Open Access Article
Open Access ArticleEnhancing curcumin's solubility and antibiofilm activity via silica surface modification†
Caio H. N.
Barros
 ,
Henry
Devlin
,
Henry
Devlin
 ,
Dishon W.
Hiebner
,
Dishon W.
Hiebner
 ,
Stefania
Vitale
,
Stefania
Vitale
 ,
Laura
Quinn
,
Laura
Quinn
 and
Eoin
Casey
and
Eoin
Casey
 *
*
School of Chemical and Bioprocess Engineering, University College Dublin, Ireland. E-mail: eoin.casey@ucd.ie
First published on 20th March 2020
Abstract
Bacterial biofilms are microbial communities in which bacterial cells in sessile state are mechanically and chemically protected against foreign agents, thus enhancing antibiotic resistance. The delivery of active compounds to the inside of biofilms is often hindered due to the existence of the biofilm extracellular polymeric substances (EPS) and to the poor solubility of drugs and antibiotics. A possible strategy to overcome the EPS barrier is the incorporation of antimicrobial agents into a nanocarrier, able to penetrate the matrix and deliver the active substance to the cells. Here, we report the synthesis of antimicrobial curcumin-conjugated silica nanoparticles (curc-NPs) as a possibility for dealing with these issues. Curcumin is a known antimicrobial agent and to overcome its low solubility in water it was grafted onto the surface of silica nanoparticles, the latter functioning as nanocarrier for curcumin into the biofilm. Curc-NPs were able to impede the formation of model P. putida biofilms up to 50% and disrupt mature biofilms up to 54% at 2.5 mg mL−1. Cell viability of sessile cells in both cases was also considerably affected, which is not observed for curcumin delivered as a free compound at the same concentration. Furthermore, proteomics of extracted EPS matrix of biofilms grown in the presence of free curcumin and curc-NPs revealed differences in the expression of key proteins related to cell detoxification and energy production. Therefore, curc-NPs are presented here as an alternative for curcumin delivery that can be exploited not only to other bacterial strains but also to further biological applications.
Introduction
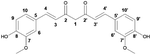
The use of nanoparticles (NPs) for antibacterial and antibiofilm purposes has gained more attention as the range of combination of materials, antimicrobial compounds and delivery systems keeps broadening. Materials in the nanoscale usually present an enhanced biological activity due to the enlarged overall surface area per unit of mass of the particle.1 Nanoparticles can either be inherently antimicrobial such as the ones made of silver,2 copper3 or zinc4 or they can function as nanocarriers5,6 or nanoplatforms for the conjugation of bioactive molecules.7–10 A more recent area for nanoparticles applications relates to their use as antibiofilm agents.11,12 This approach is more complex due to the multifaceted nature of biofilms and the existence of the extracellular polymeric substance (EPS),13–15 which is a key component of this bacterial mode of life. Biofilms are widely found in nature and consist of a microenvironment in which microorganisms are encased in a nutrient-rich organic matrix while existing in a sessile life mode.16 The transition of bacteria from planktonic to biofilm mode goes through the stages of reversible attachment to a surface, followed by irreversible attachment, maturation phases and dispersion.17 Despite the fact that the sessile and biofilm-detached bacterial phenotypes are fundamentally different from the planktonic phenotype regarding growth kinetics and cell surface properties, the intrinsic susceptibility to antibiotics is the same.18 However, the EPS matrix produced by the biofilm phenotype enhances bacterial resistance19 and eases the perpetuation of the species by providing an environment physically and chemically protected from foreign agents.14,20 Thus, biofilms represent a threat in industrial21 and biomedical environments,22 for instance.The quest for more eco-friendly and sustainable nanomaterials motivates the study of non-metallic nanoparticles often conjugated to natural products. The advantage of using natural products for this end lies on the fact that they have well-known biological activities and few cytotoxic effects in contrast with conventional antimicrobial drugs.23 Curcumin (Fig. 1) is a polyphenolic natural compound24 that fits within this category. It is the active ingredient of turmeric (Curcuma longa) and has been used for more than 2000 years as a dye, spice and medicine.25 Curcumin has been linked to antimicrobial,26 anti-inflammatory, antioxidant27 and anticancer25,28 activities. In order to exploit this biological activity and at the same time overcome its low solubility in water, nanocarrier delivery systems have been developed for curcumin. Specifically, for antimicrobial/antibiofilm applications, curcumin has been encapsulated in liposomes, solid lipid nanoparticles,29 and also linked to chitosan-conjugated nanomaterials,30 silver nanoparticles31 and in the form of microemulsions32 or nanocurcumin.33,34 The delivery of curcumin in the form of nanocomposite,35 nanoparticles,34,36 inclusion complexes37 and encapsulated in polymeric nanoparticles38 has also been explored. Shlar et al.39 recently showed how the method for curcumin delivery affects the antibacterial mode of action of the compound. Curcumin has also been reported to downregulate quorum sensing (QS) genes related to the production of biofilm virulence factors in P. aeruginosa,40 reduce oxidative stress response,41 and has been suggested as a modulin and curli protein (biofilm matrix proteins) inhibitor in in silico studies performed with E. coli.11 Furthermore, it displays an inhibition effect on alginate and exopolysaccharide production in Vibrio sp., as well as a suppression on QS-dependent swimming and swarming motility.41
The chemical modification of curcumin for better solubility or achievement of desired biological activities has been widely explored.42,43 In particular, the modification of phenolic groups in C8 and C8′ is very common due to the reactivity of this functional group.44 In this paper, the phenolic groups of curcumin were used to conjugate them to the surface of carboxylate-functionalised silica NPs and synthesise curcumin-functionalised silica NPs (curc-NPs). The antibiofilm activity of curc-NPs was then tested against a strain of Pseudomonas putida, a Gram-negative bacteria found in environmental niches45 and whose biofilms impact on water treatment. P. putida (PCL 1482) was chosen as a bacterial strain model for biofilm production46 as it is known to produce a biomolecule-rich biofilm matrix in which bacterial cells are embedded.47 Silica NPs were chosen as a nanoplatform given their low toxicity to both prokaryotes48 and eukaryotes49 and low environmental impact.50 Despite the fact that, for silica NPs, curcumin is often delivered in mesoporous silica51–53 or added to the silica matrix itself,54 here the compound is functionalised covalently only at the surface of the NPs. The NPs produced are shown to have little antibacterial effect, however a noticeable antibiofilm activity.
Materials and methods
Reagents
King B Agar, peptone (vegetable) No 1, tetracycline hydrochloride, gentamicin sulfate salt, calcium chloride (CaCl2), magnesium sulfate (MgSO4), acetic acid, ammonium hydroxide (NH4OH), ammonium bicarbonate, methanol, (3-aminopropyl)triethoxysilane (APTES), succinic anhydride, N1-(3-trimethoxysilylpropyl)diethylenetriamine (DETA), dimethylformamide (DMF), dimethyl sulfoxide (DMSO), N,N′-dicyclohexylcarbodiimide (DCC), N-hydroxysuccinimide (NHS), 4-dimethylaminopyridine (DMAP), curcumin, dithiothreitol (DTT), iodoacetic acid, acetic acid, acetonitrile, formic acid, Dowex® cation exchange resin, glycerol, Mowiol 4-88 and Trypsin (Mass Spectrometry grade) were purchased from Sigma Aldrich and utilized as supplied. Potassium phosphate dibasic was purchased from Honeywell, Fluka™. The water used in all experiments was MilliQ grade water, purified using an Elga Process Water System.Nanoparticle synthesis
Synthesis of silica NPs was based on the Stöber method55 and the subsequent functionalisation steps (amination, carboxylation) were carried out according to a procedure previously described.56 For the synthesis of curc-NPs, 80 mg of carboxylate-functionalised silica NPs, 47 mg of DCC, 14 mg of DMAP and 80 mg of curcumin were dispersed in 50 mL of dry DMF under inert atmosphere at 0 °C. The mixture was stirred to room temperature and then left under agitation for 20 h. Yellowish coloured curc-NPs were recovered by centrifugation (9000 rpm, 20 min) and washed at least three times with water via centrifugation (Scheme 1). In the case where fluorescently labelled NPs were desired, 1 mL of dye conjugate was added in the mixture of the adapted Stoeber reaction method. The dye conjugate was previously prepared by mixing 5 mg of fluorescein isothiocyanate (FITC) with 2.5 mL of ethanol and 25 μL of APTES, following a procedure described in the literature.57,58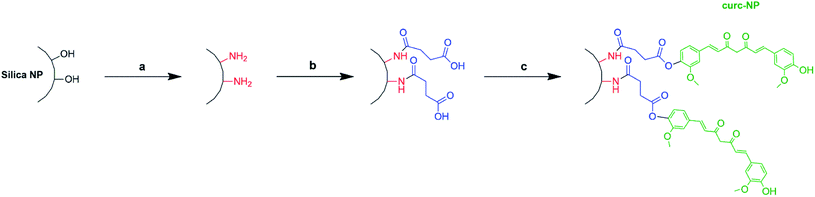 | ||
| Scheme 1 Synthetic route for production of curc-NPs (a) DETA, acetic acid 1 mmol L−1, 30 min; (b) succinic anhydride, DMF, 24h; (c) curcumin, DCC, DMAP, dry DMF, 20 h. | ||
Nanoparticle characterisation
In order to assess the success of curcumin functionalisation, UV-vis spectroscopy was used to check the typical absorption band at 425 nm. For this end, a plate reader (SpectraMax iD3, Molecular Devices) was used with a resolution of 5 nm and a spectral window from 280 to 900 nm. Dynamic Light Scattering (DLS) and zeta potential measurements were performed in a Zetasizer Nano ZS (Malvern Instruments). NPs samples in 1 mg mL−1 aqueous dispersion were analysed in a folded capillary zeta cell. For size measurements, experiments were run in duplicate with 10 runs for each measurement. For zeta potential measurements, experiments were done in triplicate with 10 scans each. Transmission electron microscopy (TEM) analysis of curc-NPs was done using a FEI Tecnai G2, with samples deposited on carbon-coated copper grids. Size distribution of nanoparticles was determined using the Fiji software. Fourier Transformed Infrared Spectroscopy (FTIR) was performed in a Bruker Vertex 70 spectrophotometer. Samples in powder form were deposited onto NaCl FTIR cards; spectra were taken with a resolution of 4 cm−1, from 4000 to 400 cm−1 and 64 scans. Dissolution Nuclear Magnetic Resonance (NMR) was used according to a previously reported method.59 Silica NPs in the powder form (5–10 mg) were dissolved in 662 μL of deuterated water (D2O) and then 38 μL of deuterated sodium hydroxide (NaOD) was added. The mixture was incubated at 37 °C overnight. The same procedure was performed for free curcumin. 1H NMR spectra were obtained in a Varian Inova 400 MHz Spectrometer using 256 scans per sample.In order to determine the density of curcumin molecules at the silica NPs surface, the keto–enol tautomerism was exploited. To 1.0 mL solutions of curcumin in water (25, 18.75, 12.5, 9.37, 6.25, 4.69, 3.13 μg mL−1), 50 μL of NaOH 2 N was added and the values of absorbance at 465 nm were taken. A sample of curc-NPs underwent the same treatment and the density of curcumin molecules was estimated using the core size determined by TEM.
