Open Access Article
Open Access ArticleEstablishing empirical design rules of nucleic acid templates for the synthesis of silver nanoclusters with tunable photoluminescence and functionalities towards targeted bioimaging applications†
Jason Y. C.
Lim‡
 a,
Yong
Yu‡
a,
Yong
Yu‡
 a,
Guorui
Jin
a,
Kai
Li
a,
Guorui
Jin
a,
Kai
Li
 a,
Yi
Lu
a,
Yi
Lu
 b,
Jianping
Xie
b,
Jianping
Xie
 c and
Yen Nee
Tan
c and
Yen Nee
Tan
 *ad
*ad
aInstitute of Materials Research and Engineering, The Agency for Science, Technology and Research (A*STAR), 2 Fusionopolis Way, #08-03, Innovis, 138634, Singapore
bDepartment of Chemistry, University of Illinois at Urbana-Champaign, Urbana, IL 61801, United States
cDepartment of Chemical and Biomolecular Engineering, National University of Singapore, 10 Kent Ridge Crescent, 119260, Singapore
dFaculty of Science, Agriculture & Engineering, Newcastle University, Newcastle Upon Tyne NE1 7RU, UK. E-mail: yennee.tan@newcastle.ac.uk
First published on 23rd July 2020
Abstract
DNA-templated silver nanoclusters (AgNCs) are an emerging class of ultrasmall (<2 nm) fluorophores with increasing popularity for bioimaging due to their facile synthesis and tunable emission color. However, design rules correlating different nucleotide sequences with the photoemission properties of AgNCs are still largely unknown, preventing the rational design of DNA templates to fine-tune the emission color, brightness and functionalities of AgNCs for any targeted applications. Herein, we report a systematic investigation to understand the empirical influences of the four basic DNA nucleotides on AgNC synthesis and their effects on photoluminescence properties. After establishing the importance of nucleotide–Ag+ binding and AgNC encapsulation within DNA tetraplex structures, we then determined the unique attributes of each individual nucleobase using different combinations of systematically varied DNA templates. Using the empirical design rules established herein, we were able to predict the photoluminescence behaviours of AgNCs templated by complex aptamer sequences with specific binding affinity to human cancer cells, and to deliberately control their emission color by rational modifications of the DNA template sequences for targeted bioimaging. Our empirical findings from this systematic experimentation can contribute towards the rational design of DNA sequences to customise the photoluminescence properties and biofunctionalities of DNA-protected AgNCs towards multicolour targeted bioimaging applications.
1. Introduction
Nucleic acids such as DNA are one of the most important biological macromolecules in nature, responsible for the storage and transmission of genetic information as well as regulation of many biological processes. Structurally consisting of only four universal nucleotides (i.e., adenine, thymine, cytosine and guanine), their specific permutations determine the conformations, and supramolecular structures, as well as the chemical and biological properties of DNA. Due to the ease of sequence tunability and well-understood base-pairing properties, DNA molecules can be synthetically designed at high precision and produced at low cost, leading to diverse application of DNA in biomedicine (e.g., biomarkers for diseases diagnostics), materials science (e.g., self-assembled DNA origami), electronics (e.g., DNA logic gates), and computing.1–4 More recently, the realization that biomolecules can also act as precursors (to form nanodots),5–10 and capping or reducing agents for the coordination of metal cations has revolutionized the field of bionanotechnology,11,12 opening up new opportunities to produce exotic biohybrid nanomaterials such as DNA-assembled metal nanostructures,13–16 DNA-/peptide-synthesized anisotropic metal nanoparticles,17–21 protein-/peptide-templated ultrasmall metal nanoclusters,22–27 and even nucleotide-derived nanodots (without metal precursors),7 which inherit not only the biocompatibility and functionality of biomolecules, but also possess unique physiochemical properties of nanomaterials for a wide range of technological applications.11,28–30Since the seminal work of using DNA molecules as stabilizers (or templates) to form photoluminescent silver nanoclusters (AgNCs) by Dickson and coworkers in 2004,31 the fundamental study and applications of DNA-templated AgNCs (DNA-AgNCs) have witnessed dramatic advancements in recent years.32–36 These DNA-AgNCs are typically smaller than 2 nm,37 approaching the Fermi wavelength of electrons. Due to strong quantum confinement, the continuous density of states present in the bulk metals are broken up into discrete energy levels which give rise to optical, electrical and chemical properties distinctly different from those of the bulk metal and metal nanoparticles of larger sizes.38 While AgNCs can also be produced using templates such as zeolites,39–41 organic polymers,42–44 thiols,45–51 sugar molecules,52 and peptides,53–55 DNA-AgNCs are especially valued for biological applications due to their biological and physicochemical properties. DNA-AgNCs are highly biocompatible, possess generally good photostability, are easy to synthesize using environmentally-benign reagents, and possess high versatility for tuning their photophysical properties, including photoemission color and brightness, across the entire range of the visible and near-IR spectrum by simply varying the nucleotide sequences of the DNA templates. They are important alternatives to traditional organic fluorophores and inorganic quantum dots as organic fluorophores often require multi-stepped chemical syntheses and are susceptible to photobleaching,56 while quantum dots can pose cytotoxicity concerns.57,58
A huge repertoire of AgNCs with widely varying photophysical properties are currently known,35 with those templated by poly-cytosine sequences59 being arguably the most thoroughly studied. However, to date, the ability to predict AgNC emission colors a priori from simply knowing the templating DNA sequence still remains unrealized. This is due to the numerous complexities preventing a full elucidation of these design rules. For example, DNA molecules can self-assemble and adopt supramolecular structures in solution, each nucleobase has different inherent affinities for silver, the great variety of geometries and atomic compositions of AgNCs,60 and even electron-transfer interactions between nucleobases and AgNCs,61 just to name a few. Traditionally, identifying suitable DNA molecules to template the formation of AgNCs with different photoluminescence colors relied on the trial-and-error screening of DNA libraries.62,63 More recently, machine learning and data mining approaches have been exploited to better predict how different templating sequences can affect the AgNC emission colors with greater precision and accuracy.64,65 Nevertheless, a systematic empirical approach to elucidate these coveted design rules by studying the effects of methodologically changing combinations of nucleobases on the DNA template has yet to be demonstrated.
