Open Access Article
Open Access ArticleFunctionalized carbon nanotube electrodes for controlled DNA sequencing†
Rameshwar L.
Kumawat
 a and
Biswarup
Pathak
a and
Biswarup
Pathak
 *ab
*ab
aDiscipline of Metallurgy Engineering and Materials Science, Indore, Madhya Pradesh 453552, India. E-mail: biswarup@iiti.ac.in
bDiscipline of Chemistry, School of Basic Sciences, Indian Institute of Technology (IIT) Indore, Indore, Madhya Pradesh 453552, India
First published on 14th July 2020
Abstract
In the last decade, solid-state nanopores/nanogaps have attracted significant attention in the rapid detection of DNA nucleotides. However, reducing the noise through controlled translocation of the DNA nucleobases is a central issue for the development of nanogap/nanopore-based DNA sequencing to achieve single-nucleobase resolution. Furthermore, the high reactivity of the graphene pores/gaps causes clogging of the pore/gap, leading to the blockage of the pores/gaps, sticking, and irreversible pore closure. To address the prospective of functionalization of the carbon nanostructure and for accomplishing this objective, herein, we have studied the performance of functionalized closed-end cap armchair carbon nanotube (CNT) nanogap-embedded electrodes, which can improve the coupling through non-bonding electrons and may provide the possibility of N/O–H⋯π interactions with the nucleotides, as single-stranded DNA is transmigrated across the electrode. We have investigated the effect of functionalizing the closed-end cap CNT (6,6) electrodes with purine (adenine, guanine) and pyrimidine (thymine, cytosine) molecules. Weak hydrogen bonds formed between the probe molecule and the target DNA nucleobase enhance the electronic coupling and temporarily stabilize the translocating nucleobase against the orientational fluctuations, which may reduce noise in the current signal during experimental measurements. The findings of our density functional theory and non-equilibrium Green's function-based study indicate that this modeled setup could allow DNA nucleotide sequencing with a better and reliable yield, giving current traces that differ by at least 1 order of current magnitude for all the four target nucleotides. Thus, we feel that the functionalized armchair CNT (6,6) nanogap-embedded electrodes may be utilized for controlled DNA sequencing.
1. Introduction
Recent advances in DNA sequencing have paved their way in personalized medicine, which is the next Frontier in health care as it can be used to detect predisposition concerning several genetic illnesses and delivers accurate treatments.1–4 To fully accomplish this, enhanced control and cost of the procedure are further needed to be improved.4,5 The progress towards faster, reliable, and cheaper sequencing has been in high demand since the “$1000 Human Genome Project” was launched.3Nanopore/nanogap-based human genome sequencing is one of the most developed technologies since it is promising in providing faster, reliable, and cheaper sequencing. Furthermore, it has the potential to bring genomic science into personalized medicine.1–10 In the past few decades, significant development and successes have been achieved. In 2007, Jin He and co-workers suggested the functionalization of gold (Au) nanoelectrodes with a cytosine probe/reader for DNA (deoxyribonucleic acid) sequencing.11 After that, in 2008, Haiying He and co-workers tested four DNA nucleobase molecules as probes and found the cytosine probe to yield robust results in terms of nucleobase distinguishability.12 Again, in 2009, Jin He and co-workers demonstrated that tunneling with double-functionalized contacts could allow the nucleobase structure of the unmodified DNA oligomers to be read with a resolution analogous to that of ion-current read-outs in the nanopores/nanogaps.13 In 2012, Pathak and co-workers investigated the effect of chemical double-functionalization on Au nanoelectrodes for enhancing nanopore-based DNA sequencing.14 They demonstrated that the chemically functionalized molecular probes are capable of temporarily forming hydrogen (H) bonds with the incoming DNA base part and the phosphate group, thus reducing the noise and further slowing down the translocating speed of the nucleobases between the Au nanoelectrodes. Furthermore, Su and co-workers reported that guanine-functionalized Au nanoelectrodes revealed promising results.15 In 2010, Lindsay and co-workers demonstrated that functionalized Au-electrodes can be used to identify single nucleobases in a DNA oligomers.16,17 However, transverse conductance across the DNA molecules located between the two Au nano-electrodes has been aggressively examined and debated. Therefore, special attention has been devoted to exploring low-dimensional (2D and 1D) materials.18–20
Low-dimensional materials, for example, graphene,1,2,4–8,10,19–31 hexagonal-boron nitride (hBN),32,33 graphene-hBN,34 molybdenum disulfide (MoS2),35,36 silicene,37 black phosphorene,38,39 boron-carbide (BC3),40 and various other low dimensional materials, have been studied for DNA sequencing owing their excellent electronic and transport properties. For the progress of pore/gap-based DNA sequencing techniques, all these materials have provided new paradigms since their atomic thickness is comparable to the DNA nucleobase spacing in single-stranded DNA (ssDNA).22,26 Mostly, graphene-based nanopore/nanogap,2,26 nanoribbon,41 and carbon nanotubes (CNTs)18 have been explored as potential nano-electrodes materials for DNA sequencing. So far, several experimental and theoretical works have been done on DNA sequencing using graphene electrodes.22,24,26,31 Traversi and co-workers experimentally demonstrated that the solid-state nanoscale-sized pore integrated with the graphene nanoribbon could be utilized for single-nucleobase sensing.29 Kim and co-workers theoretically reported that graphene nanoribbon (GNR)-based nanochannel device can distinguish between the four nucleobases.42,43 They suggested that Fano resonance could be the key factor for such detection. However, the passage of DNA through the graphene-based pores/gaps was confirmed; a significant drawback of the technique was that the DNA nucleobase adhered to the nanopore/nanogap surface. Hence, the reactivity of the graphene edge leads to the deformation of the geometry.26 Such a reactivity also leads to the clogging of the pore, leading to the blockage of the nanopores, sticking, and irreversible pore closure.2,26,44 Several theoretical works have also reported that the precise functionalization of such pores/gaps or non-perforated regions with ligands such as aryl- or alkyl-groups may minimize the clogging of DNA nucleotides and deliver better surface energy as well as diffusion performance.45,46 Prasongkit and one of the present authors tried to improve the sensitivity by chemically functionalizing both the graphene electrodes to improve the electronic coupling with the translocating target DNA base.47 Likewise, Biswas and co-workers have demonstrated that a universal reader based on hydrogen-bonding or π–π stacking can be used for DNA sequencing.48 Despite all these works, experimental and theoretical researchers are continually searching for a low-dimensional nanopore/nanogap-based functionalized electrode material that can provide molecular-level resolution with enhanced control.2,18,26,34