Bacterial growth kinetics
Using a glycerol stock, Green Fluorescent Protein (GFP)-expressing P. putida (PCL 1482) possessing a tetracyclin-resistance gene was streaked onto a King B agar plate containing tetracyclin (40 μg mL−1) and incubated at 30 °C for 24 h. A single colony was removed from the plate and used to inoculate a sterile conical flask containing 50 mL of King B media and supplemented with tetracyclin (40 μg mL−1). The culture was incubated for 16–18 h at 30 °C, 200 rpm. The OD600 of the overnight culture was adjusted to 0.001 using sterile King B media supplemented with CaCl2 (1.5 mmol L−1), MgCl2 (1.5 mmol L−1) and tetracyclin (40 μg mL−1). This bacterial mixture (150 μL) was then poured into 96-well plates and each row was assigned to a different treatment: control (addition of sterile water), free curcumin at final concentrations of 0.05 and 0.025 mg mL−1 (1% DMSO), bare NPs (5.0 and 2.5 mg mL−1), curc-NPs (5.0 and 2.5 mg mL−1). From this point onwards, curc-NPs at 5.0 mg mL−1 refers to the concentration of the entire nanoparticle (silica + curcumin) and is always compared to free curcumin at 0.05 mg mL−1 as having the same overall curcumin quantity. The same is valid for curc-NPs 2.5 mg mL−1 and free curcumin at 0.025 mg mL−1. As the addition of NPs and curcumin have an influence in the turbidity of the suspension, a number of rows were assigned to “blanks”, whose wells were supplemented with gentamycin (40 μg mL−1) to impede bacterial growth. The optical density at 600 nm (OD600) was monitored using a plate reader (SpectraMax iD3, Molecular Devices), with absorbance measurements every hour. During 24 h, the plates were shaken in between the measurements while maintaining a temperature of 30 °C.Colony forming units (CFU) counting
From the wells of the bacterial growth kinetics experiment, 50 μL were taken and diluted to a 10−7 dilution in sterile water. Following which, 50 μL of the final dilution was spread onto tetracylin-supplemented King B agar plates. After 24 h, the colonies formed were counted. This procedure was done in triplicate.Biofilm inhibition assay
The remaining bacterial suspensions were removed from the wells of the bacterial growth kinetics experiment. The biofilms formed were washed twice with water and then 200 μL of crystal violet solution (0.1% w/v) was added and the plate was left static for 20 min in the dark. The crystal violet solution was removed and the stained biofilms were washed with water five times to remove excess unbound dye. Acetic acid 30% was added (200 μL) to dissolve the stained biofilms and plates were shaken at 125 rpm in the dark for 25 min. Then, the absorbance values at 600 nm was taken using the plate reader. Once again, the wells that were supplemented with gentamycin (40 μg mL−1) were used as blanks.Pre-formed biofilms removal assay
P. putida biofilms were cultivated as described previously for 24 h. After removal of the media and washing of the biofilm with water, 200 μL of suspensions of bare NPs (5.0 mg mL−1), curc-NPs (5.0 mg mL−1) and free curcumin (0.05 mg mL−1 in 1% DMSO) were added. The 96-well plates were shaken for 24 h at 125 rpm and 30 °C and the suspensions were removed from the wells. After washing the biofilms twice with water, the same procedure previously described for crystal violet staining was carried out.MTT assay
In order to assess the viability of biofilm bacterial cells the methylthiazolydiphenyltetrazolium bromide (MTT) assay was used. The activity of bacterial reductases can be observed by the reduction of the MTT bromide to formazan, inducing a colour change to purple-blue. After biofilm growth in 96-well plates as previously described, the bacterial culture was discarded and biofilms were washed once with phosphate buffer solution (PBS) and then each well was filled with 150 μL of PBS and 50 μL of MTT solution (pre-warmed to 30 °C). After 1 h incubation, the wells were emptied and 200 μL of DMSO were added. The plate was agitated at 125 rpm for 15 min to facilitate solubilisation. Absorbance was read at 550 nm. At least 8 replicates were carried out for each condition.Confocal microscopy
For biofilm growth, a 5 mL volume of a P. putida overnight grown culture with OD600 adjusted to 1 and supplemented with CaCl2 to a final concentration of 1.5 mmol L−1 was added to a sterile 50 mL centrifuge tube containing a glass coverslip (24 mm × 50 mm) and plugged with sterile cotton wool. Tubes were incubated for 24 h at 30 °C at 100 rpm. Each biofilm-coated glass coverslip was carefully removed from the centrifuge tubes and gently rinsed three times in water. The coverslip was then placed horizontally on a sample holder and 150 μL a curc-NPs suspension (0.5 mg mL−1) was added directly to the biofilm, followed by incubation in the dark for 15 min. After incubation, the biofilms were then gently rinsed 3 times in water. Each coverslip was mounted in Mowiol 4-88 (pH 8.5) mounting medium as described previously by our group.60 Horizontal plane z-stack images were acquired with an Olympus FluorView FV1000 CLSM attached to an inverted Olympus IX81 microscope with a 60×/1.35 NA UPL SAPO oil immersion objective (Olympus Optical, Tokyo, Japan). At least 3 image stacks, with a z-step of 1 μm, from each of 3 independent experiments were acquired and used for each analysis. All image files were analysed in Fiji image processing software.61EPS extraction and quantification
For the extraction of the EPS, the procedure was adapted from the one described by Wu and Xi (2009).62P. putida bacterial overnight culture was diluted as described in previous sections, and then to each well of a sterile 24-well plate, 900 μL of this bacterial mixture was added followed by the addition of 100 μL of the appropriate concentration of the test NP/compound as to reach a final concentration of 5.0 mg mL−1 bare NPs, 5.0 mg mL−1 curc-NPs and 0.05 mg mL−1 of free curcumin in 1% DMSO. The plates were incubated for 3 days at 30 °C with shaking at 125 rpm. The media was then removed and the biofilms were washed with PBS buffer and suspended in NaCl 0.9% (1.0 mL per 4 wells; each condition was set up in 4 wells). After sonication for 2 min, Dowex® resin was added at 1% (m/v) and these mixtures were left agitating at 300 rpm in the dark for 2 h. After centrifugation (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g, 20 min, 4 °C), the supernatants were filtered using a Millipore® 0.22 μm membrane filter. The EPS suspensions were kept at −80 °C until use. This procedure was made in triplicate for each condition.
000 g, 20 min, 4 °C), the supernatants were filtered using a Millipore® 0.22 μm membrane filter. The EPS suspensions were kept at −80 °C until use. This procedure was made in triplicate for each condition.
For protein quantification, the Lowry assay63 was used and a standard curve was produced by using solutions of different concentrations of BSA. For carbohydrate quantification, the phenol–sulfuric acid method64 was chosen and glucose solutions of different concentrations were used as standards.