In this study, we report a systematic empirical investigation on the design principles of ssDNA sequences to rationally tune the photoemission colors of AgNCs. Compared to hybridized DNA templates such as DNA hairpin loops and double-stranded DNA for AgNC templating, ssDNA templates are more versatile due to their conformational flexibility which allow exposure of most if not all the nucleobases present in the DNA template for easy coordination and interaction with silver cations or AgNCs. While this ensures higher binding affinities and potentially greater AgNC stabilization by simultaneous binding to multiple nucleobases, ssDNA templates also allow better fundamental understanding of how different DNA nucleobases affect the photoluminescence of the resultant AgNCs by eliminating additional complications arising from structural features such as loops or hairpins, despite their known ability to also template photoluminescent AgNCs.66 For our empirical study, we first established the fundamental influence exerted by each nucleobase on AgNC formation using four homo-oligonucleotides containing 20 cytosines, guanines, adenines or thymines, denoted as dC20, dG20, dA20 and dT20, respectively, as templates of AgNCs. We then studied the effects of introducing a second nucleobase into the DNA sequence by systematically changing their relative proportions in the sequence. These dual-block DNA sequences contain combinations of two nucleobases with the general formula of d[X5nY(20−5n)], where n is an integer from 0 to 4 and X and Y are any two of the four possible nucleotides (A, T, C, and G). Intriguingly, we found that each of the four nucleobases exert their own characteristic influences on the photoluminescence properties of AgNCs. Thereafter, to confirm the importance of having consecutive homonucleotide sequences for formation of bright-luminescent AgNCs, we studied the effects of using interdigitated sequences containing two nucleobases, bearing the general formula d[XY]10. Importantly, we evaluate the influence of different nucleobase combinations on time-dependent variations in AgNC emissive properties – a factor hitherto not considered in the vast majority of these studies. We then validated the empirical rules we have established by predicting the photoluminescence properties of AgNCs templated by several complex long aptamer sequences (41–77 mers) containing all four nucleobases. These design rules also facilitated our design of bi-functional DNA templates by incorporating cancer cell specific aptamer sequences, leading to the formation of bright photoluminescent DNA-AgNCs with a tailored emissive color and cell-targeting capabilities for cancer diagnostics. Our systematic empirical approach can complement existing high-throughput screening and data-analytical methods to better understand how the photoluminescence properties of AgNCs can be influenced by a specific templating DNA sequence.
2. Results and discussion
2.1. Pre-requisites for photoluminsecent DNA-templated silver nanocluster formation
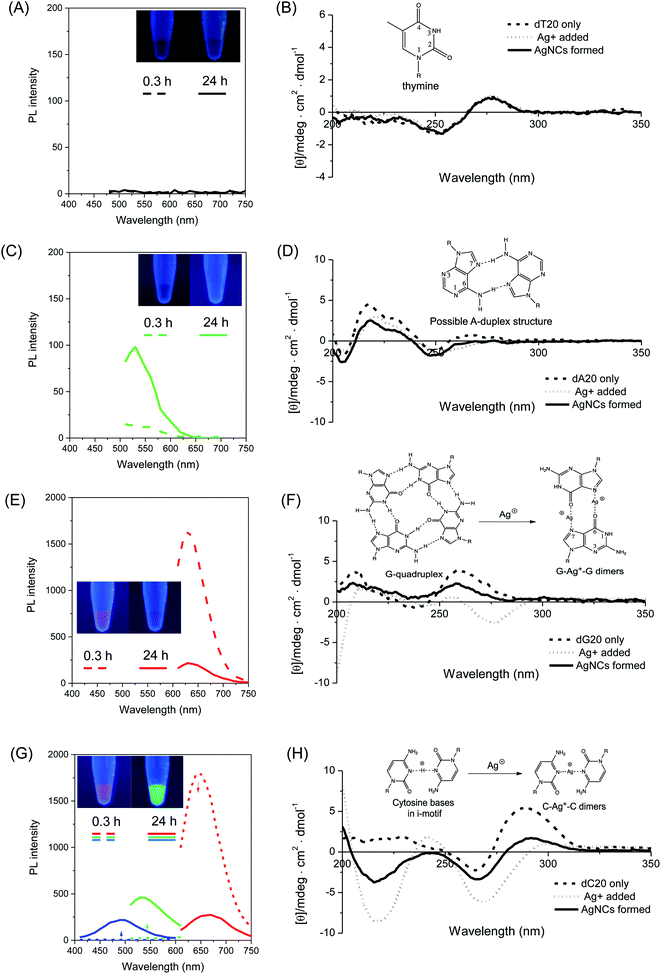
Firstly, four homo-oligonucleotides with fixed length of 20 bases, i.e. homo-adenine (dA20), homo-tymine (dT20), homo-guanine (dG20), and homo-cytosine (dC20), were used as templates to elucidate the essential structural features of DNA for the formation of photoluminescent silver nanoclusters (AgNCs). Fig. 1 shows the time evolution photoluminescence profiles (left panel) of AgNCs templated by the respective homo-oligonucleotides. In the presence of dA20 or dT20, no AgNCs were formed upon the chemical reduction of the silver nitrate precursor by sodium borohydride (NaBH4) after an initial incubation period of 20 minutes. At 24 hours, the dT20-templated sample remained non-emissive (Fig. 1A), while spectroscopically-pure (λex = 440 nm/λem = 520 nm) and weakly-luminescent green emissive AgNCs were formed in the presence of the dA20 template (Fig. 1C). As shown in Fig. 1E, the dG20 template produced red-emitting AgNCs in 20 minutes (λex = 570 nm/λem = 630 nm), whose photoluminescence (PL) intensity decreased with time and no other emitter was formed in the process. While the dC20 template also formed red-emitting AgNCs (λex = 580 nm/λem = 670 nm) initially, a time-dependent color conversion of dC20-AgNCs to green- (λex = 440 nm/λem = 540 nm) and blue-emitting (λex = 340 nm/λem = 490 nm) species was observed after 24 hours of incubation, giving detectable emission color changes (inset in Fig. 1G). Interestingly, cytosine and guanine, which formed bright emissive AgNCs in this study, were found to bind strongly to the silver precursor (Ag+), as determined by the isothermal titration calorimetric (ITC) studies reported by Sastry et al.67 The same study also showed that the binding of thymine, which did not form photoluminescent AgNCs, to Ag+ was the weakest. This highlights the importance of nucleobase–Ag+ binding in forming photoluminescent AgNCs.To investigate the functional roles of DNA templates in the synthesis of AgNCs, circular dichroism (CD) spectroscopy was employed to obtain some mechanistic insights into the structural changes of DNA templates in the process of AgNC formation. Thus, the CD spectra of the respective DNA templates were recorded at various stages of AgNC formation as shown in Fig. 1 (right panel), i.e., before (dashed line) and after addition (dotted line) of silver nitrate, as well as 20 minutes after addition of NaBH4 to form the AgNCs (solid lines). Similar to the PL spectrum, no detectable changes in the CD spectra of dT20 templates were observed upon addition of Ag+, and after NaBH4 reduction, with all three spectra practically superimposable on each other (Fig. 1B). For the dA20 template, a positive peak at ca. 220 nm was observed in its CD spectrum (Fig. 1D, dashed line), suggesting the presence of self-assembled supramolecular structures, such as adenine–adenine dimers in solution.68 Subsequently, the binding of Ag+ and its reduction by NaBH4 caused marginal, but detectable changes to its CD spectra, implying that only small structural changes in the DNA template occurred during these processes. These findings indicated that both the dT20 and dA20 sequences interact weakly with Ag+ and Ag0 at neutral pH, in accordance with our aforementioned findings that they act as poor templates for the formation of photoluminescent AgNCs.