In this regard, CNT-based electrodes have been used and functionalized with various probes for DNA-based biosensor applications.49–55 Wong et al. have demonstrated the use of functionalized CNTs for atomic force microscopy (AFM) tips, which has opened up the applications of molecular recognition and chemically sensitive imaging in chemistry and biology.49 In another study, Wong et al. have demonstrated that a CNT AFM tip can be used for imaging nanostructures50 and biological molecules such as DNA and amyloid-b-protofibrils (related to Alzheimer disease).51 Moreover, it has been reported that the CNT tip can provide higher resolution than the Si or metal-based tips. Lin et al. have also developed amperometric biosensors using CNT-nanoelectrode-based ensembles.52 Likewise, CNT-based biosensors for nucleic acid recognition processes are rapidly being developed to achieve the goal of rapid, controlled, simple, and low-cost testing of human genetic and infectious diseases.53–55 The nanometre-scale diameter of a CNT also allows the measurement of interactions at the single-molecule level.55 Furthermore, N-doped CNTs have been found to be very effective for DNA sequencing.56–58
Moreover, such low-dimensional CNTs have an atomically thin structure (chemical and physical) and unique electrical transport properties,59–65 which can allow the individual identification of DNA nucleotides based on in-plane transverse transmission and current signals, and may accomplish single-nucleobase resolution with enhanced control. Thus, to realize the potential of solid-state DNA sequencing on low-dimensional materials, to enable the reading of nucleotides at the single-molecule level, and also to enhance the readout time, a high-fidelity control of ssDNA translocation still needs to be accomplished. Motivated by these reports, we predict that CNT can be a better electrode as it may not be as reactive as the graphene edges and the presence of π-clouds in CNT may improve the coupling with the translocating DNA nucleotides. Specifically, such a curvature (benzene-like six-membered rings at the closed-end cap provides the possibility of π–π coupling with the nucleotides) in the CNT may improve the N/O–H⋯π interaction with the DNA nucleotides. In addition, suitable functionalizing groups can be attached to the CNT caps, which could lead to better electronic coupling with the incoming ssDNA nucleobases through the formation of temporary H-bonds.11–15,45,46 Such a functionalization molecular probe could be attached to only one electrode or both the electrodes. The functionalization of the CNT caps could be a better way to achieve single-nucleobase resolution, reducing the noise in transverse electric current signals and further slowing down the translocation speed of the target DNA nucleobases between the functionalized CNT electrodes. Also, the functionalized CNT-based nanogap could simplify the DNA sequencing process and allows different types of electrical measurements (such as conductance and transverse electronic current). That being the case, CNT is very promising and opens up possibilities for controlled translocation of the DNA nucleotides. Hence, the present study explores the applicability of functionalized closed-end cap armchair CNT (6,6)-based nanogap-embedded electrodes for DNA sequencing.
In this work, we have studied the structural, electronic, and transport properties of the four setups, namely, deoxyadenosine monophosphate (dAMP), deoxyguanosine monophosphate (dGMP), deoxythymidine monophosphate (dTMP), and deoxycytidine monophosphate (dCMP), when inserted inside a functionalized closed-end cap armchair CNT (6,6)-based nanogap setup. This has been done to accomplish an unambiguous distinction between all the four nucleotides in DNA sequencing. The electronic structure and binding energies are analyzed using the density functional theory (DFT). Further, current–voltage (I–V) characteristics, zero-bias transmission function, and bias-dependent transmission function for all the four nucleotides were analyzed using the non-equilibrium Green's functional combined with DFT.
2. Model and computational methods
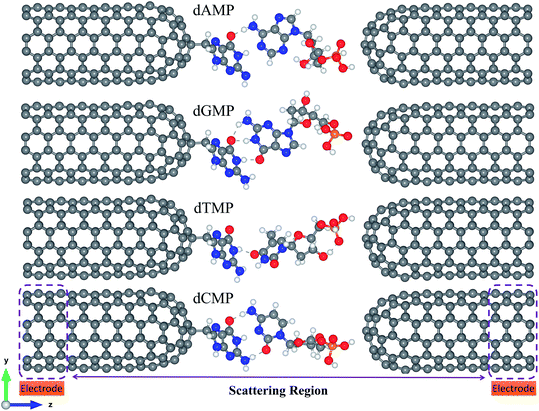
Fig. 1 shows that the proposed nanogap device consists of two semi-infinite functionalized closed-end cap armchair CNT (6,6) electrodes with a DNA nucleotide. Herein, we have taken purine or pyrimidine types DNA nucleobase molecule as the reader molecule, which acts as the molecular probe for the functionalized CNT (6,6) electrodes. We have examined all four molecular probes for sequencing the DNA nucleobases [Fig. 1 and S1 (ESI†)].The pairing geometry between the reader molecule (A/G/C/T) and the target nucleotide molecule (a target is a segment of a ssDNA including nucleobase, sugar, and phosphate group) is obtained by performing full geometry optimizations of all the four pairs using the DFT methodology with the B3LYP/6-31+G* level of theory, as implemented in the Gaussian 09 code.66 The fully optimized configurations of the reader-target (for example, guanine-DNA nucleotides) pairs are shown in Fig. S2.† Then, we placed each of these reader-target pairs in the nanogap, where the whole nanogap setup (Fig. 1 and S1†) was fully optimized using the DFT methodology, as implemented in the SIESTA code.67,68 The electrode–electrode spacing of 19.01 Å (shortest gap length between two CNT electrodes) was maintained throughout the calculations. Both the semi-infinite electrodes are periodic in the z-direction. We have considered a significant vacuum distance (along the x and y-directions) of about 28 Å, which is enough to avoid any unphysical interaction between the repetitive images of the nanogap system.
