Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Label as you fold: methyltransferase-assisted functionalization of DNA nanostructures†
Christian
Heck
 *a,
Dmitry
Torchinsky
a,
Gil
Nifker
a,
Felix
Gularek
b,
Yael
Michaeli
a,
Elmar
Weinhold
b and
Yuval
Ebenstein
*a,
Dmitry
Torchinsky
a,
Gil
Nifker
a,
Felix
Gularek
b,
Yael
Michaeli
a,
Elmar
Weinhold
b and
Yuval
Ebenstein
 *a
*a
aSchool of Chemistry/Center for Nanoscience and Nanotechnology/Center for Light-Matter Interaction, Tel Aviv University, Ramat Aviv, Tel Aviv, Israel. E-mail: check@tauex.tau.ac.il; uv@post.tau.ac.il
bInstitute of Organic Chemistry, RWTH Aachen University, D-52056 Aachen, Germany
First published on 1st October 2020
Abstract
Non-DNA labels are key components for the construction of functional DNA nanostructures. Here, we present a method to graft covalent labels onto DNA origami nanostructures in an enzymatic one-pot reaction. The DNA methyltransferase M.TaqI labels the DNA nanostructures with azide groups, which serve as universal attachment points via click chemistry. Direct labeling with fluorescent dyes is also demonstrated. The procedure yields structures with high fluorescence intensities and narrow intensity distributions. In combination with UV crosslinking it enables the creation of temperature-stable, intense fluorescent beacons.
Introduction
DNA nanotechnology is a versatile technique for the construction of complex, functional nanostructures.1–4 Non-DNA components provide additional functionality, usually in the form of chemically-modified oligonucleotides that are hybridized to the nanostructures and that are typically manufactured by a DNA synthesizing company.5,6 Here, we expand the toolbox for DNA nanotechnology by enzymatic introduction of covalent modifications. The cost for chemically modified oligonucleotides renders the synthesis of DNA nanostructures expensive if many modifications are to be incorporated. Enzymatic labeling, on the other hand, offers an effective way to introduce many modifications at once. The locations of the modifications within a DNA nanostructure are determined by the recognition sequence of the respective labeling enzyme. When modified with fluorophores, for example, such strongly-labeled structures could be used as fluorescent beacons.7 The attachment of phospholipid anchors could facilitate the encapsulation with a lipid bilayer membrane for improved bioavailability.8 The interaction between DNA origami nanostructures and enzymes is poorly studied, so far limited to restriction endonucleases and ligases.9–12 The tight packing of DNA in these structures poses a challenge for enzymatic manipulation.DNA methyltransferases are enzymes that methylate double-stranded DNA. They work sequence-specifically and the resulting methylation pattern either protects from cleavage (in bacteria) or regulates gene activity (in higher organisms).13 The methyltransferase M.TaqI methylates adenine in the TCGA motif using the natural cofactor S-adenosyl-L-methionine (AdoMet or SAM). When chemically modified cofactors are supplied, methyltransferases can also transfer other chemical moieties, which was termed “methyltransferase-directed transfer of activated groups” (mTAG).14–17 An azide-carrying cofactor, for example, enables the sequence-specific, covalent introduction of azide groups into DNA.18,19 More or less arbitrary functional groups can be attached to these universal linkage points via click chemistry.20 While the labeling of genomic DNA is an established method, for DNA origami and some other DNA nanostructures it is not straight forward. The tightly-interwoven DNA in assembled DNA origami nanostructures is too closely-packed to allow access