Proteomics
Protein digestion was performed by using 50 μL of extracted EPS, to which 2.5 μL of DTT 100 mmol L−1 were added to reduce disulphide bonds for 30 min at 60 °C. Then, 2.6 μL of iodoacetic acid 200 mmol L−1 were added. After vortexing, incubation was carried out in the dark at room temperature for 30 min. The sample was then diluted to 0.5 mL using ammonium bicarbonate buffer (50 mmol L−1 pH 8) and 1.5 μL of trypsin solution (20 μg in 20 μL of acetic acid 50 mmol L−1) was added. The digestion took place overnight at 37 °C and was stopped by addition of 5 μL of acetic acid 100%. For sample clean up, the supernatant containing fragmented proteins was isolated via centrifugation and then ZipTips® pipette tips (C18, 0.2 μL bed) were used for peptide isolation. Firstly, wetting solution was aspirated (acetonitrile 0.1% formic acid) and dispensed, followed by the equilibration solution (deionized water 0.1% formic acid). The peptides were bound by aspirating and dispensing the sample solution 7 times, followed by a washing step (1 mL of deionized water 0.1% formic acid). Finally, the peptides were collected by elution with acetonitrile 0.1% formic acid (4 μL). The resulting solution was dried out under vacuum and resuspended in 30 μL of MS grade water containing 2.5% acetonitrile and 0.5% acetic acid. Peptides were analysed on a quadrupole Orbitrap (Q-Exactive, Thermo Scientific) mass spectrometer equipped with a reversed-phase NanoLC UltiMate 3000 HPLC system (Thermo Scientific). Peptide samples were loaded onto C18 reversed phase columns (10 cm length, 75 μm inner diameter) using an injection volume of 5 μL and eluted with a linear gradient from 2 to 95% acetonitrile containing 0.5% acetic acid in 60 min at a flow rate of 250 nL min−1. The mass spectrometer was operated in data dependent mode, automatically switching between MS (precursor ion) and MS2 (fragments of the precursor ion) acquisition. Survey full scan MS spectra (m/z 350–1600) were acquired in the Orbitrap with a resolution of 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. MS2 spectra had a resolution of 17
000. MS2 spectra had a resolution of 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500. The twelve most intense ions were sequentially isolated and fragmented by higher-energy C-trap dissociation.
500. The twelve most intense ions were sequentially isolated and fragmented by higher-energy C-trap dissociation.
MaxQuant software package was used for protein identification using the proteome database of Pseudomonas putida (strain ATCC 47054/DSM 6125/NCIMB 11950/KT 2440) (extracted from Uniprot). Only proteins with a minimum number of peptides of 2 and minimum number of unique peptides of 1 were considered. Label-free quantification of the proteins was also run using the same software, in triplicate. Only proteins identified in at least two of the three replicates were considered.
Results and discussion
Nanoparticle characterisation
Curcumin was used to functionalise the surface of silica NPs through the formation of an ester bond between a carboxylate moiety at the surface of the NPs and the phenol groups present in the natural compound. For this coupling reaction NHS was used as catalyst and DCC as coupling reagent. Despite a relatively large hydrodynamic radius (measured by DLS), the NPs have core diameter of 61.5 ± 7.9 nm, as demonstrated by TEM imaging (Fig. 2A). The zeta potential is negative due to the presence of phenolate groups at the surface of the NPs, providing an extra electrostatic stability to the colloidal system, besides the steric stabilization of the curcumin molecules themselves. As curcumin has a high electronic conjugation, UV-vis absorption bands are evident (major peak at 425 nm) and were used to confirm the success of the reaction (Fig. 2B). To further confirm the functionalisation of the NPs with these compounds, dissolution 1H NMR using NaOD in D2O was used to identify the signals corresponding to aromatic protons. Curcumin is very sensitive to high pH values, so the same pre-treatment was carried out for the free compound and NMR spectra were acquired. As seen in Fig. 3, the spectrum of curc-NP contains all the signals of the curcumin (aromatic region between 6.5–7.5 ppm and methoxy protons at 3.8 ppm) as well as the signals from the succinate moiety and the aminated silane. NMR analysis thus further confirmed the conjugation of curcumin to the surface of silica NPs. The density of curcumin molecules was estimated using the absorbance of these compounds in the UV-visible region and the NPs core size obtained by TEM images (Table 1). The challenge here was to diminish as much as possible the error caused by the scattering profile of the silica, which increases with smaller wavelengths. To address this issue, the absorbance spectra of curcumin and curc-NPs were taken at pH 11, in which the enol tautomer is favoured and the absorption peak shifts from ∼425 nm to ∼465 nm (Fig. S1†). At 465 nm, the absorption is more intense and further away from an intense silica scattering, and by using silica NPs at the same concentration as a blank, the density of curcumin molecules at the silica surface could be estimated. The footprint of curcumin was estimated as 0.4 molecules per nm2 and 1.0% (w/w) contribution in mass in the silica matrix.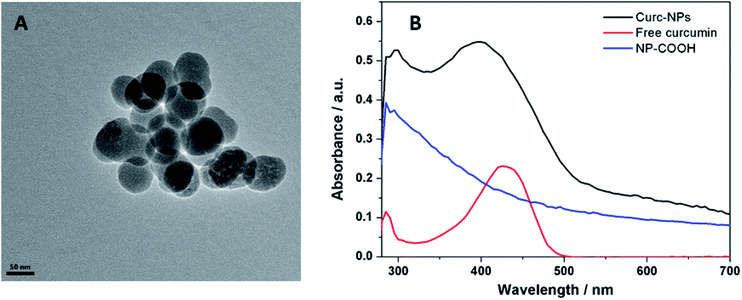 | ||
| Fig. 2 Scanning Electron Microscopy (SEM) images of curc-NPs (A) UV-vis absorption spectra of the precursor NP-COOH and free curcumin (absorption band at 425 nm) and curc-NPs (B). | ||
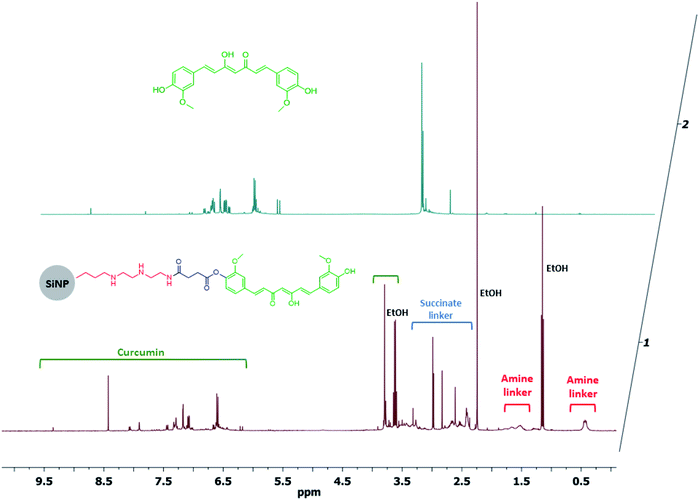 | ||
| Fig. 3 Dissolution 1H NMR spectra of curc-NPs and free curcumin in NaOD/D2O. HOD signal (4.8 ppm) was suppressed for visualisation purposes. | ||
| Size by TEM (nm) | Size by DLS (nm) | Zeta potential (mV) | % of curcumin (w/w) | Density of curcumin molecules per nm2 | |
|---|---|---|---|---|---|
| Curc-NPs | 61.5 ± 7.9 | 167.4 ± 48.7 | −44.1 ± 0.8 | 1.0 | 0.4 |
Antibacterial and antibiofilm assays
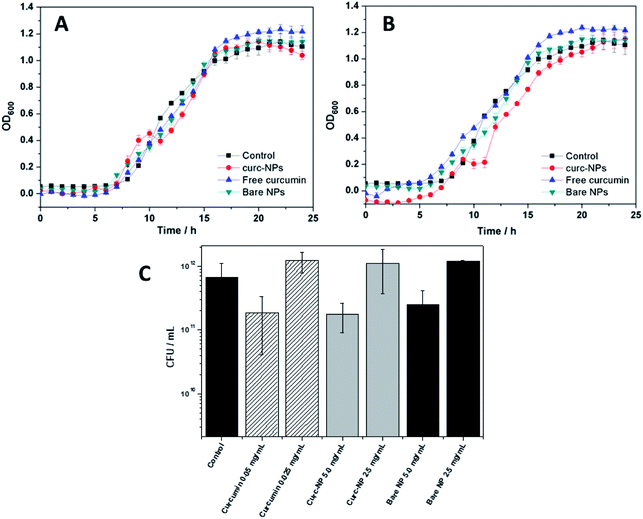
In order to assess the antibacterial and antibiofilm activities of curc-NPs, the growth of P. putida was monitored during 24 h in the presence of curc-NPs (5.0 and 2.5 mg mL−1). These concentration values were chosen in order to compare the activities of the NPs to the activity of free curcumin; the latter was used at a concentration of 0.05 and 0.025 mg mL−1 (1% DMSO), which are the equivalent concentrations of active compound conjugated to the nanoparticles, according to the estimation of surface coverage (Table 1). Bare silica NPs were also used in these assays at a concentration of 5.0 and 2.5 mg mL−1 to validate the conjugation of the active compounds. Curc-NPs had a minor effect on bacterial growth at 5.0 mg mL−1, by affecting the growth rate during the exponential phase (Fig. 4A and B) when compared to a control (addition of sterile water) and bare NPs. Free curcumin did not have any antibacterial effect on P. putida. The Colony Forming Units (CFU) remaining in the media after growth for 24 h in the presence of free curcumin, curc-NPs and bare NPs did not have a statistical difference when compared to the control (Fig. 4C). It is worth mentioning, however, that a control biofilm has an average CFU mL−1 value in the order of 1011, in contrast with 1012 CFU mL−1 for planktonic cells (data not shown).It has been reported that one of the major bactericidal mechanisms by which curcumin acts is through bacterial membrane disruption and leakage of intracellular content.65,66 In this study, even at relatively high curcumin concentrations, this effect was not significant enough to the point of being detected. Nevertheless, the higher local concentration of curcumin at the NPs surface could account for a more effective membrane disruption effect.