In stark contrast, the homo-oligonucleotide sequences which are capable of forming bright emissive AgNCs (i.e., dC20 and dG20) showed dramatic changes in their CD spectra during the AgNC formation process. For instance, the CD spectra of dG20 exhibited a positive peak at 260 nm and a negative peak at 240 nm (Fig. 1F, dashed line), indicating the parallel-stranded G-quadruplex structures where the individual oligo-guanine strands formed tetrameric structures via Hoogsteen base pairing in solution.69–72 When Ag+ was added, the characteristic peaks of the G-quadruplex structure disappeared, accompanied with the appearance of a new prominent peak at a negative ellipticity of 275 nm (Fig. 1F, dotted line). This observation strongly implies the significant disruption of the G-quadruplex structure upon Ag+ binding, probably due to the formation of guanine–Ag+–guanine dimers,73 which is consistent with the strong guanine–Ag+ binding affinity.67 Interestingly, upon chemical reduction of Ag+, the original peaks at 240 nm and 260 nm re-appeared in the CD spectrum (Fig. 1F, solid line), suggesting that the G-quadruplex structure of the dG20 template was restored to some extent in solution following the release of Ag+ from the binding sites to form AgNCs, thus freeing the guanine ligands for re-formation of the G-quadruplex structures. Owing to their hydrophobicity, the as-synthesized AgNCs are likely to be encapsulated within the hydrophobic interior of the reformed G-quadruplex scaffolds. This process most likely stabilizes the as-formed AgNCs against various photoluminescence quenching pathways in solution, resulting in the high brightness of red emissive dG20-AgNCs as observed in this study. As shown in Fig. 1H (dashed line), the CD spectrum of dC20 alone displayed a positive peak at 285 nm, which is a signature of i-motif tetraplex structures in solution.74 However, the introduction of Ag+ resulted in the loss of this characteristic peak with simultaneous appearance of two negative peaks at 220 nm and 265 nm (Fig. 1H, dotted line), suggesting the binding of cytosine bases to Ag+. Concomitantly, this process led to a severe disruption of the original i-motif structure, possibly arising from the coordination of cytosine bases to Ag+ to form cytosine–Ag+–cytosine dimers75 that tilted the bases relative to the helical axis. However, upon the chemical reduction of Ag+ by NaBH4, a partial restoration of the i-motif structure was seen (Fig. 1H, solid line), allowing encapsulation and stabilization of the as-formed AgNCs within the interior of the hydrophobic i-motif. Indeed, i-motif structures in DNA sequences with repeated C4 units have been shown to successfully template the synthesis of photoluminescent AgNCs.76
These CD spectroscopic studies have unravelled the important aspects of designing DNA templates for the formation of bright photoluminescent AgNCs. Specifically, two key pre-requisites have been established. Firstly, strong binding affinity of the nucleobase to Ag+ is necessary to hold the metal cation in close proximity for an efficient interaction with the DNA bases upon chemical reduction. This process reduces the probability of forming non-emissive silver nanoparticles of larger sizes (>2 nm). Secondly, self-assembly of DNA molecules into supramolecular structures that can interact with silver atoms and encapsulate the as-formed AgNCs within DNA cages is necessary, thereby stabilizing them against photoluminescence quenching. Indeed, recent crystallographic evidence of AgNCs templated by a DNA decamer reveals very tight binding of the AgNCs within the hydrophobic interior of the DNA structure, completely shielded from the surrounding aqueous environment.77 Thus, DNA templates that are unable to fulfill the aforementioned criteria such as dT20 and dA20 as demonstrated herein were unable to form bright emissive AgNCs in solution.
To better understand the color change mechanism of bright emissive dC20-AgNCs and photoluminescence decay of dG20-AgNCs as observed over time, as shown in Fig. 1G and E respectively, detailed structural studies of DNA templates and the atomic number of AgNCs were further investigated using CD spectroscopy and electrospray ionization (ESI) mass spectroscopy (MS). The time course CD spectra of dC20-AgNCs over 24 hours revealed that the color-conversion phenomenon is accompanied by increasing disruption of the i-motif structure of dC20 (ESI Fig. S1†), which is similar to the structural change of the dC20 template interacting with Ag+ (Fig. 1H, dotted line). This observation strongly suggests that increasing quantities of Ag+ are produced during AgNC aging. The ESI mass spectrum obtained 24 hours after the addition of NaBH4 for AgNC formation showed the presence of multiple species ranging from 6 to 10 Ag atoms per nanocluster (ESI Fig. S2†). The driving force of this conversion has been attributed to the oxidative decomposition of AgNCs by atmospheric oxygen.78–80
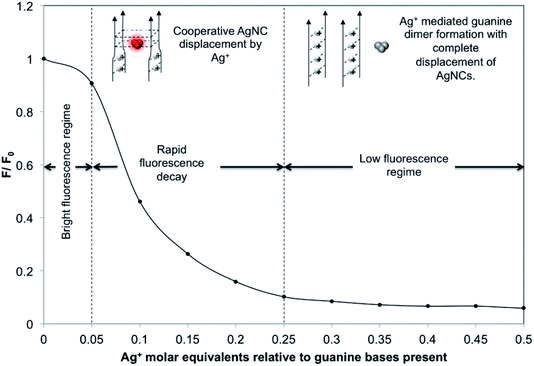
In a similar way, dG20-AgNCs may also be subjected to oxidative decomposition by oxygen in the air to generate Ag+ ions, which can in turn disrupt the G-quadruplex structure of dG20 templates. The CD spectra of dG20-AgNCs with increasing ages (ESI Fig. S3†) showed a corresponding increase in prominence of the 275 nm peak, which is the characteristic CD spectra of dG20 binding with Ag+. However, the ESI-MS results showed that the dG20-AgNCs were composed of predominantly 8 Ag atoms per cluster (ESI Fig. S4†), which is different from that observed for the dC20-AgNCs (ESI Fig. S2†). This is consistent with the observation that only a single red-emitting species was present in the solution when exposed to air (Fig. 1E). These results may suggest that the oxidative decomposition of AgNCs in the dG20 template occur more extensively than that in the dC20 template to form Ag+. Thus, AgNCs emitting at shorter wavelengths would not be formed and stabilized within the dG20 template, presumably due to the large difference in binding affinity between the as-generated Ag+ and AgNCs to the guanine bases. The effect of decomposition-generated Ag+ due to the dG20-AgNC oxidation61 induced structural change of the DNA template was supported by a titration experiment, where the PL intensity of a freshly-prepared solution of dG20-AgNCs was monitored with increasing quantities of added AgNO3 (Fig. 2). It was found that total quenching of the PL intensity of AgNCs occurred at an approximately 0.25 equivalents of Ag+ relative to the guanine bases, corresponding to one Ag+ added for every four guanine bases present in the template. Thus, for every four guanine bases that form a tetrad in the G-quadruplex structure, dimer formation involving any two of them (coordinated by 1 Ag+) can cause structural disruption to displace any photoluminescent AgNCs present within the template. Based on these results, an oxidation-driven mechanism may be proposed for the decomposition of dG20-AgNCs (inset in Fig. 2). As illustrated in this scheme, the high affinity of Ag+ to dG20 forms the guanine–Ag+–guanine dimers, which disrupt the G-quadruplex structure by changing the orientation of guanine bases relative to each other and enlarging their separation.10 This weakens the guanine–AgNC binding interaction and increases the susceptibility of AgNCs to oxidative decomposition and displacement by the released Ag+ from their binding sites.