for the enzyme. Accordingly, our initial experiments on assembled DNA origamis were unsuccessful (Fig. S1†). On the other hand, labeling of staples and scaffold before annealing is prevented since DNA methyltransferases generally label double-stranded DNA. Wireframe DNA nanostructures might be free from this restriction,21 but offer only limited functionality. The strategy we present here to solve this conundrum is to label DNA origami nanostructures in situ, during the folding process. The thermophilic methyltransferase M.TaqI is particularly suited to label DNA origami during folding because of two reasons: 1 – Its optimal working temperature lies between 50 °C and 65 °C, which is also the temperature range in which DNA origami folds.22 2 – The recommended buffer for M.TaqI (NEB CutSmart) contains 10 mM magnesium ions, promising optimal folding conditions for DNA origami. These properties of M.TaqI enabled the implementation of the in situ labeling strategy, a scheme of which is shown in Fig. 1A. For testing purposes we chose the widely-used triangular DNA origami.23 The scaffold and staple strands of the DNA origami are mixed with methyltransferase M.TaqI and the azide-donating cofactor Ado-6-azide24 and exposed to a 1 h temperature ramp from 65 °C to 50 °C. The DNA origami folds within this temperature corridor and during the process of stepwise double strand formation, M.TaqI has the chance to label those double-stranded segments which have the recognition sequence TCGA. This temperature ramp is not supposed to yield fully intact DNA origamis, but only to label the constituent DNA material. In the next step, proteinase K is added to digest the methyltransferase that otherwise could prevent a proper origami folding when still being attached to the double-stranded DNA. After 1 h incubation at 40 °C, the origami is folded in the crude mixture by a standard folding protocol (cool down from 80 °C to 20 °C over 90 min). In an overnight reaction, dibenzocyclooctyne(DBCO)-carrying functional molecules (here: the fluorophore Cy5) are attached via strain-promoted azide alkyne cycloaddition (SPAAC) to the enzymatically introduced alkynes on the DNA origami. Then, the assembled origamis are purified by the desired method (agarose gel electrophoresis, centrifugal filters, etc.).
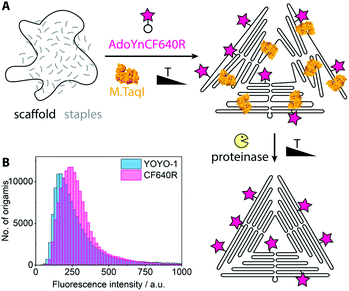
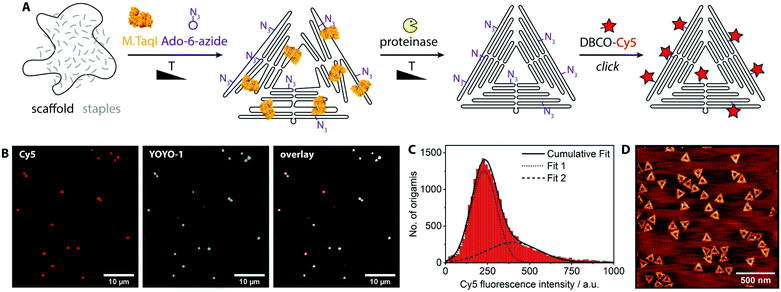 | ||
| Fig. 1 (A) Schematic labeling strategy: (1) while the DNA origami partially folds, M.TaqI labels scaffold and staples with azide groups. (2) The enzyme is digested and the DNA origami is properly folded. (3) The label of interest (here: Cy5) is attached via copper-free click reaction. (B) Fluorescence microscopy images of DNA origamis that were Cy5-labeled. Red channel: Cy5, covalent labels. Blue channel: YOYO-1, intercalating DNA stain. For a control where no enzyme had been added see Fig. S2.† (C) Histogram of the Cy5 fluorescence intensity distribution of ca. 500 fields of view after spot intensities were analyzed by an image analysis software; the data were fitted by two Gaussians, the centers of which are at 229 a.u. (Fit 1) and 400 a.u. (Fit 2). (D) AFM image of the assembled and labeled DNA origamis. | ||
Results and discussion
