12–19 CrossRef CAS PubMed.
- R. Ramirezvargas, N. Serranosilva, Y. E. Navarronoya, R. Alcantarahernandez, M. Lunaguido, F. Thalasso and L. Dendooven, Water Sci. Technol., 2015, 72, 990–997 CrossRef CAS PubMed.
- L. Ye, M.-F. Shao, T. Zhang, A. H. Y. Tong and S. Lok, Water Res., 2011, 45, 4390–4398 CrossRef CAS PubMed.
- T. Kindaichi, T. Ito and S. Okabe, Appl. Environ. Microbiol., 2004, 70, 1641–1650 CrossRef CAS PubMed.
- D. R. Speth, M. H. I. T. Zandt, S. Guerrerocruz, B. E. Dutilh and M. S. M. Jetten, Nat. Commun., 2016, 7, 11172–11181 CrossRef CAS PubMed.
- D. Kim and S. Kim, Water Res., 2006, 40, 887–894 CrossRef CAS PubMed.
- R. A. Simm, D. S. Mavinic and W. D. Ramey, J. Environ. Eng. Sci., 2011, 5, 365–376 CrossRef.
- Z. Huang, P. B. Gedalanga, P. Asvapathanagul and B. H. Olson, Water Res., 2010, 44, 4351–4358 CrossRef CAS PubMed.
- J. Zhang, P. Wu, B. Hao and Z. Yu, Bioresour. Technol., 2011, 102, 9866–9869 CrossRef CAS.
- J. Dolinsek, I. Lagkouvardos, W. Wanek, M. Wagner and H. Daims, Appl. Environ. Microbiol., 2013, 79, 2027–2037 CrossRef CAS PubMed.
- Y. Liang, D. Li, H. Zeng, C. Zhang and J. Zhang, Bioresour. Technol., 2015, 196, 741–745 CrossRef CAS PubMed.
- L. Ye, D. Li, J. Zhang and H. Zeng, Bioresour. Technol., 2019, 276, 190–198 CrossRef CAS PubMed.
- A. Karkman, K. Mattila, M. Tamminen and M. Virta, Biotechnol. Bioeng., 2011, 108, 2876–2883 CrossRef CAS PubMed.
- N. Segata, J. Izard, L. Waldron, D. Gevers, L. Miropolsky, W. S. Garrett and C. Huttenhower, Genome Biol., 2011, 12, 1–18 CrossRef.
- J. Gao, G. Liu, H. Li, L. Xu, L. Du and B. Yang, Bioprocess Biosyst. Eng., 2016, 39, 1115–1127 CrossRef CAS PubMed.
- L. Shi, Y. Huang, M. Zhang, Y. Yu, Y. Lu and F. Kong, Sci. Total Environ., 2017, 598, 77–86 CrossRef CAS PubMed.
- A. O. Sliekers, S. C. M. Haaijer, M. H. Stafsnes, J. G. Kuenen and M. S. M. Jetten, Appl. Microbiol. Biotechnol., 2005, 68, 808–817 CrossRef CAS PubMed.
- S. Park, W. Bae and B. E. Rittmann, Environ. Sci. Technol., 2010, 44, 335–342 CrossRef CAS PubMed.
- G. Liu and J. Wang, Chemosphere, 2015, 141, 19–25 CrossRef CAS PubMed.
- C. M. Fitzgerald, P. Camejo, J. Z. Oshlag and D. R. Noguera, Water Res., 2015, 70, 38–51 CrossRef CAS PubMed.
- N. A. Keene, S. R. Reusser, M. J. Scarborough, A. L. Grooms, M. Seib, J. Santo Domingo and D. R. Noguera, Water Res., 2017, 121, 72–85 CrossRef CAS PubMed.
- X. Zhang, Y. Liang, Y. Ma, J. Du, L. Pang and H. Zhang, Ecol. Eng., 2016, 93, 104–111 CrossRef.
- A. Bartroli, J. Perez and J. Carrera, Environ. Sci. Technol., 2010, 44, 8930–8935 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra07628j |
| This journal is © The Royal Society of Chemistry 2020 |