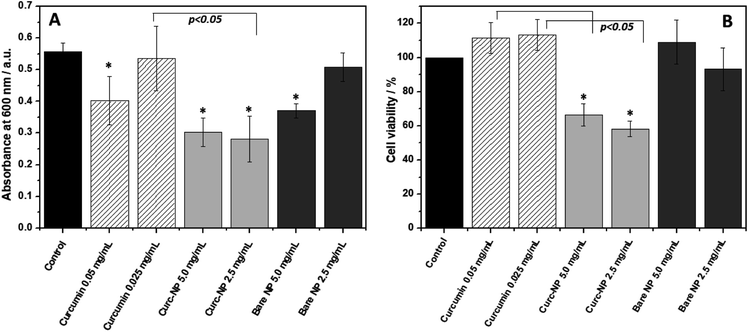
When looking at biofilm formation, a different scenario takes place (Fig. 5A). The biofilms formed in the presence of curc-NPs have significantly less biomass when compared to the controls and to the bare NPs at a concentration of 2.5 mg mL−1 (50% and 55%, respectively). Free curcumin at an equivalent concentration of 0.025 mg mL−1 was not able to hinder biofilm formation. Interestingly, free curcumin at 0.05 mg mL−1 had a minor antibiofilm effect, however the increase in concentration of curc-NPs did not result in an elevated antibiofilm activity. When looking at the bacterial cell viability inside the biofilms formed (Fig. 5B), again curc-NPs at 2.5 mg mL−1 shows an improved bactericidal effect when compared to the control, free curcumin and bare NPs; it has diminished cell viability to about 55%. These results suggest that the attachment of curcumin to the silica surface is improving its biological activity. The increase in concentration from 2.5 to 5.0 mg mL−1 does not yield a better efficacy, and this could be possibly due to a lack of stability of a more concentrated colloidal suspension, which ends up leading to some precipitation of curc-NPs at the bottom of the wells.
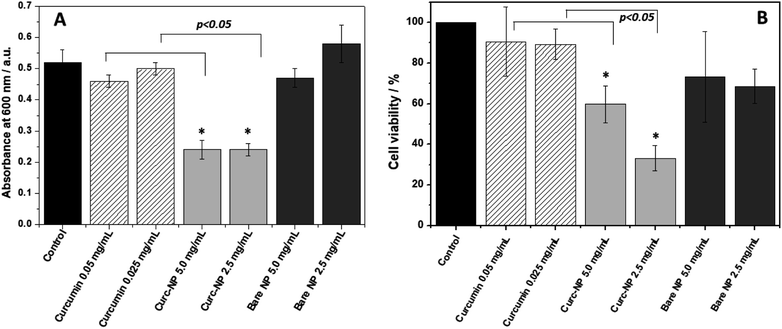
The performance of the conjugated nanoparticles was also tested using pre-formed 24 h biofilms (Fig. 6). After 24 h growth and biofilm formation, the culture media was discarded and the biofilms formed were washed with water. These biofilms were exposed to the NPs and to the free active compound at the same concentration as in the bacterial growth kinetics experiment. Curc-NPs were able to disrupt the pre-formed biofilms to a significant extent (54% biomass reduction on average). Once again, curc-NPs at 5.0 mg mL−1 did not present enhanced activity when compared to it at 2.5 mg mL−1. On the other hand, free curcumin was not able to decrease the biofilm biomass when compared to the water control at any concentration. The cell viability of biofilms exposed to curc-NPs at 2.5 mg mL−1 decreased to 33%. This again shows that although not very active against planktonic bacteria, curc-NPs are efficient against sessile bacteria in the biofilm environment. The active curcumin concentration range used here and its associated findings are in line with other studies in the field; for example, in a study of curcumin against P. aeruginosa, only at a concentration of 0.037 mg mL−1 was it able to display bactericidal activity.65 In a study on the synergistic effects of curcumin with standard antibiotics, the average minimum inhibitory concentrations (MIC) of curcumin against Gram-negative clinically isolated strains was of 0.1269 mg mL−1.67 For methicillin-resistant Staphylococcus aureus, the MIC of curcumin may vary from 0.250 mg mL−1 to 0.00097 mg mL−1 when used synergistically with oxacillin.68 Therefore, this work presents biological activity results which are found within the expected curcumin concentration range, however a significative enhance in activity is obtained when this molecule is grafted onto silica NPs.
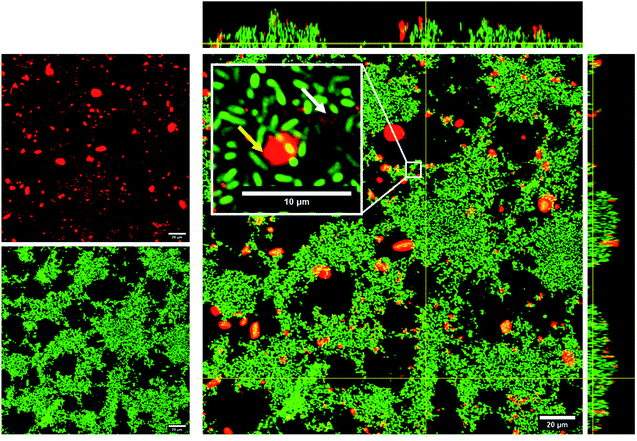
The improved biofilm disintegration effect seen for conjugated NPs in comparison with their free form counterparts might relate to the fact that the silica NPs are transported into the biofilms, thus increasing the local concentration of the active compound within the biofilm. The accumulation of curc-NPs in the biofilms is noticeable by confocal microscopy, in which fluorescently labelled NPs were used to determine the fate and extent of aggregation when in contact with P. putida biofilms. As seen in Fig. 7, curc-NPs form larger aggregates of around 2–3 μm in the presence of the biofilm and culture media (even being monodispersed as shown in Table 1). Regardless of this effect, the NPs can penetrate into the biofilms to a certain degree, as shown in the inset of the confocal image in Fig. 7, which could enhance the permeation of curcumin into the innermost layers of the biofilm. The hydrophobic character of this compound in their free form is probably an obstacle for the effective permeation throughout the network of hydrophilic biomolecules (proteins, polysaccharides, eDNA). In addition to this effect, the fact that curcumin is linked to a succinic anhydride moiety via an ester bond makes them prone to be released by the cleavage of this bond by lipases. This class of enzymes are usually abundant in many biofilms species,69–71 including in P. putida,72 and often have a low specificity towards their substrates. In fact, a number of studies have reported enhanced antibiofilm effect via bacterial lipase-triggered release of antibiotics.73,74
Proteomic profiling
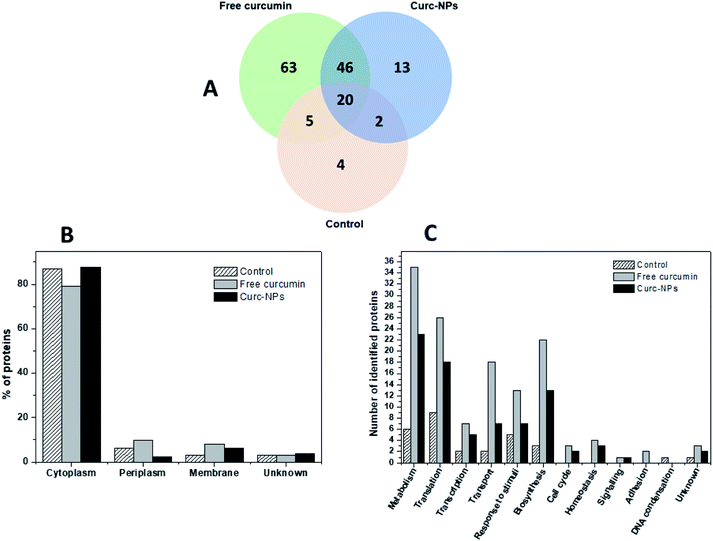
Considering the broad range of proteins produced by bacterial cells in the biofilm mode and that foreign agents could trigger some responses in the microbial protein expression, we aimed to gain some mechanistic insights on the action of curcumin both in its free form and conjugated to silica NPs. In order to accomplish this, P. putida biofilms were grown for 3 days in the presence of the test compounds/nanoparticles and then the EPS matrix was extracted and underwent trypsin digestion and peptide identification using liquid chromatography-mass spectrometry (LC-MS/MS). Using the MaxQuant server, the proteins produced in each condition could be identified. Therefore, a full proteomic profiling of P. putidawas obtained from the early stages of biofilm formation (before bacterial attachment) up to a point in which it is considered a mature biofilm (72 h). The Venn diagram in Fig. 8A shows a general view of the proteins identified in each condition. The exposure to either free curcumin or curc-NPs resulted in the expression of 122 proteins that were not produced in the control experiment. This suggests that curcumin induced a stress to the bacterial system that led to an overactivation of the protein expression machinery of the cells. The cellular localisation profile did not change significantly amongst conditions (Fig. 8B), however when looking at the cellular processes in which these proteins are involved in, interesting differences and patterns arise. At a widespread glance, all cellular processes were overloaded, including the production of proteins related to response to stimuli, cellular transport and biosynthesis. Also, some proteins related to some cellular processes were found that were not identified at all in control conditions, such as the ones involved in cell signalling, cell cycle and homeostasis.Some proteins stand out as being exclusively expressed under one condition or another. For instance, under the stress caused by free curcumin, bacterial cells were found to produce a putative xenobiotic reductase (Uniprot entry: Q88MU0), which is involved in the metabolism of a wide range of substrates. The xenobiotic reductase A (XenA) from P. putida has been reported as essential for the degradation of quinolines via the 8-hydroxycoumarin pathway.75 XenA degrades olefinic substrates by breaking the double bond of α,β-unsaturated carbonyl compounds using NADH or NADPH as electron donors.76 As curcumin contains two α,β-unsaturated carbonyl groups in its structure, it can be inferred that the production of this xenobiotic reductase is one of the defence mechanisms of the bacterial cells against curcumin. The putative xenobiotic reductase reported here has a 36% identity (by BLAST) with NADPH-dependent curcumin/dihydrocurcumin reductase (CurA) reported by Hassaninasab et al.77 who postulated a possible NADPH-dependent curcumin inactivation mechanism by E. coli (Scheme 2). It is thus possible that the same occurs for P. putida exposed to curcumin.