For single-molecule fluorescence measurements, the covalently labeled DNA origami nanostructures were stained with the intercalating dye YOYO-1 and adsorbed onto chemically activated coverslips.25 The fluorescence microscopy images show a nearly perfect overlap between the DNA stain (YOYO-1, blue) and the introduced fluorophores (Cy5, red) (Fig. 1B). A control sample where the enzyme had been left out (Fig. S2†) shows no intense Cy5 signals. A large data set of ca. 500 fields of view was analyzed by an in-house image analysis program to assess the fluorescence intensity distribution of the Cy5 labels.26 The resulting histogram is shown in Fig. 1C. The data can be approximated by two Gaussians, where the smaller peak is at about double the intensity of the larger one. While the main peak represents individual DNA origamis, the smaller one likely originates from two DNA origamis having adsorbed next to each other: since the optical resolution is ∼300 nm, it may occur that two triangles are closer than that and will appear as a single spot with twice the fluorescence intensity. (See Fig. S3† for a more detailed view on the fit.) The triangle design is based on the 7249 base-long M13mp18 scaffold, which contains 12 TCGA motifs. Since these are palindromic, each motif theoretically can be labeled twice. Taking into account that some motifs are at cross over positions and likely not accessible to the enzyme, the total number of expected labels on the triangle will be below 24. A recent study showed that enzymes act in a binary on/off fashion on assembled DNA origami nanostructures, with the number of accessible sites depending on the type of enzyme, type of DNA origami, location on the origami, structural strain and mismatches in the local environment of the recognition site.9 In a follow-up, local and global mechanical fluctuations were highlighted as key determinants for enzyme accessibility, with high fluctuations favoring access.10 This matches our observation that elevated temperature and the yet-unfolded state of the origami increase the labeling efficiency. Notably, also the methyltransferase M.EcoRI was observed to prefer relaxed over tense DNA double helices as substrates.27 The narrow fluorescence intensity distribution observed in our experiments indicates most origamis to carry a similar, if not the same number of labels. When fluorescence microscopy samples were prepared so that remnant single Cy5 molecules would also adsorb to the glass surface, the fluorescence intensity of the labeled origamis was at least eight times as intense as the single dye molecules (Fig. S4†). This indicates that each origami carried several dye molecules. Atomic force microscopy (AFM, Fig. 1D) confirmed that the DNA origamis were well-folded.Azide groups provide a high flexibility in terms of functionalities that can be attached. However, the labeling process can be streamlined when the DNA methyltransferase directly grafts the group of interest onto the DNA. For example, cofactors modified with fluorophores can also be processed by some methyltransferases.28 Thereby the click chemistry step can be omitted and the reaction mix can be directly purified after the origami assembly. Fig. 2 shows the reaction scheme and a fluorescence intensity histogram for direct labeling of a triangular origami with M.TaqI and a cofactor analogue containing the dye CF640R.29 The histogram further shows the intensity distribution of the intercalating dye YOYO-1. Exemplary fluorescence microscopy images can be found in the ESI (Fig. S5†). Due to the different labeling modes of the two dyes, the intensity distributions are not expected to be identical. However, the narrow intensity distributions are good indications for homogeneous labeling.
It lies in the nature of DNA methyltransferases that they don't discriminate between scaffold and staple strands within their palindromic recognition sequences. This means that also the scaffold strand is covalently labeled in the enzymatic approach, which is not possible with conventional methods. Selective labeling of only the scaffold strand may be achieved by supplying methylated staple strands.