2.2. Effects of combining two different nucleotides on the photoluminescence of AgNCs
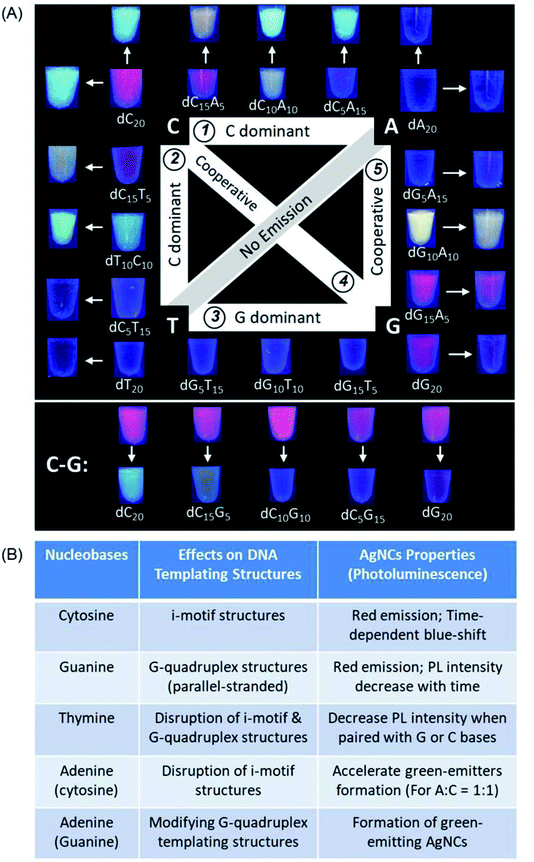
After the pre-requsites for forming ultrasmall AgNCs by homo-oligonucleotides have been set, systematic investigation was extended to a series of DNA templates containing any combination of two types of nucleobases with a general formula of 5′-[dX5nY(20−5n)]-3′ (where X and Y represent any two of the four possible nucleobases and n is an integer from 0 to 4) to synthesize AgNCs with fine-tuned properties. Fig. 3A shows all the possible combinations of two nucleotides in a 20 nt DNA template (see the complete sequences in ESI Table S1†) and the digital photos of the corresponding DNA-AgNCs under UV light, except for the A–T series which failed to produce any emissive species.Based on the above systematic investigations, we have summarized a set of empirical rules to highlight the influence of each nucleotide in a series of 20-mer long DNA templates on the photoluminescence properties of AgNCs (Fig. 3B). First, cytosine when present as the main component of any two-nucleotide combination (>50%) produces red-emitting AgNCs, which subsequently change to green and blue-emitting species. Second, guanine when present as the main component of any two-nucleotide combination (>50%), produces red-emitting AgNCs with PL intensity decaying with time. Third, both cytosine and guanine in the same sequence show co-dominant effects on the photoluminescence properties of the as-synthesized AgNCs as these two nucleobases bind strongly to Ag+ and are able to form distinct supramolecular structures. Fourth, adenine exerts strong influences on the luminescence of AgNCs when paired with cytosine or guanine, especially in near equal quantities: both dC10A10 and dG10A10 sequences show notable formation of distinct bright-luminescent green emitters. Finally, thymine alone cannot produce luminescent AgNCs, but can disrupt the supramolecular structure of the DNA template. The combination of thymine and guanine leads to reduced photoluminescence intensity of the resultant AgNC. Contrastingly, thymine–cytosine combinations still allow bright AgNCs to be formed, albeit favouring the production of green emitters.