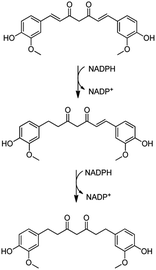 | ||
| Scheme 2 Proposed curcumin metabolism for inactivation via xenobiotic reductase pathway. Adapted from Hassaninasab et al.77 | ||
Additionally, an outer membrane A (OmpA) family protein (Uniprot entry: Q88MR7) was also identified under curcumin stress. OmpA is a pathogenicity-related protein comprised of N-terminal eight stranded β barrel domain that is embedded in the bacterial membrane.78 The expression levels of OmpA are often dependent on external factors such culture conditions and presence of certain chemicals. The exposure to phenol, for instance, may induce an overexpression of these proteins as reported by Zhang et al.79 using E. coli as a bacterial model. Curcumin, being a phenolic compound, might fit in the category of compounds that upregulate OmpA expression.
The growth of P. putida bacterial cultures in biofilm-forming conditions in the presence of curc-NPs also led to the expression of certain proteins which were not seen for the other two conditions. Adenylosuccinate synthetase (PurA – Uniprot entry: Q88D8) is an example of this protein group; it is involved in purine biosynthesis, being usually overexpressed in stress conditions. Santos et al.80 reported the upregulation of PurA of P. putida KT2440 under phenol stress conditions. Additionally, the expression of a probable efflux pump periplasmatic linker (TtgA – Uniprot entry: Q88N30) denotes a bacterial response to the presence of a harmful compound. Efflux pumps are directly correlated to drug resistance and are responsible for the expulsion of antibiotics from the bacterial cytoplasm to the exterior of the cell.81 For P. putida GM73, expression of genes correlated to efflux pumps-related proteins were linked to toluene resistance, for instance.82 Interestingly, the presence of arginine deiminase (Uniprot entry: Q88P52) was also detected. This enzyme contributes with the maintenance of arginine reserves of the cell, which is converted into adenosine 5′-triphosphate (ATP) when normal respiration is not taking place.83 Arginine deiminase's activity is usually coupled with catabolic ornithine carbamoyltransferase, especially when in conditions of carbon depletion. This last enzyme was also identified in conditions of exposure to both free curcumin and curc-NPs in our experiments. The fact that the expression of cytoplasmic proteins was elevated in the presence of not only free curcumin but also curc-NPs suggests that curcumin is entering the bacterial cell, even when delivered while covalently conjugated to silica nanoparticles. This could indicate curcumin release from the nanoparticles by the aforementioned enzymatic lipolytic mechanism, once the nanoparticles are too large to enter the bacterial cell. A full list of the proteins identified in the biofilm EPS of P. putida PCL 1482 expressed under the three conditions tested (control, exposure to free curcumin and exposure to curc-NPs) can be found in Table S1.†
When looking at the relative amounts of protein expressed by P. putida under each condition revealed by a label-free-quantification (LFQ) data treatment, other distinctive features emerge which further confirms the observations made thus far (Table 2). For biofilms grown under the stress of both free curcumin and curc-NPs, the enzymes arginine deiminase and catabolic ornithine carbamoyltransferase appear as two of the 8 most abundant identified proteins. This evidence confirms that the presence of curcumin in its free form or conjugated to silica nanoparticles acts as a nutrient depletion and/or a respiratory stress factor which leads to an overproduction of reserves of arginine for the production of ATP. It is also noteworthy that a putative adhesion protein (Uniprot entry: Q88PP2) stands as the fifth and sixth most abundant protein in control and free curcumin conditions, respectively, but is not found in P. putida exposed to curc-NPs. Adhesion proteins have a large contribution on biofilm formation and maintenance across a variety of bacterial strains.84,85 Therefore, the fact that it was not found in the biofilm EPS of P. putida under curc-NPs stress conditions goes in line with the fact the curc-NPs had a superior performance in terms of antibiofilm activity when compared to curcumin in its free form. If by any reason curc-NPs were able to inhibit adhesion proteins expression, this would ultimately impact the formation and establishment of biofilms.