Some of the most-promising applications of DNA nanostructures lie in the biological and medical field.30,31 There, DNA nanostructures are often exposed to environments with low or no magnesium ion content and to higher temperatures, which both have the potential to denature them. In order to provide robust fluorescent beacons for diagnostic purposes, we stabilized the triangular DNA origamis by crosslinking. The origami shown in Fig. 1 and 2 were prepared with staple strands containing additional thymines at staple crossovers and ends which enable crosslinking by UV irradiation (“UV welding”).32 When preparing the crosslinked fluorescent beacons, bleaching was prevented by adding the fluorescent dyes only after the UV irradiation (Fig. 3). To test their resilience, the crosslinked and fluorescently labeled structures were exposed to a heat shock of 65 °C for 30 min. The structures stabilized by UV retained most of their fluorescence intensity (upper histograms in Fig. 3), as opposed to a control batch that has not been exposed to UV (lowest histogram in Fig. 3 and Fig. S6†). Subsequent AFM analysis showed the triangles to be intact but deformed despite the crosslinking step, so further optimization of the crosslinking conditions will be pursued (Fig. S7†). Compared to conventional materials, DNA origami-based beacons are attractive due to their precise physicochemical properties, biocompatibility and multifunctional programmability.30
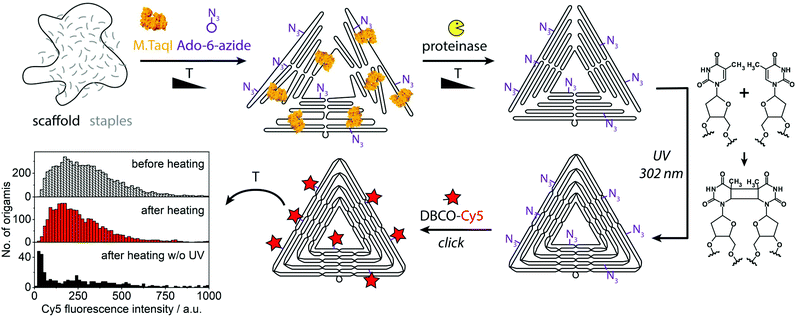 | ||
| Fig. 3 Assembly of UV-crosslinked fluorescent beacons. The histogram shows fluorescence intensity distributions of a crosslinked sample before and after it was kept at 65 °C for 30 min and of a control that was heated without having been crosslinked. For a histogram of the control before heating see Fig. S6.† | ||
Conclusions
In summary, we demonstrated the covalent labeling of DNA origami nanostructures by the DNA methyltransferase M.TaqI. DNA origami were directly labeled with fluorophores or with azide groups followed by attachment of fluorophores through copper-free click chemistry. Both yielded reproducibly intense fluorescent beacons, estimated to carry at least eight fluorophores per origami. UV crosslinking enabled beacons that retained a high degree of their fluorescence even after prolonged heat treatment.Functionalization of the DNA origami is dependent on the position and accessibility of M.TaqI's recognition sequence TCGA. Thus, the advantages of this covalent labeling approach come at the price of limited control over the position and number of the introduced labels. This is especially true when compared to the traditional hybridization of modified staple strands, whose incorporation can be programmed with nanometer precision. However, if the latter is realized by modified internal staples, many different chemically modified staples have to be synthesized in parallel. If it is to be realized by staple extensions to which ‘reporter strands’ of identical sequence are hybridized, then losses might occur as even long reporter strands might detach over time.
Azide groups provide a high degree of flexibility in terms of the group to be attached. Even temperature-sensitive biomolecules such as proteins may be linked to origami via the two-step click chemistry route described in the first section of the paper. At least ten different DNA methyltransferases are known to be compatible with chemically modified cofactors.33 They offer potential to either label alternative positions, or, when combined, to increase the number of labels per DNA origami. The latter is especially true for M.SssI: its recognition sequence ‘CG’ is present 325 times in the commonly-used p7249 scaffold. Since this enzyme is not stable at the standard folding temperatures, DNA origami folding would have to be shifted to lower temperatures, e.g. by DNA-selective denaturing agents.34 In the past years there was significant progress in upscaling scaffold and staple production for DNA origami with the help of enzymatic systems.35–37 The enzymatic labeling strategy presented here could enable the concomitant modification with functional non-DNA components in such biotechnological upscaling approaches.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Gal Radovsky for AFM measurements. C. H. is grateful for the University of Potsdam-Tel Aviv University Postdoc Exchange Fellowship and the Ratner Postdoc Fellowship. Y. E. acknowledges funding from ERC Consolidator grant 817811; Y. E. and E. W. acknowledge funding from a joint Israeli German R&D Nanotechnology grant (German Federal Ministry of Education and Research and Israeli Innovation Authority 13GW0282B and NATI 61976).References
- F. Hong, F. Zhang, Y. Liu and H. Yan, Chem. Rev., 2017, 117, 12584–12640 CrossRef CAS.