2.3. Prediction of the photoluminescence colors of aptamer-templated AgNCs for targeted cancer cell imaging
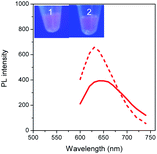
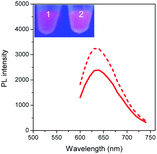
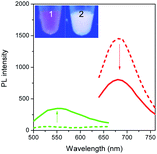
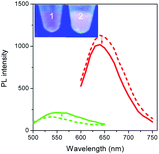
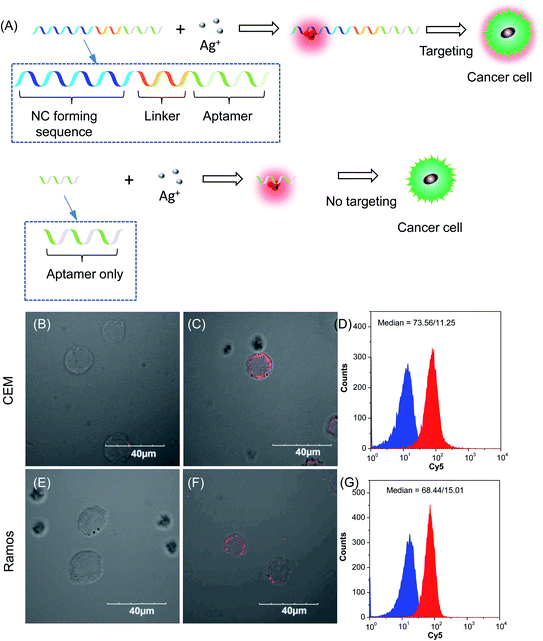
To validate our empirical design rules for predicting the emission color of AgNCs templated by arbitrary DNA sequences, we selected two ssDNA aptamers (sgc 8 and TDO 5 which are known to bind to the human cancer cell lines CEM and Ramos respectively)82 as DNA templates for AgNC synthesis. The sgc 8 and TDO 5 aptamers have 41 and 47 nucleotides respectively, and both sequences contain guanine as the majority base with consecutive guanine sequences present (Table 1). Thus according to our empirical rules, both sgc 8 and TDO 5 aptamers would be expected to produce red-emissive AgNCs. To modulate the emission colors of AgNCs templated by the selected aptamers, a T10C20 sequence was appended to the 3′-terminus of each aptamer (see the complete DNA sequences in Table 1). The T10 sequence was introduced as a linker to isolate the NC forming site from the cancer cell targeting sequence, while the C20 sequence was introduced to increase the content of cytosine in the modified aptamer such that it is now the dominant base present in the sequence (i.e., 42.3% for sgc 8-T10C20 and 41.6% for TDO 5-T10C20). As such, the AgNCs templated by both modified aptamer sequences are expected to show a time-dependent photoluminescence color change. However, as the ratio of cytosine to guanine bases present in sgc 8-T10C20 is higher than that for TDO 5-T10C20, a greater extent of color change is expected for the former. As predicted, the AgNCs synthesized using sgc 8 and TDO 5 indeed showed red emission (λem = 645 nm for sgc 8 and 635 nm for TDO 5) that decayed with time, over 24 hours, which is a characteristic of guanine dominance in the sequence (Table 1). The two modified aptamers indeed caused the change of the emission colors of AgNCs from red (item 1 and dashed lines) to yellow (item 2 and solid lines) with time, due to the formation of green emitters, with a smaller extent of conversion seen for the TDO 5-T10C20 as predicted.For cellular bioimaging and cancer diagnostic applications, the ability of aptamer sequences to specifically target cancer cells is of critical importance. However, the use of unmodified aptamers (e.g. sgc 8 and TDO 5) to template AgNCs may result in the loss of their specific targetting properties due to structural changes associated with AgNC templation and ligation. To address this issue, we hypothesised that introducing an additional AgNC forming sequence which was separated from these aptamers by a linker sequence could preserve the targeting properties, yet retain the ability to form bright-emissive AgNCs for bioimaging (Fig. 4A). To test this hypothesis, we further evaluated the targeting properties of the AgNCs synthesized using original unmodified aptamers (sgc 8 and TDO 5) and modified aptamers (sgc 8-T10C20 and TDO 5-T10C20) as model templates by confocal laser scanning microscopy and flow cytometry. CEM and Ramos cells were mixed with AgNCs produced by the respective aptamers, i.e., sgc 8 and sgc 8-T10C20 for CEM, and TDO 5 and TDO 5-T10C20 for Ramos respectively. The corresponding cellular images were examined after four hours of incubation. As shown in Fig. 4B and E, AgNCs synthesized using unmodified aptamers (sgc 8 in B and TDO 5 in E) did not attach to the cell surface, possibly due to the loss of targeting ability during the formation of AgNCs. Those AgNCs synthesized using modified aptamers (sgc 8-T10C20 and TDO 5-T10C20), however, attach very well on the surface of respective cancer cells (Fig. 4C and F). These results further confirmed the specific targeting properties of AgNCs formed by the modified aptamers for respective cells because of the inclusion of linker sequences that can isolate the NC formation sequence from the targeting sequence to prevent the loss of aptamer specificity. The specific targeting properties of AgNCs synthesized with modified aptamers were further confirmed by flow cytometry. As shown in Fig. 4D and G, the median PL values of CEM and Ramos cells incubated with respective AgNCs templated by modified aptamers (sgc 8-T10C20 or TDO 5-T10C20 (in red)) were about 6.5-fold and 4.6-fold that of respective cells incubated with AgNCs templated by unmodified aptamers (in blue).
3. Conclusion
In summary, we have conducted systematic investigations of DNA templates (homo-oligonucleotides, combinatorial C–G/C–T/G–A/G–T/C–G/A–T series, interdigitated DNA sequences, and aptamer–NC forming sequences) to unravel the guiding principles in forming AgNCs with fine-tuned photoluminescence properties and functionality. It is revealed that strong base–Ag+ binding and formation of stable supramolecular structures such as i-motifs and G-quadruplexes are two important pre-requisites for forming bright photoluminescent AgNCs. In particular, cytosine and guanine are able to form bright-emissive AgNCs, albeit with very different photoemission behaviors. The former exhibits oxidative color conversion from red to cyan with time, while the latter favors the formation of single red-emitting AgNCs. Altering the DNA sequence with adenine and thymine bases changes the supramolecular structure of DNA templates. The inclusion of adenine bases in DNA templates encourages the formation of green-emitters due to the unique adenine–AgNC bonding interactions, while thymine adversely influences the PL intensity of the resultant AgNCs. In addition, the properties of AgNCs are greatly influenced by the relative base–Ag+ binding affinities, where a large disparity (e.g. C/T and C/A) results in a dominance of the stronger-binding base, while a smaller disparity (e.g. C/G and G/A) allows significant mutual influence and co-dominance. Consecutive sequences of cytosine and/or guanine are also required to form bright emissive AgNCs with strong PL intensity, where these criteria can be used to rationally design suitable DNA sequences for AgNC formation. Based on the empirical rules established from this study, we can predict the luminescence color of AgNCs templated by two arbitrarily selected cancer-cell targeting aptamers. This predictive property was also extended to the modified aptamer sequences, preserving both their specific cellular binding properties and ability to template the formation of photoluminescent AgNCs. Our systematic empirical approach to uncover the design principles of nucleic acid templates for AgNC synthesis complements the existing methodologies towards the ultimate goal of controlling both the photoluminescence and functionalities of DNA-protected AgNCs for multicolor targeted bioimaging applications.4. Experimental section
4.1. Materials and instrumentation
Silver nitrate (AgNO3, 99.9%) and sodium borohydride (NaBH4, 98%) were purchased from Sigma-Aldrich and used without further purification. All DNA sequences (Sigma Aldrich) were purified by desalting by the manufacturer and used as received without any further purification. Ultrapure water (18 MΩ, prepared from a Millipore Elix 3 purification system) was used throughout the experiment as solvent. Photoluminescence emission spectra were measured on a TECAN infinite M200 plate reader (Tecan Trading AG, Switzerland) using a 384-well black polystyrene microplate (Corning Incorporated, USA) as a sample carrier. All CD measurements were performed using a JASCO J-810 spectrometer in the wavelength range of 200 nm to 350 nm using a 0.1 cm quartz cuvette. Unless otherwise stated, the concentration of DNA used for CD spectroscopy was 3.00 μM, and all AgNC syntheses were carried out as described before dilution for CD measurements.4.2. Synthesis of silver nanoclusters (AgNCs)
AgNCs were synthesized in the manner reported by Dickson and co-workers31 based on the optimized base![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ag+
Ag+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BH4− mole ratio of 2
BH4− mole ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. A typical synthesis of dC20-templated AgNCs is as follows: AgNO3 (4.5 μL, 2.00 mM) was added to the dC20-DNA (9.0 μL, 0.10 mM) followed by water (32.0 μL) and vigorously mixed by vortexing for 30 seconds. After standing for 20 minutes, a freshly-prepared aqueous solution of NaBH4 (4.5 μL, 2.00 mM) was added and mixed by vortexing for a further 30 seconds, then the resulting solution was left to incubate at room temperature for 20 minutes. The photoluminescence of the AgNC solution (50.0 μL) was monitored at regular time intervals, and the solution was kept at room temperature in the dark to prevent photo-reduction of Ag+ between photoluminescence measurements.