We have used GGA-PBE (generalized gradient approximation with Perdew–Burke–Ernzerhof) approximation for the exchange and correlation functional.69 Norm-conserved Troullier-Martins pseudopotentials are used to describe the interactions between the core and the valence electrons.70 We have used the mesh cut-off value of 200 Ry for real space integration and DZP (double-zeta polarized) basis sets, including polarization orbitals for all the atoms.34–40,69
We have considered Γ-point for the sampling of the Brillouin zone due to the large cell size. The structures were relaxed by the conjugate-gradient (CG) algorithm using the tolerance in density matrix difference of 0.0001 and the atomic forces were lesser than 0.01 eV Å−1.
The binding energy (Eb) was calculated using the following equation:
| Eb = [ECNT+DNA − (ECNT + EDNA)] | (1) |
The electronic transport properties have been studied using the Landauer-Buttiker approach. We have used the NEGF technique combined with DFT (NEGF + DFT), as implemented in the TranSIESTA code.67,71 The basis set and the real-space integration used in the electronic transport calculation is the same as that used for geometry relaxation. The transverse electric current is calculated according to the following equation:
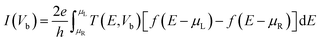 | (2) |
3. Results and discussion
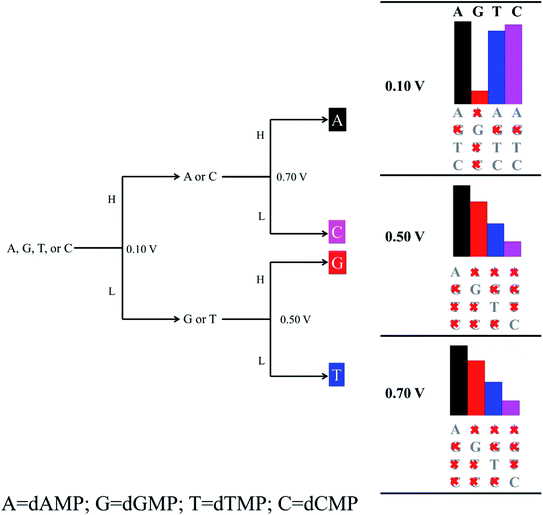
As the ssDNA is translocated through the functionalized CNT-based nanogap by a driving electric field, the probing molecule (A/G/T/C) will interact with each nucleotide while translocating over the nanogap setup by forming weak H-bonds with the reader nucleobase and the nucleobase part of the target DNA nucleotide molecule. The DNA nucleobases appearing in a natural DNA have a characteristic capability to selectively bind to their corresponding complementary nucleobase molecules.11–14 Hence, we have considered all the four nucleobase molecules (i.e., A, G, T, and C) as the reader/probe molecules in our study. Firstly, we have compared the interaction strength of the DNA nucleobases towards the electrode. The interaction strength between the CNT (pristine/functionalized) and the target DNA nucleotide molecules were investigated by analyzing the binding energy (Eb), as defined in eqn (1) (Model and computational methods). The calculated binding energy values are given in Table S1.† From Table S1,† we found that the functionalized CNT-based nanogap has high Eb values compared to the pristine CNT-based nanogap, which is important for electrical measurements. Further, the Eb values of the reader-target nucleobase pairs formed temporarily in the nanogap were investigated for all the four reader-target nucleotide systems. The interaction between the O and H bond is stronger than that of the N and H bond because of the higher electronegativity of the oxygen atom. The Eb of dGMP is higher than that of dAMP because dGMP is involved in four hydrogen bonds with two oxygen atoms (Fig. S3b†). In the case of dCMP, the higher Eb is due to three hydrogen bonds (2 O–H and 1 N–H; Fig. S3d†). We compared the dAMP and dTMP; dAMP forms two hydrogen bonds (O–H and N–H; Fig. S3a†) while dTMP forms only one hydrogen bond (O–H; Fig. S3c†); therefore, dTMP shows lower Eb. The Eb value of the reader-target nucleobase pairs formed temporarily in the nanogap can also be correlated to the number of H-bonds formed in between the reader-target base pairs. For example, in the case of the guanine–guanine base pair, the Eb was −0.58 eV per H-atom. Nevertheless, the formation of the H-bond is subject to the orientations of the target molecule. The calculated Eb values show that the formation of temporary H-bonding can stabilize the DNA molecules for a short time as it passes through the nanogap. Thus, H-bonds formed between the reader molecule and the target DNA nucleobase enhance the electronic coupling and stabilize the translocating nucleobase against orientational fluctuations, thus significantly reducing the noise in the transverse electronic current signal.11–14,42 From the calculated Eb values, we perceived that the guanine-reader gives better electronic coupling compared to the other readers (A/T/C) and pristine CNT. This indicates that the guanine-reader could be a better reader molecule for controlled and rapid DNA sequencing. Furthermore, from the point of view of a DNA sequencing device, we have studied the I–V characteristics and transmission function for the detection of all the four nucleotides using the NEGF + DFT approach.Next, we investigated the current–voltage (I–V) characteristics for the nanogap setup using four different probes (Fig. 1 and S1†). The figure of merit for identifying all the four DNA nucleotides is their accompanying currents (I), which should differ by at least 1 order of current magnitude. We have tabulated our recognition chart in Table 1. For example, at a bias voltage of 0.10 V, using either C or T as a probe, we can distinguish the purine-based nucleotides (dAMP and dGMP) from the pyrimidine-based nucleotides (dCMP and dTMP). However, dAMP cannot be distinguished from dGMP (Table 1, Fig. S4†). Similarly, dCMP cannot be distinguished from dTMP. Likewise, using the A probe, we can distinguish dAMP and dGMP. However, dCMP cannot be distinguished from dTMP. In contrast, when the G is used as a probe, we can only distinguish dGMP but dAMP, dCMP, and dTMP cannot be distinguished from each other (Table 1; Fig. 2).
| Probe | V = 0.10 V | V = 0.50 V | V = 0.70 V |
|---|---|---|---|
| A | dAMP|dGMP|dCMP, dTMP | dAMP, dGMP|dCMP|dTMP | dAMP, dGMP|dCMP|dTMP |
| G | dAMP, dCMP, dTMP|dGMP | dAMP|dGMP|dTMP|dCMP | dAMP|dGMP|dTMP|dCMP |
| T | dAMP, dGMP|dCMP, dTMP | dAMP, dGMP, dTMP, dCMP | dAMP, dCMP, dTMP|dGMP |
| C | dAMP, dGMP|dCMP, dTMP | dAMP, dCMP|dGMP, dTMP | dAMP, dGMP, dTMP, dCMP |
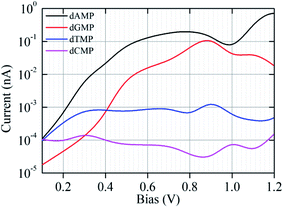 | ||
| Fig. 2 The I–V characteristic curves (plotted on a semi-logarithmic scale) for the nanogap setup functionalized with a G probe for the four different target nucleotides (dAMP, dGMP, dTMP, and dCMP). | ||
It is observed that with increasing applied bias voltage, the detection properties of the nanogap setup change. For example, at 0.50 V, the A probe delivers different current signals, and we can distinguish dCMP and dTMP. However, dAMP and dGMP remain indistinguishable from each other. Using the C probe, we can distinguish the dAMP and dCMP set from the dGMP and dTMP set. However, using the T probe, we cannot distinguish any among the four DNA nucleotides. Interestingly, using the G probe at 0.50 V, we can distinguish all the four nucleobases. Further, at 0.7 V, using the C probe, we cannot distinguish any among the four DNA nucleotides. However, using the T probe, we can distinguish dGMP but dAMP, dCMP, and dTMP cannot be distinguished. Likewise, using the A probe, we can distinguish dAMP and dGMP from dCMP and dTMP but dAMP and dGMP cannot be distinguished.
We found that the functionalization of the CNT-electrodes with the G probe facilitates the identification of the four DNA nucleotides (Fig. 2). This can be accomplished by performing three sequencing runs at different applied bias voltages (i.e., at 0.10 V, 0.50 V, and 0.70 V). We have presented a flowchart (Fig. 3), which gives an illustration of the individual identification of all the four target nucleotides. At 0.10 V, it would result in a sequence of current traces that are categorized into two effortless distinguishable trends known as high (H) and low (L) current values. High current value is expected if A (i.e., dAMP) or C (i.e., dCMP) is the target nucleotide and low current value is expected if G (i.e., dGMP) or T (i.e., dTMP) is the target nucleotide. Herein, we have noted that at 0.10 V, dGMP can be easily identified (Table 1); however, the other three nucleotides cannot be distinguished. Therefore, we require a high bias voltage to resolve the remaining ambiguity related to all the four nucleotides. At 0.50 V, it will be possible to distinguish all the four nucleotides (Table 1, Fig. 2). The difference between the two types of nucleotides is of about 1 order of current magnitude, which should make the distinction strong. Moreover, our proposed systems show the ability to identify the dAMP and dGMP nucleotides, which is very important for nanogap DNA sequencing. Hence, at 0.50 V, all the four nucleotides become distinguishable with their respective current traces. Besides, the current difference of all the four nucleotides (dAMP, dGMP, dTMP, and dCMP) is more than 1 order of current magnitude. Similarly, at 0.70 V, it also allows the current signals for the dAMP, dGMP, dTMP, and dCMP nucleotides to differ by at least 1 order of current magnitude. This leads to an easy distinction between the four nucleotides, where the high current values correspond to the dAMP nucleotide, while the low current values correspond to the dCMP nucleotide. More precisely, dAMP and dCMP give the highest and lowest current signals, respectively, whereas dGMP and dTMP can also be identified. However, we caution that the V-dependent conductance ordering could change if the experimental measurements pick up all the possible rotation angles of the nucleotides while translocating through the nanogap. All these possibilities are discussed in the later part of the manuscript.
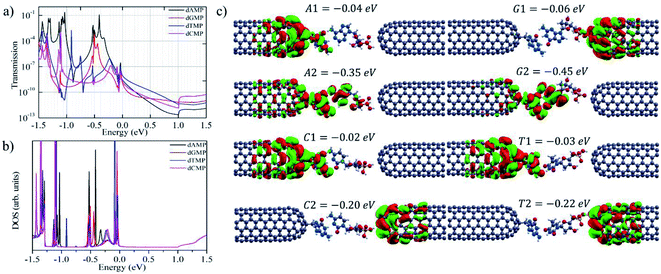
After demonstrating the most important ability of our proposed nanogap system in DNA sequencing, we studied the zero-bias transmission function and bias dependent transmission for all the four nucleotides.12,14,32,36–38,42 Herein, we focus on the most promising nanogap setup involving the guanine base as a probe. Fig. 4a and b shows the zero-bias transmission function together with the zero-bias density of states (DOS) when located inside the functionalized closed-end cap CNT (6,6) electrodes. It is noted that the DOS peak matches with the positions of the transmission function peaks. The associated molecular orbitals (MOs), with transmission peaks near the Fermi level, are presented in Fig. 4. We can observe in Fig. 4 that for the four DNA nucleotides, the Fermi level is aligned closely to the highest occupied molecular orbitals (HOMOs); in contrast, the lowest unoccupied molecular orbitals (LUMOs) are aligned far from the Fermi level. Near the Fermi region, we found that for the given nanogap setup, the transmission function has a similar shape for the four nucleotides, which can be due to the transmission peaks near the Fermi region and within the MO gap, which is far from the specific MOs that are typically associated with the contact states localized on the guanine probe and the CNT (6,6) electrodes (Fig. 4c). The different nucleotide types influence the transmission function at zero-bias. The size of purines is larger than that of pyrimidines and the number of hydrogen atoms available for interaction is also different in these two groups. Thus, the size of the target DNA nucleobase and, thereby, the gap between the nucleobase and the CNT (6,6) electrode decide the transmission magnitude through the overlap between the states localized on the nucleobase molecule and the states on the CNT (6,6) electrode. Consequently, from the transmission functions at zero-bias, one can conclude that the dissimilarity in the physical and chemical structures between the purine and pyrimidine nucleotides affects the electronic coupling strength of the nucleotides with the CNT (6,6) electrodes. As a result, they lead to the prospect of distinguishing the two different groups of DNA nucleotides under bias. The zero-bias transmission function and the corresponding DOS using the adenine, cytosine, and thymine bases as probes are presented in Fig. S5.†
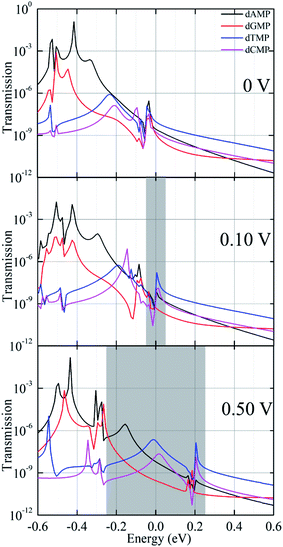
Next, to achieve a deeper understanding of the above discussed I–V characteristic features, we analyzed the bias-dependent transmission function for the four target DNA nucleotides while they were inserted inside the guanine probe functionalized CNT (6,6) electrodes (Fig. 5 and S6†). It is important to note that under the applied bias, the system is in the non-equilibrium state; hence, it has less relation with the binding energy. Therefore, once the system is in the non-equilibrium state, the most important parameter is the movement of the HOMO/LUMO peak associated with the electrode and the molecule. This means that the molecular states present in the system will play a significant role in the I–V signals.12,14,19,32,36–38,42 This is the reason as to why we have explored the bias-dependent transmission function for all the four nucleotides. Herein, when the bias is increased from 0 to 0.50 V, the transmission function peaks linked with the contact states enter the bias region and contribute to the increase in the current magnitude across the nanogap setup. The energy levels in the guanine-probe increase as the chemical potential of the left electrode as the bias is increased. Also, the position of the transmission peaks moves from high energy to low energy closely following the potential of the left electrode and the right electrode. The HOMO (A1, T1, and C1) states of dAMP, dTMP, and dCMP localized on the guanine probe and the CNT (6,6) electrodes are closest to the Fermi level, which in turns yields the highest current magnitude in the low bias region (0.1 to 0.2 V). Furthermore, for the purine nucleotides (dAMP and dGMP), when compared to the pyrimidine nucleotides (dTMP and dCMP), the HOMO (A2 and G2) states localized on the nucleobase part as well as on the guanine probe enter the bias region and contribute to the increase in the current magnitude across the nanogap setup. This could be the reason that purine nucleotides (dAMP and dGMP) afford higher current magnitude and pyrimidines (dTMP and dCMP) lower the current magnitude with increased bias (0.4 to 1.2 V). This is very much in agreement with our I–V curves as well. Further, to understand the bias-dependence conductance in the I–V curve (Fig. 2), we have analyzed the bias-dependent transmission spectra (Fig. 5 and S6†). At zero-bias transmission (Fig. 4), the HOMO peak (−0.06 eV) of dGMP appears close to the Fermi level, while the HOMO peak is at −0.04 eV for dAMP. For dCMP, the HOMO peak is at −0.02 eV and it is at −0.03 eV for dTMP. Hence, at 0.1 V bias, we get better current for dAMP, dTMP, and dCMP. However, the trend changes as we increase the voltage. In the case of dGMP, the induced resonance state G1 at E = −0.06 eV shifts significantly as we increase the voltage, which thereby increases the current. From the molecular orbital picture (Fig. 4), this state can be purely identified to be an induced state on the CNT (6,6) electrode rather than being a purely molecular state. Thus, we concluded that the molecular states on the nucleotides, the localized states on the guanine probe, and CNT (6,6) electrodes play a significant role in the transmission. Depending on the variation in the transmission function for the four DNA nucleotides, the trends in current follow specific variations in the lower and higher bias window, thus facilitating the principle capability of unambiguous identification of all the four DNA nucleotides.
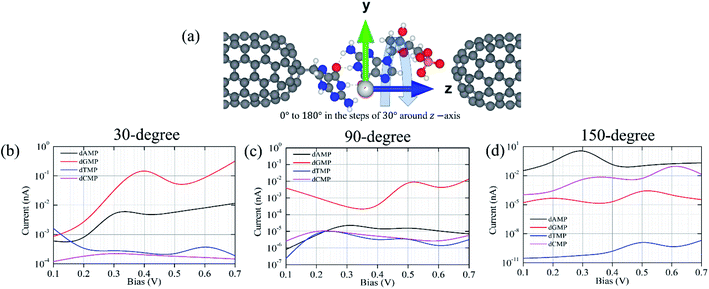
In addition, we have considered various configurations of the target nucleotides through rotation in the xy-plane around the z-axis from 0° to 180° with the steps of 30°, as shown in Fig. 6. This has been done to account for the fluctuating effects of the nucleotides while they translocate through the guanine probe functionalized CNT (6,6) nanogap, assuming that during the experimental measurements, it may pick up all the possible rotation angles. The calculated transmission functions are shown in Fig. S7.† When a nucleotide rotates, the resonance peak position shifts downwards/upwards with respect to the Fermi energy. Consequently, the magnitude of the transmission value changes. This is understandable as the extent of coupling can be different due to the orientations and alignments of the nucleotide. Further, we calculated the I–V curves when the DNA nucleotides are rotated inside the nanogap (for 30°, 90°, and 150°), as shown in Fig. 6.
Fig. 6 shows the I–V curves with different configurations for all the four nucleotides. It can be seen that the rotation of all the four nucleotides leads to fluctuation in the current signals. The calculated I–V curves for the original configuration (0° rotation; Fig. 2) is slightly different from the other configurations. However, in all the cases, the purine-based nucleotides (dAMP, dGMP) give higher currents compared to pyrimidine-based nucleotides, except in the case of the 150° rotated configuration where dCMP also shows a high current value. Fig. 7 shows the calculated current values in the 0.1–0.7 V bias range for all the nucleotides. Interestingly, in the lower bias window (0.1–0.3 V), dAMP and dTMP nucleotides can be easily distinguished due to the high and low current values, respectively. In fact, the dTMP nucleotide can be easily distinguished in the 0.1–0.7 V bias window due to the low current values. Interestingly, dCMP and dGMP can also be distinguished at 0.4 V bias though the difference in the current values is not much. Therefore, if the experimental measurements pick up all the possible rotation angles, then it may not be easy to distinguish all the four nucleotides distinctly, though dTMP and dAMP can be easily distinguished based on the highest and/or lowest current values from the other two nucleotides (dCMP, dGMP) in the lower bias region.
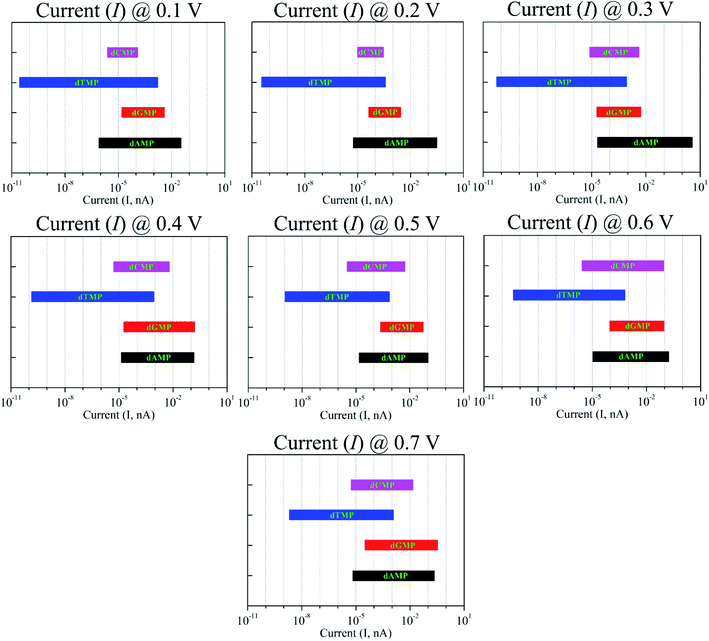 | ||
| Fig. 7 Current variation due to the nucleotide rotation (at 0°, 30°, 90°, and 150°) in the xy-plane around the z-axis under the bias voltages in the range of 0.1–0.7 V. | ||
Furthermore, we have made additional efforts to investigate the effect of different types and radii of the CNT electrodes on the DNA sequencing properties of the introduced nanogap. For this, we investigated two more nanogap setups: (i) one with armchair CNT (4,4) and (ii) another with zigzag (6,0) electrodes (detailed in Fig. S8†). Both these electrodes were also functionalized with the guanine probe as in the case of the armchair CNT (6,6) system. The calculated transmissions for all the three setups are shown in Fig. S9,† which shows that all the three setups show a very similar trend at the Fermi energy for the four nucleotides. Overall, there is a shift in the position and also a change in the width of the peak. This could be due to the device lengths (4.48 nm for CNT (6,6), 5.51 nm for CNT (4,4), and 5.59 nm for CNT (6,0)), coupling strengths, and electronic structures of the electrodes. However, at the Fermi level energy, there is not much change, irrespective of the type of CNT electrode used. Therefore, based on these results, one can say that it may be possible to identify all the four nucleotides using different types and radii of CNT electrodes though the trend can be different.
4. Conclusion
In conclusion, we have shown that the functionalized closed-end cap CNT (6,6)-based nanogap with the specific molecular probe could lead to an enhancement in the sensitivity of the target nucleobase detection in a nanogap-translocating DNA sequence. Through the formation of temporary H-bonds, the electronic coupling could be enhanced and the incoming nucleotide would be temporarily stabilized between the two electrodes, which potentially allows for less orientational fluctuations, thus the reducing noise in the current signals. Further, the functionalized closed-end CNT (6,6) nanogap provides more time for each nucleobase and allows different types of electrical measurements. From the transmission function and the I–V characteristics of the target nucleotides, we have shown that the guanine probe could lead to a significant improvement in the sensitivity of nucleotide detection in a nanogap-translocating DNA sequence. We find a considerable difference in the current signals for all the four nucleotides, which is more than 1 order of magnitude at two different voltages (0.5 and 0.7 V). This leads to an easy distinction between the purine and pyrimidine type nucleobases. More specifically, dAMP and dCMP afford higher and lower current signals, though dGMP and dTMP can be distinguished from the former two nucleotides with about 1 order of current magnitude. Bias-dependent transmission function reveals the molecular states contributing to the I–V signals, which plays a significant role in the DNA detection process. This is because of the electronic coupling of the target nucleobase with these states as well as the localized state on the guanine probe and electrodes that provide the molecular characteristics, as recorded through the I–V curve. Therefore, we believe that the functionalized closed-end cap CNT (6,6)-based nanogap electrodes may be utilized for controlled DNA sequencing.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We thank IIT Indore for the lab and computing facilities. This work is supported by DST-SERB, (Project Number: EMR/2015/002057) New Delhi, CSIR [Grant number: 01(2886)/17/EMR (II)] and (Project Number: CRG/2018/001131), and SPARC/2018-2019/P116/SL. R. L. K thanks MHRD for research fellowships. We would like to thank Dr Vivekanand Shukla for fruitful discussion throughout this work.References
- K. K. Saha, M. Drndić and B. K. Nikolić, Nano Lett., 2012, 12, 50–55 CrossRef CAS PubMed
.
- S. J. Heerema and C. Dekker, Nat. Nanotechnol., 2016, 11, 127–136 CrossRef CAS PubMed
.
- E. C. Hayden, Nature, 2014, 507, 294–295 CrossRef PubMed
.
- L. J. Steinbock and A. Radenovic, Nanotechnology, 2015, 26, 074003 CrossRef CAS PubMed
.
- E. R. Mardis, Nature, 2011, 470, 198–203 CrossRef CAS PubMed
.
- B. Rabbani, M. Tekin and N. Mahdieh, J. Hum. Genet., 2014, 59, 5–15 CrossRef CAS PubMed
.
- J. Shendure and E. L. Aiden, Nat. Biotechnol., 2012, 30, 1084–1094 CrossRef CAS PubMed
.
- B. M. Venkatesan and R. Bashir, Nat. Nanotechnol., 2011, 6, 615–624 CrossRef CAS PubMed
.