- N. C. Seeman and H. F. Sleiman, Nat. Rev. Mater., 2018, 3, 17068 CrossRef CAS.
- A. Kuzyk, R. Jungmann, G. P. Acuna and N. Liu, ACS Photonics, 2018, 5, 1151–1163 CrossRef CAS.
- C. Heck, Y. Kanehira, J. Kneipp and I. Bald, Angew. Chem., Int. Ed., 2018, 57, 7444–7447 CrossRef CAS.
- J. J. Schmied, M. Raab, C. Forthmann, E. Pibiri, B. Wünsch, T. Dammeyer and P. Tinnefeld, Nat. Protoc., 2014, 9, 1367–1391 CrossRef CAS.
- M. Madsen and K. V. Gothelf, Chem. Rev., 2019, 119, 6384–6458 CAS.
- A. Gopinath, E. Miyazono, A. Faraon and P. W. K. Rothemund, Nature, 2016, 535, 401–405 CrossRef CAS.
- S. D. Perrault and W. M. Shih, ACS Nano, 2014, 8, 5132–5140 CrossRef CAS.
- A. Stopar, L. Coral, S. Di Giacomo, A. F. Adedeji and M. Castronovo, Nucleic Acids Res., 2018, 46, 995–1006 CrossRef CAS.
- A. Suma, A. Stopar, A. W. Nicholson, M. Castronovo and V. Carnevale, Nucleic Acids Res., 2020, 48, 4672–4680 CrossRef.
- S. Ramakrishnan, B. Shen, M. A. Kostiainen, G. Grundmeier, A. Keller and V. Linko, ChemBioChem, 2019, 20, 2818–2823 CrossRef CAS.
- S. Ramakrishnan, L. Schärfen, K. Hunold, S. Fricke, G. Grundmeier, M. Schlierf, A. Keller and G. Krainer, Nanoscale, 2019, 11, 16270–16276 RSC.
- A. Jeltsch, ChemBioChem, 2002, 3, 274–293 CrossRef CAS.
- G. Lukinavičius, V. Lapiene, Z. Staševskij, C. Dalhoff, E. Weinhold and S. Klimašauskas, J. Am. Chem. Soc., 2007, 129, 2758–2759 CrossRef.
- A. A. Wilkinson, E. Jagu, K. Ubych, S. Coulthard, A. E. Rushton, J. Kennefick, Q. Su, R. K. Neely and P. Fernandez-Trillo, ACS Cent. Sci., 2020, 6, 525–534 CrossRef CAS.
- C. Heck, Y. Michaeli, I. Bald and Y. Ebenstein, Curr. Opin. Biotechnol., 2019, 55, 151–158 CrossRef CAS.
- H. Sharim, A. Grunwald, T. Gabrieli, Y. Michaeli, S. Margalit, D. Torchinsky, R. Arielly, G. Nifker, M. Juhasz, F. Gularek, M. Almalvez, B. Dufault, S. Sen Chandra, A. Liu, S. Bhattacharya, Y. W. Chen, E. Vilain, K. R. Wagner, J. Pevsner, J. Reifenberger, E. T. Lam, A. R. Hastie, H. Cao, H. Barseghyan, E. Weinhold and Y. Ebenstein, Genome Res., 2019, 29, 646–656 CrossRef CAS.
- M. H. Lauer, C. Vranken, J. Deen, W. Frederickx, W. Vanderlinden, N. Wand, V. Leen, M. H. Gehlen, J. Hofkens and R. K. Neely, Chem. Sci., 2017, 8, 3804–3811 RSC.