1. A typical synthesis of dC20-templated AgNCs is as follows: AgNO3 (4.5 μL, 2.00 mM) was added to the dC20-DNA (9.0 μL, 0.10 mM) followed by water (32.0 μL) and vigorously mixed by vortexing for 30 seconds. After standing for 20 minutes, a freshly-prepared aqueous solution of NaBH4 (4.5 μL, 2.00 mM) was added and mixed by vortexing for a further 30 seconds, then the resulting solution was left to incubate at room temperature for 20 minutes. The photoluminescence of the AgNC solution (50.0 μL) was monitored at regular time intervals, and the solution was kept at room temperature in the dark to prevent photo-reduction of Ag+ between photoluminescence measurements.
4.3. Cellular uptake assay
For confocal microscopy experiments, CEM and Ramos cells obtained from ATCC USA were cultured in the RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and 100 IU mL−1 penicillin–streptomycin and incubated at 37 °C in a humidified incubator containing 5% wt/vol CO2. After 80% confluence, cells were fixed with 4% paraformaldehyde and seeded in an 8 well-chamber (LAB-TEK, Chambered Coverglass System) at 1 × 105 cells per mL with incubation of AgNCs at a Ag+ concentration of 180 μM for 4 h and then the samples were observed under a confocal microscope (Olympus Fluoview FV1000). The images obtained were analyzed using Olympus FV10-ASW imaging software. For flow cytometry measurements, CEM and Ramos cells (2 × 105 cells per well in 24-well plates) were fixed with 4% paraformaldehyde and co-incubated with AgNCs overnight. After incubation, the cells were washed with PBS thrice to remove the free AgNCs, and then the cells were resuspended in 0.5 mL of 1× PBS for flow cytometry measurements. The Cy5 channel (λem = 570 − 750 nm) was selected to collect the photoluminescence signal.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the Institute of Materials Research and Engineering (IMRE), A*STAR under the Biomimetic and Biomedical Materials Program (IMRE/17-1P1404 & IMRE/14-1C0420) in collaboration with the Newcastle University (RSA/CCEAMD5010), National University of Singapore, and University of Illinois at Urbana-Champaign. The authors would like to thank Dr Xiaodi Su (IMRE) and Dr Li Huey Tan (UIUC) for their suggestions in improving the manuscript.References
- Y.-J. Chen, B. Groves, R. A. Muscat and G. Seelig, Nat. Nanotechnol., 2015, 10, 748–760 CrossRef CAS PubMed.
- F. Hong, F. Zhang, Y. Liu and H. Yan, Chem. Rev., 2017, 117, 12584–12640 CrossRef CAS PubMed.
- H. M. Pendergraff, P. M. Krishnamurthy, A. J. Debacker, M. P. Moazami, V. K. Sharma, L. Niitsoo, Y. Yu, Y. N. Tan, H. M. Haitchi and J. K. Watts, Mol. Ther.--Nucleic Acids, 2017, 8, 158–168 CrossRef CAS PubMed.
- N. C. Seeman and H. F. Sleiman, Nat. Rev. Mater., 2017, 3, 17068 CrossRef.
- H. V. Xu, X. T. Zheng, C. Wang, Y. Zhao and Y. N. Tan, ACS Appl. Nano Mater., 2018, 1, 2062–2068 CrossRef CAS.
- H. V. Xu, X. T. Zheng, Y. Zhao and Y. N. Tan, ACS Appl. Mater. Interfaces, 2018, 10, 19881–19888 CrossRef CAS PubMed.
- X. T. Zheng, Y. C. Lai and Y. N. Tan, Nanoscale Adv., 2019, 1, 2250–2257 RSC.
- Y. Choi, X. T. Zheng and Y. N. Tan, Mol. Syst. Des. Eng., 2020, 5, 67–90 RSC.
- H. V. Xu, Y. Zhao and Y. N. Tan, ACS Appl. Mater. Interfaces, 2019, 11, 27233–27242 CrossRef CAS PubMed.
- J. Wu, Y. Fu, Z. He, Y. Han, L. Zheng, J. Zhang and W. Li, J. Phys. Chem. B, 2012, 116, 1655–1665 CrossRef CAS PubMed.
- H. V. Xu, X. T. Zheng, B. Y. L. Mok, S. A. Ibrahim, Y. Yu and Y. N. Tan, J. Mol. Eng. Mater., 2016, 04, 1640003 CrossRef CAS.
- Y. Yu, B. Y. L. Mok, X. J. Loh and Y. N. Tan, Adv. Healthcare Mater., 2016, 5, 1844–1859 CrossRef CAS PubMed.
- J. Chao, Y. Lin, H. Liu, L. Wang and C. Fan, Mater. Today, 2015, 18, 326–335 CrossRef CAS.