- D. Branton, D. W. Deamer, A. Marziali, H. Bayley, S. A. Benner, T. Butler, M. D. Ventra, S. Garaj, A. Hibbs, X. Huang, S. B. Jovanovich, P. S. Krstic, S. Lindsay, X. S. Ling, C. H. Mastrangelo, A. Meller, J. S. Oliver, Y. V. Pershin, J. M. Ramsey, R. Riehn, G. V. Soni, V. Tabard-Cossa, M. Wanunu, M. Wiggin and J. A. Schloss, Nat. Biotechnol., 2008, 26, 1146–1153 CrossRef CAS PubMed
.
- T. Nelson, B. Zhang and O. V. Prezhdo, Nano Lett., 2010, 10, 3237–3242 CrossRef CAS PubMed
.
- J. He, L. Lin, P. Zhang and S. Lindsay, Nano Lett., 2007, 7, 3854–3858 CrossRef CAS PubMed
.
- H. He, R. H. Scheicher and R. Pandey, J. Phys. Chem. C, 2008, 112, 3456–3459 CrossRef CAS
.
- J. He, L. Lin, H. Liu, P. Zhang, M. Lee, O. F. Sankey and S. M. Lindsay, Nanotechnology, 2009, 20, 075102 CrossRef PubMed
.
- B. Pathak, H. Löfås, J. Prasongkit, A. Grigoriev, R. Ahuja and R. H. Scheicher, Appl. Phys. Lett., 2012, 100, 023701 CrossRef
.
- W. Su, R. Dong, X. Yan, H. Wang and H. Liu, J. Comput. Theor. Nanosci., 2010, 7, 1885–1888 CrossRef CAS
.
- S. Huang, J. He, S. Chang, P. Zhang, F. Liang, S. Li, M. Tuchband, A. Fuhrmann, R. Ros and S. Lindsay, Nat. Nanotechnol., 2010, 5, 868–873 CrossRef CAS PubMed
.
- S. Chang, S. Huang, J. He, F. Liang, P. Zhang, S. Li, X. Chen, O. Sankey and S. Lindsay, Nano Lett., 2010, 10, 1070–1075 CrossRef CAS PubMed
.
- X. Chen, I. Rungger, C. D. Pemmaraju, U. Schwingenschlögl and S. Sanvito, Phys. Rev. B: Condens. Matter Mater. Phys., 2012, 85, 115436 CrossRef
.
- G. F. Schneider, S. W. Kowalczyk, V. E. Calado, G. Pandraud, H. W. Zandbergen, L. M. K. Vandersypen and C. Dekker, Nano Lett., 2010, 10, 3163–3167 CrossRef CAS PubMed
.
- S. Garaj, S. Liu, J. A. Golovchenko and D. Branton, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 12192–12196 CrossRef CAS PubMed
.
- J. Prasongkit, A. Grigoriev, B. Pathak, R. Ahuja and R. H. Scheicher, Nano Lett., 2011, 11, 1941–1945 CrossRef CAS PubMed
.
- H. S. Kim and Y. H. Kim, Biosens. Bioelectron., 2015, 69, 186–198 CrossRef CAS PubMed
.
- M. Wanunu, Phys. Life Rev., 2012, 9, 125–158 CrossRef PubMed
.
- J. Kudr, S. Skalickova, L. Nejdl, A. Moulick, B. Ruttkay–Nedecky, V. Adam and R. Kizek, Electrophoresis, 2015, 36, 2367–2379 CrossRef CAS PubMed
.
- C. Dekker, Nat. Nanotechnol., 2007, 2, 209–215 CrossRef CAS PubMed
.
- N. Yang and X. Jiang, Carbon, 2017, 115, 293–311 CrossRef CAS
.
- J. Shendure and H. Ji, Nat. Biotechnol., 2008, 26, 1135–1145 CrossRef CAS PubMed
.
- H. W. Ch. Postma, Nano Lett., 2010, 10, 420–425 CrossRef CAS PubMed
.
- F. Traversi, C. Raillon, S. M. Benameur, K. Liu, S. Khlybov, M. Tosun, D. Krasnozhon, A. Kis and A. Radenovic, Nat. Nanotechnol., 2013, 8, 939–945 CrossRef CAS PubMed
.
- C. A. Merchant, K. Healy, M. Wanunu, V. Ray, N. Peterman, J. Bartel, M. D. Fischbein, K. Venta, Z. Luo, A. T. C. Johnson and M. Drndić, Nano Lett., 2010, 10, 2915–2921 CrossRef CAS PubMed
.
- S. Garaj, W. Hubbard, A. Reina, J. Kong, D. Branton and J. A. Golovchenko, Nature, 2010, 467, 190–193 CrossRef CAS PubMed
.
- S. Thomas, A. C. Rajan, M. R. Rezapour and K. S. Kim, J. Phys. Chem. C, 2014, 118, 10855–10858 CrossRef CAS
.
- S. Liu, B. Lu, Q. Zhao, L. Li, T. Gao, Y. Chen, Y. Zhang, Z. Liu, Z. Fan, F. Yang, L. You and D. Yu, Adv. Mater., 2013, 25, 4549–4554 CrossRef CAS PubMed
.
- V. Shukla, N. K. Jena, A. Grigoriev and R. Ahuja, ACS Appl. Mater. Interfaces, 2017, 9, 39945–39952 CrossRef CAS PubMed
.
- A. B. Farimani, K. Min and N. R. Aluru, ACS Nano, 2014, 8, 7914–7922 CrossRef CAS PubMed
.
- K. Liu, J. Feng, A. Kis and A. Radenovic, ACS Nano, 2014, 8, 2504–2511 CrossRef CAS PubMed
.
- R. G. Amorim and R. H. Scheicher, Nanotechnology, 2015, 26, 154002 CrossRef PubMed
.
- R. L. Kumawat, P. Garg, S. Kumar and B. Pathak, ACS Appl. Mater. Interfaces, 2018, 11, 219–225 CrossRef PubMed
.
- R. L. Kumawat and B. Pathak, J. Phys. Chem. C, 2019, 123, 22377–22383 CrossRef CAS
.
- R. L. Kumawat, P. Garg, G. Bhattacharyya and B. Pathak, ACS Appl. Electron. Mater., 2020, 2, 1218–1225 CrossRef CAS
.
- S. K. Min, W. Y. Kim, Y. Cho and K. S. Kim, Nat. Nanotechnol., 2011, 6, 162–165 CrossRef CAS PubMed
.
- C. Rajan, M. R. Rezapour, J. Yun, Y. Cho, W. J. C. Seung, K. Min, G. Lee and K. S. Kim, ACS Nano, 2014, 8, 1827–1833 CrossRef PubMed
.
- Y. Choa, S. K. Mina, W. Y. Kim and K. S. Kim, Phys. Chem. Chem. Phys., 2011, 13, 14293–14296 RSC
.
- G. F. Schneider, Q. Xu, S. Hage, S. Luik, J. N. Spoor, S. Malladi, H. Zandbergen and C. Dekker, Nat. Commun., 2013, 4, 1–7 Search PubMed
.
- X. Li, X. Wang, L. Zhang, S. Lee and H. Dai, Science, 2008, 319, 1229–1232 CrossRef CAS PubMed
.
- A. J. Alpert, J. Chromatogr. A, 1986, 359, 85–97 CrossRef CAS
.
- J. Prasongkit, A. Grigoriev, B. Pathak, R. Ahuja and R. H. Scheicher, J. Phys. Chem. C, 2013, 117, 15421–15428 CrossRef CAS
.
- S. Biswas, S. Sen, J.-O. Im, S. Biswas, P. Krstic, B. Ashcroft, C. Borges, Y. Zhao, S. Lindsay and P. Zhang, ACS Nano, 2016, 10, 11304–11316 CrossRef CAS PubMed
.
- S. S. Wong, E. Joselevich, A. T. Woolley, C. L. Cheung and C. M. Lieber, Nature, 1998, 394, 6688 CrossRef PubMed
.
- S. S. Wong, A. T. Woolley, T. W. Odom, J. L. Huang, P. Kim, D. V. Vezenov and C. M. Lieber, Appl. Phys. Lett., 1998, 73, 3465 CrossRef CAS
.
- S. S. Wong, J. D. Harper, P. T. Lansbury and C. M. Lieber, J. Am. Chem. Soc., 1998, 120, 603 CrossRef CAS
.
- Y. H. Lin, F. Lu, Y. Tu and Z. F. Ren, Nano Lett., 2004, 4, 191 CrossRef CAS
.
- J. J. Gooding, Electroanalysis, 2002, 14, 1149 CrossRef CAS
.
- N. R. Wilson and J. V. Macpherson, Nat. Nanotechonol., 2009, 4, 483–491 CrossRef CAS
.
- A. Qureshi, W. P. Kang, J. L. Davidson and Y. Gurbuz, Diamond Relat. Mater., 2009, 18, 1401–1420 CrossRef CAS
.
- V. Meunier and P. S. Krstic, J. Chem. Phys., 2008, 128, 041103 CrossRef
.
- H. S. Kim, S. J. Lee and Y. H. Kim, Small, 2014, 10, 774–781 CrossRef CAS
.
- S. W. Jung, H. S. Kim, A. E. Cho and Y.-H. Kim, ACS Appl. Mater. Interfaces, 2018, 10, 18227–18236 CrossRef CAS
.
- Z. Jiang, M. Mihovilovic, J. Chan and D. Stein, J. Phys. Condens. Matter, 2010, 22, 454114 CrossRef
.
- H. Liu, J. He, J. Tang, H. Liu, P. Pang, D. Cao, P. Krstic, S. Joseph, S. Lindsay and C. Nuckolls, Science, 2010, 327, 64–67 CrossRef CAS
.
- L. Liu, C. Yang, K. Zhao, J. Li and H. C. Wu, Nat. Commun., 2013, 4, 2989 CrossRef
.
- T. Ito, L. Sun, R. R. Henriquez and R. M. Crooks, Acc. Chem. Res., 2004, 37, 937–945 CrossRef CAS
.
- B. Arash, Q. Wang and N. Wu, J. Nanotechnol. Eng. Med., 2012, 3, 020902 CrossRef
.
- S. Fan, M. G. Chapline, N. R. Franklin, T. W. Tombler, A. M. Cassell and H. Dai, Science, 1999, 283, 512–514 CrossRef CAS PubMed
.
- L. Sun and R. M. Crooks, J. Am. Chem. Soc., 2000, 122, 12340–12345 CrossRef CAS
.
-
M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson and H. Nakatsuji, GAUSSIAN 09, Revision A.1., Gaussian, Inc., Wallingford CT, 2009 Search PubMed
.
- P. Ordejón, E. Artacho and J. M. Soler, Phys. Rev. B: Condens. Matter Mater. Phys., 1996, 53, 10441–10444 CrossRef PubMed
.
- M. Brandbyge, J. L. Mozos, P. Ordejón, J. Taylor and K. Stokbro, Phys. Rev. B: Condens. Matter Mater. Phys., 2002, 65, 165401 CrossRef
.
- C. Lee, W. Yang and R. G. Parr, Phys. Rev. B: Condens. Matter Mater. Phys., 1988, 37, 785–789 CrossRef CAS PubMed
.
- N. Troullier and J. L. Martins, Phys. Rev. B: Condens. Matter Mater. Phys., 1991, 43, 1993–2006 CrossRef CAS PubMed
.
-
S. Dutta, Electronic Transport in Mesoscopic Systems, Cambridge University Press, Cambridge, 1995 Search PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available: Binding energy values, optimized configurations for A, T, and C probe functionalized CNT (6,6) nanogap setup with all the four nucleotides; reader-target base pair geometries, energetically most stable configurations for all the four nucleotides with the G probe; I–V characteristics, transmission function, and DOS curves for the A, T, and C probe functionalized CNT (6,6) nanogap setup with all the four nucleotides; bias-dependent transmission curves; transmission curve as a function of angles; schematics of different types of radii and CNT electrodes and the corresponding transmission function curves. See DOI: 10.1039/d0na00241k |
| This journal is © The Royal Society of Chemistry 2020 |