- E. Kriukiene, V. Labrie, T. Khare, G. Urbanavičiute, A. Lapinaite, K. Koncevičius, D. Li, T. Wang, S. Pai, C. Ptak, J. Gordevičius, S. C. Wang, A. Petronis and S. Klimašauskas, Nat. Commun., 2013, 4, 2190 CrossRef.
- N. J. Agard, J. M. Baskin, J. A. Prescher, A. Lo and C. R. Bertozzi, ACS Chem. Biol., 2006, 1, 644–648 CrossRef CAS.
- N. P. Agarwal, M. Matthies, B. Joffroy and T. L. Schmidt, ACS Nano, 2018, 12, 2546–2553 CrossRef CAS.
- J. P. J. Sobczak, T. G. Martin, T. Gerling and H. Dietz, Science, 2012, 338, 1458–1461 CrossRef CAS.
- P. W. K. Rothemund, Nature, 2006, 440, 297–302 CrossRef CAS.
- G. Lukinavičius, M. Tomkuvienè, V. Masevičius and S. Klimašauskas, ACS Chem. Biol., 2013, 8, 1134–1139 CrossRef.
- D. Torchinsky, Y. Michaeli, N. R. Gassman and Y. Ebenstein, Chem. Commun., 2019, 55, 11414–11417 RSC.
- D. Torchinsky and Y. Ebenstein, Nucleic Acids Res., 2016, 44, e17 CrossRef.
- M. Endo, Y. Katsuda, K. Hidaka and H. Sugiyama, J. Am. Chem. Soc., 2010, 132, 1592–1597 CrossRef CAS.
- A. Grunwald, M. Dahan, A. Giesbertz, A. Nilsson, L. K. Nyberg, E. Weinhold, T. Ambjörnsson, F. Westerlund and Y. Ebenstein, Nucleic Acids Res., 2015, 43, e117 CrossRef.
- T. Gilboa, C. Torfstein, M. Juhasz, A. Grunwald, Y. Ebenstein, E. Weinhold and A. Meller, ACS Nano, 2016, 10, 8861–8870 CrossRef CAS.
- Y. Ke, C. Castro and J. H. Choi, Annu. Rev. Biomed. Eng., 2018, 20, 375–401 CrossRef CAS.
- D. Jiang, Z. Ge, H. J. Im, C. G. England, D. Ni, J. Hou, L. Zhang, C. J. Kutyreff, Y. Yan, Y. Liu, S. Y. Cho, J. W. Engle, J. Shi, P. Huang, C. Fan, H. Yan and W. Cai, Nat. Biomed. Eng., 2018, 2, 865–877 CrossRef CAS.
- T. Gerling, M. Kube, B. Kick and H. Dietz, Sci. Adv., 2018, 4, eaau1157 CrossRef CAS.
- J. Deen, C. Vranken, V. Leen, R. K. Neely, K. P. F. Janssen and J. Hofkens, Angew. Chem., Int. Ed., 2017, 56, 5182–5200 CrossRef CAS.
- A. Kopielski, A. Schneider, A. Csáki and W. Fritzsche, Nanoscale, 2015, 7, 2102–2106 RSC.
- F. Praetorius, B. Kick, K. L. Behler, M. N. Honemann, D. Weuster-Botz and H. Dietz, Nature, 2017, 552, 84–87 CrossRef CAS.
- C. Ducani, C. Kaul, M. Moche, W. M. Shih and B. Högberg, Nat. Methods, 2013, 10, 647–652 CrossRef CAS.
- D. Minev, R. Guerra, J. Y. Kishi, C. Smith, E. Krieg, K. Said, A. Hornick, H. M. Sasaki, G. Filsinger, B. J. Beliveau, P. Yin, G. M. Church and W. M. Shih, Nucleic Acids Res., 2019, 47, 11956–11962 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0nr03694c |
| This journal is © The Royal Society of Chemistry 2020 |