| Uniprot entry | Control | |
|---|---|---|
| 1 | Q88K29 | Nucleic acid cold-shock chaperone |
| 2 | Q88QP8 | Elongation factor Tu-A |
| 3 | Q88NY2 | Glutamate/aspartate ABC transporter-periplasmic binding protein |
| 4 | Q88DU2 | Chaperone protein DnaK |
| 5 | Q88PP2 | Putative surface adhesion protein |
| 6 | Q88N55 | 60 kDa chaperonin |
| 7 | Q88QN8 | Elongation factor G 1 |
| 8 | Q88FB9 | Chaperone protein HtpG |
| 9 | Q88PK1 | Nucleoside diphosphate kinase |
| 10 | Q88QL8 | 30S ribosomal protein S5 |
| 11 | Q88BX2 | ATP synthase subunit alpha |
| 12 | Q88FB3 | Succinate--CoA ligase [ADP-forming] subunit alpha |
| 13 | Q88QN2 | 50S ribosomal protein L2 |
| 14 | Q88DU1 | Protein GrpE |
| 15 | Q88DE8 | 30S ribosomal protein S6 |
| 16 | Q88KJ1 | Trigger factor |
| 17 | Q88Q10 | 50S ribosomal protein L21 |
| 18 | Q88QM0 | 50S ribosomal protein L6 |
| 19 | Q88Q27 | Serine hydroxymethyltransferase 2 |
| 20 | Q88QL2 | 30S ribosomal protein S4 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| Exposure to free curcumin at 0.05 mg mL −1 | ||
| 1 | Q88K29 | Nucleic acid cold-shock chaperone |
| 2 | Q88QP8 | Elongation factor Tu-A |
| 3 | Q88N55 | 60 kDa chaperonin |
| 4 | Q88P53 | Ornithine carbamoyltransferase, catabolic |
| 5 | Q88NY2 | Glutamate/aspartate ABC transporter-periplasmic binding protein |
| 6 | Q88PP2 | Putative surface adhesion protein |
| 7 | Q88BX2 | ATP synthase subunit alpha |
| 8 | Q88P52 | Arginine deiminase |
| 9 | Q88Q10 | 50S ribosomal protein L21 |
| 10 | Q88LL5 | Acyl carrier protein |
| 11 | Q88DU1 | Protein GrpE |
| 12 | Q88DU2 | Chaperone protein DnaK |
| 13 | Q88QL3 | 30S ribosomal protein S13 |
| 14 | P0A157 | 50S ribosomal protein L7/L12 |
| 15 | Q88QN2 | 50S ribosomal protein L2 |
| 16 | Q88FB9 | Chaperone protein HtpG |
| 17 | Q88QL2 | 30S ribosomal protein S4 |
| 18 | Q88KJ1 | Trigger factor |
| 19 | Q88QN9 | 30S ribosomal protein S7 |
| 20 | Q88QL9 | 50S ribosomal protein L18 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| Exposure to curc-NPs at 5.0 mg mL −1 | ||
| 1 | Q88QP8 | Elongation factor Tu-A |
| 2 | Q88K29 | Nucleic acid cold-shock chaperone |
| 3 | Q88P53 | Ornithine carbamoyltransferase, catabolic |
| 4 | Q88N55 | 60 kDa chaperonin |
| 5 | Q88P52 | Arginine deiminase |
| 6 | Q88BX2 | ATP synthase subunit alpha |
| 7 | Q88LL5 | Acyl carrier protein |
| 8 | Q88NY2 | Glutamate/aspartate ABC transporter-periplasmic binding protein |
| 9 | Q88QL3 | 30S ribosomal protein S13 |
| 10 | Q88P78 | Putative DNA-binding protein HU, form N |
| 11 | Q88QN9 | 30S ribosomal protein S7 |
| 12 | Q88FB3 | Succinate--CoA ligase [ADP-forming] subunit alpha |
| 13 | Q88DU2 | Chaperone protein DnaK |
| 14 | Q88DU1 | Protein GrpE |
| 15 | Q88QN8 | Elongation factor G 1 |
| 16 | P0A157 | 50S ribosomal protein L7/L12 |
| 17 | Q88F97 | Electron transfer flavoprotein subunit alpha |
| 18 | Q88FB2 | Succinate--CoA ligase [ADP-forming] subunit beta |
| 19 | Q88Q10 | 50S ribosomal protein L21 |
| 20 | Q88DE8 | 30S ribosomal protein S6 |
Conclusions
In summary, in this paper the natural product curcumin was used to functionalise the surface of silica NPs through covalent modification, via coupling reaction involving curcumin's phenolic groups and a carboxylate moiety linked to the silica's surface. Both curcumin as a free compound and curcumin-functionalised NPs (curc-NPs) were shown to have little to no antibacterial activity towards our model strain of P. putida. However, the antibiofilm activity of curcumin was significantly enhanced when conjugated to the silica surface. Exposure of both developing and mature biofilms to curc-NPs led to a decrease in the biofilm biomass seen by crystal violet staining, and to a decrease in the number of viable cells inside the biofilms (verified by MTT assays), which ultimately means a reduction in the capacity of the microbial environment of maintaining the biofilm phenotype. These results suggest that the conjugation of curcumin molecules onto NPs might increase their effective solubility and thus enhance their biological activity. Additionally, the higher local concentration of curcumin delivered into the biofilms might have played a role for the increased antibiofilm capacity.The identification of proteins in the P. putida EPS after exposure to both free curcumin and curc-NPs gave interesting insights on how the cells respond to the presence of curcumin delivered in two distinct manners. The expression of a putative xenobiotic reductase indicates that the bacterial cells might inactivate the curcumin molecule in a mechanism in which the α,β-unsaturation bonds are broken. The expression of arginine deiminase and catabolic ornithine carbamoyltransferase also denotes that the cells undergo a process similar to nutrient starvation under curcumin stress. Interestingly, a putative adhesion protein which was expressed in high amounts for the bacteria under free curcumin stress was not seen for the bacteria under curc-NPs stress, which might have facilitated biofilm disruption by curc-NPs in comparison with free curcumin.
This work is a demonstration of how the conjugation of an active compound to a silica surface can modify its biological activity due to changes in solubility, local molecule concentration and drug delivery. Further validation of this method will be needed to test the efficacy of not only curc-NPs but also analogous nanoparticles (silica conjugated to water insoluble natural products) towards biofilms of other species, especially pathogenic and antibiotic resistant strains. Considering the low toxicity of both silica and natural products, this combination can potentially be appealing from a biomedical and/or environmental perspective.
Funding source
This research was supported by Science Foundation Ireland (SFI) under grant number 15/IA/3008.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to acknowledge Prof. Kenneth Dawson from Centre for BioNano Interactions (CBNI – University College Dublin) for providing the use of the Malvern Zetasizer equipment.Notes and references
- V. J. Mohanraj and Y. Chen, Nanoparticles – A review, TJPR, 2006, 5, 561–573 Search PubMed.
- C. H. N. Barros, S. Fulaz, D. Stanisic and L. Tasic, Biogenic nanosilver against multidrug-resistant, Antibiotics, 2018, 1, 1–24 Search PubMed.
- O. Rubilar, M. Rai, G. Tortella and M. Cristina, Biogenic nanoparticles: copper, copper oxides, copper sulphides, complex copper nanostructures and their applications, Biotechnol. Lett., 2013, 35, 1365–1375 CrossRef CAS PubMed.
- A. Sirelkhatim, S. Mahmud and A. Seeni, Review on Zinc Oxide Nanoparticles: Antibacterial Activity and Toxicity Mechanism, Nano-Micro Lett., 2015, 7, 219–242 CrossRef CAS PubMed.
- S. Subramaniam, N. Thomas, H. Gustafsson, M. Jambhrunkar, S. P. Kidd and C. A. Prestidge, Rifampicin-loaded mesoporous silica nanoparticles for the treatment of intracellular infections, Antibiotics, 2019, 8, 5–7 CrossRef PubMed.
- J. Mayara, F. De Almeida, E. Damasceno and L. Mafra, pH-Dependent release system of isoniazid carried on nanoparticles of silica obtained from expanded perlite, Appl. Surf. Sci., 2019, 489, 297–312 CrossRef.
- I. Pal, D. Bhattachary, R. K. Kar, D. Zarena, A. Bhunia and H. S. Atreya, A peptide-nanoparticle system with improved efficacy against multidrug resistant bacteria, Sci. Rep., 2019, 9, 1–11 CrossRef CAS PubMed.
- L. Mei, Z. Lu, W. Zhang, Z. Wu, X. Zhang, Y. Wang, Y. Luo, C. Li and Y. Jia, Bioconjugated nanoparticles for attachment and penetration into pathogenic bacteria, Biomaterials, 2013, 34, 10328–10337 CrossRef CAS PubMed.
- Z. Chen, H. Ji, C. Liu, W. Bing, Z. Wang and X. Qu, A multinuclear metal complex based DNase-mimetic artificial enzyme: matrix cleavage for combating bacterial biofilms, Angew. Chem., Int. Ed., 2016, 55, 10732–10736 CrossRef CAS PubMed.
- J. Li, K. Zhang, L. Ruan, S. F. Chin, N. Wickramasinghe, H. Liu, V. Ravikumar, J. Ren, H. Duan, L. Yang and M. B. Chan-park, Block copolymer nanoparticles remove biofilms of drug-resistant Gram-positive bacteria by nanoscale bacterial debridement, Nano Lett., 2018, 18, 4180–4187 CrossRef CAS PubMed.
- A. K. Singh, P. Prakash, R. Singh, N. Nandy, Z. Firdaus, M. Bansal, R. K. Singh, A. Srivastava, J. K. Roy, B. Mishra and R. K. Singh, Curcumin quantum dots mediated degradation of bacterial biofilms, Front. Microbiol., 2017, 8, 1–17 CAS.
- P. C. Naha, Y. Liu, G. Hwang, Y. Huang, S. Gubara, V. Jonnakuti, A. Simon-soro, D. Kim, L. Gao, H. Koo and D. P. Cormode, Dextran-coated iron oxide nanoparticles as biomimetic catalysts for localized and pH-activated biofilm disruption, ACS Nano, 2019, 13, 4960–4971 CrossRef CAS PubMed.
- S. Fulaz, S. Vitale, L. Quinn and E. Casey, Nanoparticle – bio film interactions: the role of the EPS matrix, Trends Microbiol., 2019, 1–12 Search PubMed.
- H.-C. Flemming, EPS—then and now, Microorganisms, 2016, 4, 41 CrossRef PubMed.
- H. C. Flemming and J. Wingender, The biofilm matrix, Nat. Rev. Microbiol., 2010, 8, 623–633 CrossRef CAS PubMed.
- S. Satpathy, S. Kumar, S. Pattanaik and S. Raut, Review on bacterial bio fi lm: An universal cause of contamination, Biocatal. Agric. Biotechnol., 2016, 7, 56–66 CrossRef.
- K. Sauer, A. K. Camper, G. D. Ehrlich, J. W. Costerton and D. G. Davies, Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm, J. Bacteriol., 2002, 184, 1140–1154 CrossRef CAS PubMed.
- L. Gal and J. Guzzo, Biofilm-detached cells, a transition from a sessile to a planktonic phenotype: a comparative study of adhesion and physiological characteristics in Pseudomonas aeruginosa, FEMS Microbiol. Lett., 2009, 290, 135–142 Search PubMed.