- A. Heuer-Jungemann and T. Liedl, Trends Chem., 2019, 1, 799–814 CrossRef.
- S. Julin, S. Nummelin, M. A. Kostiainen and V. Linko, J. Nanopart. Res., 2018, 20, 119 CrossRef PubMed.
- N. Li, Y. Shang, Z. Han, T. Wang, Z.-G. Wang and B. Ding, ACS Appl. Mater. Interfaces, 2019, 11, 13835–13852 CrossRef CAS PubMed.
- L.-L. Li, P. Wu, K. Hwang and Y. Lu, J. Am. Chem. Soc., 2013, 135, 2411–2414 CrossRef CAS PubMed.
- Y. N. Tan, J. Y. Lee and D. I. C. Wang, J. Am. Chem. Soc., 2010, 132, 5677–5686 CrossRef CAS PubMed.
- Z. Wang, L. Tang, L. H. Tan, J. Li and Y. Lu, Angew. Chem., Int. Ed., 2012, 51, 9078–9082 CrossRef CAS PubMed.
- J. Wu, L. H. Tan, K. Hwang, H. Xing, P. Wu, W. Li and Y. Lu, J. Am. Chem. Soc., 2014, 136, 15195–15202 CrossRef CAS PubMed.
- J. Zong, S. L. Cobb and N. R. Cameron, Biomater. Sci., 2017, 5, 872–886 RSC.
- A. Aires, E. Lopez-Martinez and A. L. Cortajarena, Biosensors, 2018, 8, 110 CrossRef CAS PubMed.
- F. Pu, X. Ran, M. Guan, Y. Huang, J. Ren and X. Qu, Nano Res., 2018, 11, 3213–3221 CrossRef CAS.
- Y. Yu, J. Geng, E. Y. X. Ong, V. Chellappan and Y. N. Tan, Adv. Healthcare Mater., 2016, 5, 2528–2535 CrossRef CAS PubMed.
- Y. Yu, S. Y. New, J. Xie, X. Su and Y. N. Tan, Chem. Commun., 2014, 50, 13805–13808 RSC.
- Y. Zhou, J. Wang, G. Yang, S. Ma, M. Zhang and J. Yang, Anal. Methods, 2019, 11, 733–738 RSC.
- Y. Yu, W. D. Lee and Y. N. Tan, Mater. Sci. Eng., C, 2020, 109, 110525 CrossRef CAS PubMed.
- E. Assah, W. Goh, X. T. Zheng, T. X. Lim, J. Li, D. Lane, F. Ghadessy and Y. N. Tan, Colloids Surf., B, 2018, 169, 214–221 CrossRef CAS PubMed.
- B. Shen, V. Linko, K. Tapio, M. A. Kostiainen and J. J. Toppari, Nanoscale, 2015, 7, 11267–11272 RSC.
- X. T. Zheng, W. L. Goh, P. Yeow, D. P. Lane, F. J. Ghadessy and Y. N. Tan, Sens. Actuators, B, 2019, 279, 79–86 CrossRef CAS.
- J. T. Petty, J. Zheng, N. V. Hud and R. M. Dickson, J. Am. Chem. Soc., 2004, 126, 5207–5212 CrossRef CAS PubMed.
- Y. S. Ang, W. W. E. Woon and L.-Y. L. Yung, Nucleic Acids Res., 2018, 46, 6974–6982 CrossRef CAS PubMed.
- Y. Chen, M. L. Phipps, J. H. Werner, S. Chakraborty and J. S. Martinez, Acc. Chem. Res., 2018, 51, 2756–2763 CrossRef CAS PubMed.
- J. C. Léon, D. González-Abradelo, C. A. Strassert and J. Müller, Chem.–Eur. J., 2018, 24, 8320–8324 CrossRef PubMed.
- S. Y. New, S. T. Lee and X. D. Su, Nanoscale, 2016, 8, 17729–17746 RSC.
- J. M. Obliosca, C. Liu and H.-C. Yeh, Nanoscale, 2013, 5, 8443–8461 RSC.
- S. Choi, R. M. Dickson and J. Yu, Chem. Soc. Rev., 2012, 41, 1867–1891 RSC.
- J. Yang, E. Sargent, S. Kelley and J. Y. Ying, Nat. Mater., 2009, 8, 683–689 CrossRef CAS PubMed.
- Ö. Dag, E. J. Henderson, W. Wang, J. E. Lofgreen, S. Petrov, P. M. Brodersen and G. A. Ozin, J. Am. Chem. Soc., 2011, 133, 17454–17462 CrossRef PubMed.
- G. De Cremer, B. F. Sels, J.-i. Hotta, M. B. J. Roeffaers, E. Bartholomeeusen, E. Coutiño-Gonzalez, V. Valtchev, D. E. De Vos, T. Vosch and J. Hofkens, Adv. Mater., 2010, 22, 957–960 CrossRef CAS PubMed.
- M. El-Roz, I. Telegeiev, N. E. Mordvinova, O. I. Lebedev, N. Barrier, A. Behilil, M. Zaarour, L. Lakiss and V. Valtchev, ACS Appl. Mater. Interfaces, 2018, 10, 28702–28708 CrossRef CAS PubMed.
- L. Shang and S. Dong, Chem. Commun., 2008,(9), 1088–1090 RSC.
- Z. Yuan, N. Cai, Y. Du, Y. He and E. S. Yeung, Anal. Chem., 2014, 86, 419–426 CrossRef CAS PubMed.
- J. Zheng and R. M. Dickson, J. Am. Chem. Soc., 2002, 124, 13982–13983 CrossRef CAS PubMed.
- I. Chakraborty, S. Mahata, A. Mitra, G. De and T. Pradeep, Dalton Trans., 2014, 43, 17904–17907 RSC.
- C. P. Joshi, M. S. Bootharaju, M. J. Alhilaly and O. M. Bakr, J. Am. Chem. Soc., 2015, 137, 11578–11581 CrossRef CAS PubMed.
- R. Yu, N. Lin, W. Yu and X. Y. Liu, CrystEngComm, 2015, 17, 7986–8010 RSC.
- Z. Wu, E. Lanni, W. Chen, M. E. Bier, D. Ly and R. Jin, J. Am. Chem. Soc., 2009, 131, 16672–16674 CrossRef CAS PubMed.
- J. Yan, H. Su, H. Yang, S. Malola, S. Lin, H. Häkkinen and N. Zheng, J. Am. Chem. Soc., 2015, 137, 11880–11883 CrossRef CAS PubMed.