- D. Sharma, L. Misba and A. U. Khan, Antibiotics versus biofilm: an emerging battleground in microbial communities, Antimicrob. Resist. Infect. Contr., 2019, 3, 1–10 Search PubMed.
- S. Fulaz, S. Vitale, L. Quinn and E. Casey, Nanoparticle–Biofilm Interactions: The Role of the EPS Matrix, Trends Microbiol., 2019, 1–12 Search PubMed.
- S. Galié, C. García-gutiérrez, E. M. Miguélez, C. J. Villar, F. Lombó and G. Di Bonaventura, Biofilms in the food Industry: Health aspects and control methods, Front. Microbiol., 2018, 9, 1–18 CrossRef PubMed.
- S. Veerachamy, T. Yarlagadda and G. Manivasagam, Bacterial adherence and biofilm formation on medical implants: A review, Proc. Inst. Mech. Eng., Part H, 2014, 228, 1083–1099 CrossRef PubMed.
- S. Garc, H. Elizondo-castillo, M. Arruebo, G. Mendoza and S. Irusta, Evaluation of the antimicrobial activity and cytotoxicity of different components of natural origin present in essential oils, Molecules, 2018, 8, 1–18 Search PubMed.
- K. B. Pandey and S. I. Rizvi, Plant polyphenols as dietary antioxidants in human health and disease, Oxid. Med. Cell. Longevity, 2009, 2, 270–278 CrossRef PubMed.
- H. Hatcher, R. Planalp, J. Cho, F. M. Torti and S. V. Torti, Curcumin: From ancient medicine to current clinical trials, Cell. Mol. Life Sci., 2008, 65, 1631–1652 CrossRef CAS PubMed.
- S. Z. Moghadamtousi, H. A. Kadir, P. Hassandarvish, H. Tajik, S. Abubakar and K. Zandi, A Review on antibacterial, antiviral, and antifungal activity of curcumin, BioMed Res. Int., 2014, 2014, 1–12 CrossRef PubMed.
- S. J. Hewlings, Curcumin: A review of its' effects on human health, Foods, 2017, 1–11 Search PubMed.
- R. Wilken, M. S. Veena, M. B. Wang and E. S. Srivatsan, Curcumin: A review of anti-cancer properties and therapeutic activity in head and neck squamous cell carcinoma, Mol. Cancer, 2011, 1–19 Search PubMed.
- P. Jourghanian, S. Ghaffari, M. Ardjmand, S. Haghighat and M. Mohammadnejad, Sustained Release Curcumin Loaded Solid Lipid Nanoparticles, Adv. Pharm. Bull., 2016, 6, 17–21 CrossRef CAS PubMed.
- S. A. Gaware, K. A. Rokade and S. N. Kale, Journal of Drug Delivery Science and Technology Silica-chitosan nanocomposite mediated pH-sensitive drug delivery, J. Drug Delivery Sci. Technol., 2019, 49, 345–351 CrossRef CAS.
- Z. Song, Y. Wu, H. Wang and H. Han, Synergistic antibacterial effects of curcumin modi fied silver nanoparticles through ROS-mediated pathways, Mater. Sci. Eng., C, 2019, 99, 255–263 CrossRef CAS PubMed.
- C. Liu, W. Lee and W. Wu, Photodynamic inactivation against Pseudomonas aeruginosa by curcumin microemulsions, RSC Adv., 2016, 6, 63013–63022 RSC.
- T. Huang, P. Chen and C. Lee, Curcumin nanoparticles are a promising anti-bacterial and anti-inflammatory agent for treating periprosthetic joint infections, Int. J. Nanomed., 2004, 14, 469–481 Search PubMed.
- R. K. Basniwal, H. S. Buttar, V. K. Jain and N. Jain, Curcumin nanoparticles: preparation, characterization, and antimicrobial study, J. Agric. Food Chem., 2011, 9, 2056–2061 Search PubMed.
- R. Dhivya, J. Ranjani, J. Rajendhran and M. Rajasekaran, pH responsive curcumin/ZnO nanocomposite for drug delivery, Adv. Mater. Lett., 2015, 6, 505–512 CrossRef CAS.
- M. Rajasekaran and J. Annaraj, A lucid build-up of nanostructured curcumin, quercetin and their interaction with DNA, J. Nanosci. Nanotechnol., 2014, 14, 4874–4879 CrossRef CAS PubMed.
- F. Odeh, H. Nsairat, W. Alshaer and S. Alsotari, Remote loading of curcumin-in-modified β-cyclodextrins into liposomes using a transmembrane pH gradient, RSC Adv., 2019, 9, 37148–37161 RSC.
- J. Annaraj, D. Raman, D. Kuberapandian and M. Rajasekaran, Studies on the enhanced biological applications of PVA loaded nanocurcumin, J. Nanosci. Nanotechnol., 2014, 2, 490–495 Search PubMed.
- I. Shlar, S. Droby, R. Choudhary and V. Rodov, The mode of antimicrobial action of curcumin depends on the delivery system: monolithic, RSC Adv., 2017, 7, 42559–42569 RSC.
- H. A. P. Bais, Curcumin, a known phenolic from Curcuma longa, attenuates the virulence of Pseudomonas aeruginosa PAO1 in whole plant and animal pathogenicity models, J. Agric. Food Chem., 2008, 56, 1955–1962 CrossRef PubMed.
- S. Sethupathy, K. Ganesh, S. Ananthi and S. Mahalingam, Proteomic analysis reveals modulation of iron homeostasis and oxidative stress response in Pseudomonas aeruginosa PAO1 by curcumin inhibiting quorum sensing regulated virulence factors and biofilm production, J. Proteomics, 2016, 145, 112–126 CrossRef CAS PubMed.
- V. Lopes-rodrigues, A. Oliveira, M. Correia-da-silva and M. Pinto, A novel curcumin derivative which inhibits P-glycoprotein, arrests cell cycle and induces apoptosis in multidrug resistance cells, Bioorg. Med. Chem., 2017, 25, 581–596 CrossRef CAS PubMed.
- C. Airoldi, C. Zona, E. Sironi, L. Colombo, M. Messa, D. Aurilia, M. Gregori, M. Masserini, M. Salmona, F. Nicotra and B. La, Curcumin derivatives as new ligands of Aβ peptides, J. Biotechnol., 2011, 156, 317–324 CrossRef CAS PubMed.
- A. ManusSheng Biao Wan, H. Yang, Z. Zhou, Q. Cindy Cui, D. Chen, J. Kanwar, I. Mohammad, Q. Ping Dou and T. H. Chancript, Evaluation of curcumin acetates and amino acid conjugates as proteasome inhibitors, Int. J. Mol. Med., 2012, 26, 447–455 Search PubMed.
- S. Peter, P. Oberhettinger, L. Schuele, A. Dinkelacker, W. Vogel, D. Dörfel, D. Bezdan, S. Ossowski, M. Marschal, J. Liese and M. Willmann, Genomic characterisation of clinical and environmental Pseudomonas putida group strains and determination of their role in the transfer of antimicrobial resistance genes to Pseudomonas aeruginosa, BMC Genomics, 2017, 18, 1–11 CrossRef PubMed.
- M. Klausen, M. Gjermansen, J. Kreft and T. Tolker-nielsen, Dynamics of development and dispersal in sessile microbial communities: examples from Pseudomonas aeruginosa and Pseudomonas putida model biofilms, FEMS Microbiol. Lett., 2006, 1, 1–11 CrossRef PubMed.
- W. Chang, M. Van De Mortel, L. Nielsen, G. N. De Guzman, X. Li and L. J. Halverson, Alginate production by Pseudomonas putida creates a hydrated microenvironment and contributes to biofilm architecture and stress tolerance under water-limiting conditions, J. Bacteriol., 2007, 189, 8290–8299 CrossRef CAS PubMed.
- D. N. Williams, S. H. Ehrman and T. R. P. Holoman, Evaluation of the microbial growth response to inorganic nanoparticles, J. Nanobiotechnol., 2006, 8, 1–8 Search PubMed.
- M. Cheng, In vivo toxicologic study of larger silica nanoparticles in mice, Int. J. Nanomed., 2017, 12, 3421–3432 CrossRef PubMed.
- H. Al-obaidi, S. M. King, M. J. Bowes, M. J. Lawrence, A. F. Drake, M. A. Green and P. J. Dobson, Fate of silica nanoparticles in simulated primary wastewater treatment, Environ. Sci. Technol., 2009, 43, 8622–8628 CrossRef PubMed.
- Z. Kong, H. Kuo, A. Johnson, L. Wu and K. L. B. Chang, Curcumin-loaded mesoporous silica nanoparticles markedly enhanced cytotoxicity in hepatocellular carcinoma cells, Int. J. Mol. Sci., 2019, 20 Search PubMed.
- C. Chen, W. Sun, X. Wang, Y. Wang and P. Wang, Rational design of curcumin loaded multifunctional mesoporous silica nanoparticles to enhance the cytotoxicity for targeted and controlled drug release, Mater. Sci. Eng., C, 2018, 85, 88–96 CrossRef CAS PubMed.
- N. S. Elbialy, S. Faisal, B. Fahad and A. Noorwali, Microporous and Mesoporous Materials Multifunctional curcumin-loaded mesoporous silica nanoparticles for cancer chemoprevention and therapy, Microporous Mesoporous Mater., 2020, 291, 1–11 CrossRef.
- R. K. Gangwar, G. B. Tomar, V. A. Dhumale, S. Zinjarde, R. B. Sharma and S. Datar, Curcumin conjugated silica nanoparticles for improving bioavailability and its anticancer applications, J. Agric. Food Chem., 2013, 61, 9632–9637 CAS.
- W. Stöber, A. Fink and E. Bohn, Controlled growth of monodisperse silica spheres in the micron size range, J. Colloid Interface Sci., 1968, 69, 62–69 CrossRef.
- M. Zanoni, O. Habimana, J. Amadio and E. Casey, Antifouling activity of enzyme-functionalized silica nanobeads, Biotechnol. Bioeng., 2016, 113, 501–512 CrossRef CAS PubMed.
- D. R. Hristov, L. Rocks, P. M. Kelly, S. S. Thomas, A. S. Pitek, P. Verderio, E. Mahon and K. A. Dawson, Tuning of nanoparticle biological functionality through controlled surface chemistry and characterisation at the bioconjugated nanoparticle surface, Sci. Rep., 2015, 5, 1–8 Search PubMed.
- M. Montalti, L. Prodi, E. Rampazzo and N. Zaccheroni, Dye-doped silica nanoparticles as luminescent organized systems for nanomedicine, Chem. Soc. Rev., 2014, 43, 4243–4268 RSC.
- F. Meder, S. S. Thomas, L. W. Fitzpatrick, A. Alahmari, S. Wang, J. G. Beirne, G. Vaz, G. Redmond and K. A. Dawson, Labeling the structural integrity of nanoparticles for advanced in situ Tracking in bionanotechnology, ACS Nano, 2016, 10, 4660–4671 CrossRef CAS PubMed.
- S. Fulaz, D. Hiebner, C. H. N. Barros, H. Devlin, S. Vitale, L. Quinn and E. Casey, Ratiometric Imaging of the in Situ pH Distribution of Biofilms by Use of Fluorescent Mesoporous Silica Nanosensors, ACS Appl. Mater. Interfaces, 2019, 11, 32679–32688 CrossRef CAS PubMed.
- J. Schindelin, I. Arganda-carreras, E. Frise, V. Kaynig, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J. Tinevez, D. J. White, V. Hartenstein, P. Tomancak and A. Cardona, Fiji - an Open Source platform for biological image analysis, Nat. Methods, 2012, 9, 676–682 CrossRef CAS PubMed.
- J. Wu and C. Xi, Evaluation of different methods for extracting extracellular DNA from the biofilm matrix, Appl. Environ. Microbiol., 2009, 75, 5390–5395 CrossRef CAS PubMed.
- O. H. Lowry, N. J. Rosebrough, A. L. Farr and R. J. Randall, Protein measurement with the Folin phenol reagent., J. Biol. Chem., 1951, 193, 265–275 CAS.
- M. Dubois, K. A. Gilles, J. K. Hamilton, P. A. Rebers and F. Smith, Colorimetric method for determination of sugars and related substances, Anal. Chem., 1956, 28, 350–356 CrossRef CAS.
- P. Tyagi, M. Singh, H. Kumari, A. Kumari and K. Mukhopadhyay, Bactericidal activity of curcumin I is associated with damaging of bacterial membrane, PLoS One, 2015, 1–15 Search PubMed.
- E. Y. Sukandar, N. F. Kurniati, K. Puspatriani and H. P. Adityas, Antibacterial activity of curcumin in combination with tetracycline against Staphylococcus aureus by disruption of cell wall, Res. J. Med. Plant, 2018, 12, 1–8 CAS.
- A. Kali, D. Bhuvanechwar, P. M. V. Charles and K. S. Seetha, Antibacterial synergy of curcumin with antibiotics against biofilm producing clinical bacterial isolates, J. Basic Clin. Pharm., 2016, 7, 93–96 CrossRef CAS PubMed.
- S. Mun, D. Joung, Y. Kim, O. Kang, S. Kim, Y. Seo, Y. Kim, D. Lee, D. Shin, K. Kweon and D. Kwon, Phytomedicine Synergistic antibacterial effect of curcumin against methicillin-resistant Staphylococcus aureus, European Journal of Integrative Medicine, 2013, 20, 714–718 CAS.
- J. Saising, S. Singdam, M. Ongsakul and S. P. Voravuthikunchai, Lipase, protease, and biofilm as the major virulence factors in staphylococci isolated from acne lesions, BioSci. Trends, 2012, 6, 160–164 CrossRef CAS PubMed.
- G. G. Willsey and M. J. Wargo, Extracellular lipase and protease production from a model drinking water bacterial community Is functionally robust to absence of individual members, Plos one, 2015, 1–15 Search PubMed.
- P. Tielen, H. Kuhn, F. Rosenau, K. Jaeger, H. Flemming and J. Wingender, Interaction between extracellular lipase LipA and the polysaccharide alginate of Pseudomonas aeruginosa, BMC Microbiol., 2013, 13, 1 CrossRef PubMed.
- H. Fatima, N. Khan, A. U. Rehman and Z. Hussain, Production and partial characterization of lipase from Pseudomonas putida, Ferment. Technol., 2014, 4, 1–7 Search PubMed.
- Y. Liu, Y. Ren, Y. Li, L. Su, Y. Zhang, F. Huang, J. Liu and H. J. Busscher, Nanocarriers with conjugated antimicrobials to eradicate pathogenic biofilms evaluated in murine in vivo and human ex vivo infection models, Acta Biomater., 2018, 79, 331–343 CrossRef CAS PubMed.
- M. Chen, J. Wei, S. Xie, X. Tao, Z. Zhang, P. Ran and X. Li, Bacterial biofilm destruction by size/surface charge-adaptive micelles, Nanoscale, 2019, 11, 1410–1422 RSC.
- J. J. Griese, R. P. Jakob, S. Schwarzinger and H. Dobbek, Xenobiotic reductase A in the degradation of quinoline by Pseudomonas putida 86: physiological function, structure and mechanism of 8-Hydroxycoumarin Reduction, J. Mol. Biol., 2006, 361, 140–152 CrossRef CAS PubMed.
- O. Spiegelhauer, F. Dickert, S. Mende, D. Niks, R. Hille, M. Ullmann and H. Dobbek, Kinetic Characterization of Xenobiotic Reductase A from Pseudomonas putida 86, Biochemistry, 2009, 48, 11412–11420 CrossRef CAS PubMed.
- A. Hassaninasab, Y. Hashimoto, K. Tomita-yokotani and M. Kobayashi, Discovery of the curcumin metabolic pathway involving a unique enzyme in an intestinal microorganism, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 6615–6620 CrossRef CAS PubMed.
- A. W. Confer and S. Ayalew, The OmpA family of proteins: Roles in bacterial pathogenesis and immunity, Vet. Microbiol., 2013, 163, 207–222 CrossRef CAS PubMed.
- D. Z. Hui and L. X. Lin, Characterization of outer membrane proteins of Escherichia coli in response to phenol stress, Curr. Microbiol., 2011, 62, 777–783 CrossRef PubMed.
- P. M. Santos, D. Benndorf and I. Sá-correia, Insights into Pseudomonas putida KT2440 response to phenol-induced stress by quantitative proteomics, Proteomics, 2004, 4, 2640–2652 CrossRef CAS PubMed.
- J. Sun, Z. Deng and A. Yan, Biochemical and Biophysical Bacterial multidrug efflux pumps: Mechanisms, physiology and pharmacological exploitations, Biochem. Biophys. Res. Commun., 2014, 453, 254–267 CrossRef CAS PubMed.
- K. Kim, S. Lee, K. Lee and D. Lim, Isolation and characterization of toluene-sensitive mutants from the toluene-resistant bacterium Pseudomonas putida GM73, J. Bacteriol., 1998, 180, 3692–3696 CrossRef CAS.
- A. Mercenier, J. Simon, C. Vander Wauven, D. Haas, V. Stalon, L. De Microbiologie, F. Sciences and U. L. De Bruxelles, Regulation of enzyme synthesis in the arginine deiminase pathway of Pseudomonas aeruginosa, J. Bacteriol., 1980, 144, 159–163 CrossRef CAS PubMed.
- T. Roger, M. Bhakoo and Z. Zhang, Bacterial adhesion and biofilms on surfaces, Prog. Nat. Sci., 2008, 18, 1049–1056 CrossRef.
- W. M. Dunne, Bacterial adhesion: Seen any good biofilms lately?, Clin. Microbiol. Rev., 2002, 15, 155–166 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0na00041h |
| This journal is © The Royal Society of Chemistry 2020 |