- Q. Yao, Y. Yu, X. Yuan, Y. Yu, D. Zhao, J. Xie and J. Y. Lee, Angew. Chem., Int. Ed., 2015, 54, 184–189 CrossRef CAS PubMed.
- J. Zheng, C. Zhang and R. M. Dickson, Phys. Rev. Lett., 2004, 93, 077402 CrossRef PubMed.
- S. Pal, R. Varghese, Z. Deng, Z. Zhao, A. Kumar, H. Yan and Y. Liu, Angew. Chem., Int. Ed., 2011, 50, 4176–4179 CrossRef CAS PubMed.
- Y. Cui, Y. Wang, R. Liu, Z. Sun, Y. Wei, Y. Zhao and X. Gao, ACS Nano, 2011, 5, 8684–8689 CrossRef CAS PubMed.
- S. Roy, A. Baral and A. Banerjee, ACS Appl. Mater. Interfaces, 2014, 6, 4050–4056 CrossRef CAS PubMed.
- J. Yu, S. A. Patel and R. M. Dickson, Angew. Chem., Int. Ed., 2007, 46, 2028–2030 CrossRef CAS PubMed.
- W. Gong, P. Das, S. Samanta, J. Xiong, W. Pan, Z. Gu, J. Zhang, J. Qu and Z. Yang, Chem. Commun., 2019, 55, 8695–8704 RSC.
- R. Hardman, Environ. Health Perspect., 2006, 114, 165–172 CrossRef PubMed.
- F. M. Winnik and D. Maysinger, Acc. Chem. Res., 2013, 46, 672–680 CrossRef CAS PubMed.
- C. M. Ritchie, K. R. Johnsen, J. R. Kiser, Y. Antoku, R. M. Dickson and J. T. Petty, J. Phys. Chem. C, 2007, 111, 175–181 CrossRef CAS PubMed.
- S. M. Copp, D. Schultz, S. Swasey, J. Pavlovich, M. Debord, A. Chiu, K. Olsson and E. Gwinn, J. Phys. Chem. Lett., 2014, 5, 959–963 CrossRef CAS PubMed.
- S. Walczak, K. Morishita, M. Ahmed and J. Liu, Nanotechnology, 2014, 25, 155501 CrossRef PubMed.
- C. I. Richards, S. Choi, J.-C. Hsiang, Y. Antoku, T. Vosch, A. Bongiorno, Y.-L. Tzeng and R. M. Dickson, J. Am. Chem. Soc., 2008, 130, 5038–5039 CrossRef CAS PubMed.
- S. M. Swasey, S. M. Copp, H. C. Nicholson, A. Gorovits, P. Bogdanov and E. G. Gwinn, Nanoscale, 2018, 10, 19701–19705 RSC.
- S. M. Copp, P. Bogdanov, M. Debord, A. Singh and E. Gwinn, Adv. Mater., 2014, 26, 5839–5845 CrossRef CAS PubMed.
- S. M. Copp, A. Gorovits, S. M. Swasey, S. Gudibandi, P. Bogdanov and E. G. Gwinn, ACS Nano, 2018, 12, 8240–8247 CrossRef CAS PubMed.
- P. R. O'Neill, L. R. Velazquez, D. G. Dunn, E. G. Gwinn and D. K. Fygenson, J. Phys. Chem. C, 2009, 113, 4229–4233 CrossRef.
- S. Shukla and M. Sastry, Nanoscale, 2009, 1, 122–127 RSC.
- Ö. Persil, C. T. Santai, S. S. Jain and N. V. Hud, J. Am. Chem. Soc., 2004, 126, 8644–8645 CrossRef PubMed.
- J. Kypr, I. Kejnovská, D. Renčiuk and M. Vorlíčková, Nucleic Acids Res., 2009, 37, 1713–1725 CrossRef CAS PubMed.
- W. Li, L. Liu, Y. Fu, Y. Sun, J. Zhang and R. Zhang, Photochem. Photobiol. Sci., 2013, 12, 1864–1872 RSC.
- Y. Fu, J. Zhang, X. Chen, T. Huang, X. Duan, W. Li and J. Wang, J. Phys. Chem. C, 2011, 115, 10370–10379 CrossRef CAS.
- G. Tao, Y. Chen, R. Lin, J. Zhou, X. Pei, F. Liu and N. Li, Nano Res., 2018, 11, 2237–2247 CrossRef CAS.
- S. M. Swasey, L. E. Leal, O. Lopez-Acevedo, J. Pavlovich and E. G. Gwinn, Sci. Rep., 2015, 5, 10163 CrossRef CAS PubMed.
- D. Zikich, K. Liu, L. Sagiv, D. Porath and A. Kotlyar, Small, 2011, 7, 1029–1034 CrossRef CAS PubMed.
- A. Ono, S. Cao, H. Togashi, M. Tashiro, T. Fujimoto, T. Machinami, S. Oda, Y. Miyake, I. Okamoto and Y. Tanaka, Chem. Commun., 2008,(39), 4825–4827 RSC.
- B. Sengupta, K. Springer, J. G. Buckman, S. P. Story, O. H. Abe, Z. W. Hasan, Z. D. Prudowsky, S. E. Rudisill, N. N. Degtyareva and J. T. Petty, J. Phys. Chem. C, 2009, 113, 19518–19524 CrossRef CAS.
- C. Cerretani, H. Kanazawa, T. Vosch and J. Kondo, Angew. Chem., Int. Ed., 2019, 58, 17153–17157 CrossRef CAS PubMed.
- M. R. Carro Temboury, V. Paolucci, E. N. Hooley, L. Latterini and T. Vosch, Analyst, 2016, 141, 123–130 RSC.
- S. Choi, S. Park, K. Lee and J. Yu, Chem. Commun., 2013, 49, 10908–10910 RSC.
- X. Yuan, Z. Luo, Q. Zhang, X. Zhang, Y. Zheng, J. Y. Lee and J. Xie, ACS Nano, 2011, 5, 8800–8808 CrossRef CAS PubMed.
- D. J. E. Huard, A. Demissie, D. Kim, D. Lewis, R. M. Dickson, J. T. Petty and R. L. Lieberman, J. Am. Chem. Soc., 2019, 141, 11465–11470 CrossRef CAS PubMed.
- X. Chen, M. C. Estévez, Z. Zhu, Y.-F. Huang, Y. Chen, L. Wang and W. Tan, Anal. Chem., 2009, 81, 7009–7014 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0na00381f |
| ‡ J. Y. C. L. and Y. Y. contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |