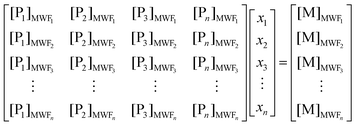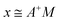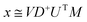Open Access Article
Open Access ArticleCharacterization of the Fe metalloproteome of a ubiquitous marine heterotroph, Pseudoalteromonas (BB2-AT2): multiple bacterioferritin copies enable significant Fe storage†
Michael G.
Mazzotta
 *,
Matthew R.
McIlvin
*,
Matthew R.
McIlvin
 and
Mak A.
Saito
and
Mak A.
Saito
 *
*
Marine Chemistry and Geochemistry Department, Woods Hole Oceanographic Institution, Woods Hole, Massachusetts, USA. E-mail: mgm@whoi.edu; msaito@whoi.edu
First published on 15th April 2020
Abstract
Fe is a critical nutrient to the marine biological pump, which is the process that exports photosynthetically fixed carbon in the upper ocean to the deep ocean. Fe limitation controls photosynthetic activity in major regions of the oceans, and the subsequent degradation of exported photosynthetic material is facilitated particularly by marine heterotrophic bacteria. Despite their importance in the carbon cycle and the scarcity of Fe in seawater, the Fe requirements, storage and cytosolic utilization of these marine heterotrophs has been less studied. Here, we characterized the Fe metallome of Pseudoalteromonas (BB2-AT2). We found that with two copies of bacterioferritin (Bfr), Pseudoalteromonas possesses substantial capacity for luxury uptake of Fe. Fe![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C in the whole cell metallome was estimated (assuming C
C in the whole cell metallome was estimated (assuming C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P stoichiometry ∼51
P stoichiometry ∼51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to be between ∼83 μmol
1) to be between ∼83 μmol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol Fe
mol Fe![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C, ∼11 fold higher than prior marine bacteria surveys. Under these replete conditions, other major cytosolic Fe-associated proteins were observed including superoxide dismutase (SodA; with other metal SOD isoforms absent under Fe replete conditions) and catalase (KatG) involved in reactive oxygen stress mitigation and aconitase (AcnB), succinate dehydrogenase (FrdB) and cytochromes (QcrA and Cyt1) involved in respiration. With the aid of singular value decomposition (SVD), we were able to computationally attribute peaks within the metallome to specific metalloprotein contributors. A putative Fe complex TonB transporter associated with the closely related Alteromonas bacterium was found to be abundant within the Pacific Ocean mesopelagic environment. Despite the extreme scarcity of Fe in seawater, the marine heterotroph Pseudoalteromonas has expansive Fe storage capacity and utilization strategies, implying that within detritus and sinking particles environments, there is significant opportunity for Fe acquisition. Together these results imply an evolved dedication of marine Pseudoalteromonas to maintaining an Fe metalloproteome, likely due to its dependence on Fe-based respiratory metabolism.
C, ∼11 fold higher than prior marine bacteria surveys. Under these replete conditions, other major cytosolic Fe-associated proteins were observed including superoxide dismutase (SodA; with other metal SOD isoforms absent under Fe replete conditions) and catalase (KatG) involved in reactive oxygen stress mitigation and aconitase (AcnB), succinate dehydrogenase (FrdB) and cytochromes (QcrA and Cyt1) involved in respiration. With the aid of singular value decomposition (SVD), we were able to computationally attribute peaks within the metallome to specific metalloprotein contributors. A putative Fe complex TonB transporter associated with the closely related Alteromonas bacterium was found to be abundant within the Pacific Ocean mesopelagic environment. Despite the extreme scarcity of Fe in seawater, the marine heterotroph Pseudoalteromonas has expansive Fe storage capacity and utilization strategies, implying that within detritus and sinking particles environments, there is significant opportunity for Fe acquisition. Together these results imply an evolved dedication of marine Pseudoalteromonas to maintaining an Fe metalloproteome, likely due to its dependence on Fe-based respiratory metabolism.
Significance to metallomicsFe is an essential nutrient to marine heterotrophic bacteria that catalyze the decay of biogenic particles derived from phototrophic organisms. Pseudoalteromonas associates with detrital matter and marine snow, making it an ideal model microbe for exploring biochemistries relevant to carbon export. We have used metalloproteomics to identify the Fe reservoirs within this bacterium, which includes two active copies of the bacterioferritin protein, enabling significant Fe storage, and contributing to ∼72% of the Fe metallome. Incorporation of Fe into reactive oxygen stress mitigation and respiratory enzymes was also identified. Together, our results indicate the substantial storage capacity for Fe within this marine heterotrophic bacterium, despite the scarcity of Fe in seawater. |
Introduction
Fe is an essential element to nearly all living organisms and limited in availability within the surface ocean due to the low solubility of Fe3+ in oxic seawater.1 Field incubation experiments have demonstrated that Fe limitation controls primary productivity in major regions of the oceans and in turn can affect the biological pump of carbon from the surface to the deep ocean.2–5 This biological pump removes carbon through the fixation of CO2 by marine phototrophs, many of which utilize sophisticated metalloproteins throughout their metabolism. The biogenic particles that result from these phototrophic organisms are then consumed by microbially mediated degradation and zooplanktonic members of the community, also facilitated by metalloenzymes, representing an additional and valuable component of carbon-cycling within the marine environment.6,7 Thus, understanding the microbial metalloenzymes that mediate these processes facilitates understanding global biogeochemical changes.8Mechanisms for sequestering scarce bioactive trace metals, such as Fe, are critical to carbon cycling.9,10 Fe limitation is known to be important in controlling photosynthetic activity in the epipelagic zone (0–200 m), but there is increasing interest in the challenges in Fe nutrition faced by heterotrophic bacteria in the epipelagic and mesopelagic zones (200–1000 m), where Fe sources can vary including, dust, sinking particles, coastal sediment and hydrothermal vents.2,11–13 Fe serves roles in catalysis and electron transport within autotrophic and heterotrophic microbes, however the partitioning of these trace metals into their respective metalloenzymes is a question outstanding.14,15 Notably, pelagic heterotrophic bacteria require greater quotas of Fe per unit biomass in comparison to phytoplankton.16–20 The strategies of heterotrophic Fe uptake have been probed at the molecular level, ascertaining the structural details of organism-specific siderophores,21–27 and additionally at the biological level in an effort to understand Fe transport processes and taxonomic specificity.28–31 Despite the understandings gained on the mechanisms of Fe uptake and transport, the marine heterotrophic bacterial cellular metallome remains underexplored.
Pseudoalteromonas is a ubiquitous marine heterotroph, a genus that was proposed within the γ-subgroup of the Proteobacteria class, isolated in both the open ocean and coastal marine environment.32 Numerous metagenomic–transcriptomic studies have characterized the cosmopolitan nature of this bacterium from global sampling campaigns.33,34 Observed as a constituent of phytoplankton blooms and sinking particles in field and mesocosm experiments,35–38 these organisms have been characterized as fast-growing copiotrophic bacteria adapted to organic-rich conditions, in contrast to oligotrophic microbes that grow under low substrate concentration with higher affinities for specific carbon substrates.39,40Alteromonadales have been isolated from the surface of particles within sediment traps and are often associated with detritus and other marine snow sources, making them an ideal model microbe for exploring biochemistries relevant to carbon export.41,42 Understanding how trace metals influence Pseudoalteromonas, including their organic matter degradation capabilities and their metalloenzyme reactivity and metal selectivity, is useful in understanding the biogeochemical constraints on these microbes.43–45
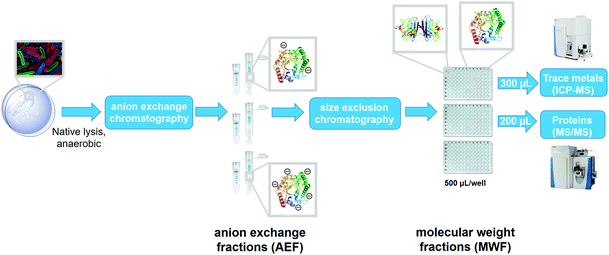
This study examines the connections between the biogeochemistry of marine heterotrophic bacteria and Fe through the investigation of the cellular metalloproteome of Pseudoalteromonas. This method employs two dimensions of chemical separation using anion exchange and size exclusion chromatography on cytosolic metalloproteins after native (non-denaturing) lysis methods (Fig. 1).46–48 The resulting 384 sub-samples of the purified lysate were characterized by proteomics and inductively coupled plasma-mass spectrometry (ICP-MS) to identify protein and elemental distributions within each sample, which together elucidates the major soluble metalloprotein reservoirs. This manuscript focuses on the Fe metallome of Pseudoalteromonas, while a future manuscript will describe the additional metals.
Experimental
Culturing of Pseudoalteromonas
Marine broth medium was prepared by microwave sterilization49 of 800 mL of 0.2 μm-filtered Vineyard Sound seawater with the addition of 0.2 μm-filter-sterilized solutions of 4 g peptone (Fisher Scientific) and 0.8 g yeast extract (BD Difco). Marine agar plates were prepared with the same composition as that of the marine broth described herein, with the inclusion of 12 g granulated agar (Fisher Scientific) and sterilization achieved by autoclaving for 30 min. Cultures of Pseudoalteromonas sp. BB2-AT2 were revived from a 20% glycerol stock of BB2-AT2 (provided by Kay Bidle, Rutgers University) and used to inoculate approximately 1 mL of marine broth. The inoculated solution was allowed to incubate at 23 °C for 3 hours, then streaked onto a marine agar plate with 50 μL inoculum and incubated at 23 °C overnight. Pellets of BB2-AT2 were grown in volume 500 mL of marine broth medium by inoculation at 23 °C with selection of a single colony from a marine agar plate. Growth curves were obtained with a SpectraMax Me5 unit (Molecular Devices) through absorbance measurements at 600 nm pipetting aliquots into a Corning clear 96 well plate at each time point. Samples were analyzed at 23 °C with a kinetic run over a period of 18 hours with a read interval of 30 minutes, with aliquots added at every timepoint. To validate this approach these plate reader growth rates were successfully intercompared with growth measurements using a Shimadzu UV-1601 spectrophotometer in a quartz cuvette (b = 1 cm) with a 2.5 mL aliquot of inoculated medium (ESI†). Pellets were harvested in mid-exponential phase by centrifugation at 8000 rpm for 20 minutes using an Eppendorf 5810R centrifuge at 3 °C, the solution was decanted, and the pellet washed with 0.2 μm-filtered seawater (4 mL × 3). Pellets of biomass were stored at −80 °C until chromatographic analysis was performed.Cellular metal quota analysis
The measured quotas reflect contributions of intracellular and extracellular metals. Biomass from triplicate 10 mL cultures were centrifuged at 8000 rpm for 20 minutes at 3 °C at mid-exponential phase of growth. The cell pellet was washed by resuspension in ∼1 mL filtered seawater, transferred to an acid-cleaned microfuge tube and centrifuged again for 15 min at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C. The whole cell pellet was digested in 800 μL of 5% trace metal grade HNO3 (Optima) containing 1 ppb Indium (In) then cells were resuspended multiple times. After digesting for 7 days at room temperature, solids were removed by centrifugation, and supernatants were collected for quota analysis. Process blank digestions containing acid, but no cells, were performed in parallel. Digests were diluted by a factor of 10 with additional 5% nitric acid (also containing 1 ppb In), transferred to acid-washed 96 deepwell plates (Thermo Scientific) before being analyzed on an ICAP-Q inductively coupled plasma-mass spectrometer (Thermo) with an SC-4 DX FAST autosampler (Elemental Scientific, Inc.). Metal concentrations were calibrated to a multi-element standard curve (Spex, Certiprep) over a range of 1–50 ppb. Phosphorus concentrations were also measured by ICPMS and calibrated to a separate standard curve ranging from 100–1500 ppb using a stock solution of Na3PO4. Samples were analyzed in KED mode using helium as a collision gas after a 45 s sample uptake window. Mass windows were scanned 3 times during measurements. In was used to correct for variation in sample delivery and plasma suppression between samples. Blanks were subtracted from measured concentrations. Technical replicates (n = 3, 3, 2) were averaged for each biological triplicates (ESI†). Averages of biological replicates and their standard deviation were then calculated. Cellular metal quota data can be accessed via https://www.bco-dmo.org/dataset/808598.
000 rpm at 4 °C. The whole cell pellet was digested in 800 μL of 5% trace metal grade HNO3 (Optima) containing 1 ppb Indium (In) then cells were resuspended multiple times. After digesting for 7 days at room temperature, solids were removed by centrifugation, and supernatants were collected for quota analysis. Process blank digestions containing acid, but no cells, were performed in parallel. Digests were diluted by a factor of 10 with additional 5% nitric acid (also containing 1 ppb In), transferred to acid-washed 96 deepwell plates (Thermo Scientific) before being analyzed on an ICAP-Q inductively coupled plasma-mass spectrometer (Thermo) with an SC-4 DX FAST autosampler (Elemental Scientific, Inc.). Metal concentrations were calibrated to a multi-element standard curve (Spex, Certiprep) over a range of 1–50 ppb. Phosphorus concentrations were also measured by ICPMS and calibrated to a separate standard curve ranging from 100–1500 ppb using a stock solution of Na3PO4. Samples were analyzed in KED mode using helium as a collision gas after a 45 s sample uptake window. Mass windows were scanned 3 times during measurements. In was used to correct for variation in sample delivery and plasma suppression between samples. Blanks were subtracted from measured concentrations. Technical replicates (n = 3, 3, 2) were averaged for each biological triplicates (ESI†). Averages of biological replicates and their standard deviation were then calculated. Cellular metal quota data can be accessed via https://www.bco-dmo.org/dataset/808598.
Metalloproteomic analysis
The genome of BB2-AT2 was downloaded from NCBI, sequenced by JCVI (https://www.ncbi.nlm.nih.gov/sra/?term=BB2AT2; accessed December 11, 2019); previously classified as Alteromonas, upon BLAST analysis more closely matches the taxon Pseudoalteromonas. For metalloproteomic analysis, all plasticware used was acid-washed unless otherwise mentioned. Buffers were prepared in Milli-Q water with Tris base (Fisher BioReagents) and NaCl (Sigma Aldrich, trace metal basis) with removal of extraneous metal contaminants from solution by Chelex prepared accordingly.50 The pellet of BB2-AT2 was suspended in approximately 6 mL of 50 mM Tris buffer (pH 8.8) under anaerobic conditions (<1 ppm O2) in an N2-filled anaerobic chamber (Coy Laboratory Products Inc.) using a Fisherbrand Model 120 sonic dismembrator, sonicated for 40 s at 2 s on/off intervals while on ice. The resulting solution was then diluted to approximately 30 mL with additional 50 mM Tris buffer (pH 8.8) and centrifuged at 4000 rpm at 3 °C for 15–20 min. The resulting supernatant was decanted and then purified by anion exchange chromatography using an Agilent 1100 series HPLC. A single-use GE HiTrap Q HP anion exchange column was conditioned prior to use by washing the column with 50 mM Tris buffer (pH 8.8) for 5 min at a flow rate of 1 mL min−1, then 50 mM Tris buffer containing 1 M sodium chloride (pH 8.8) for 5 min at a flow rate of 1 mL min−1 and finally with 50 mM Tris buffer (pH 8.8) for 5 min at a flow rate of 1 mL min−1. The supernatant was then loaded onto the column via a 60 mL loading syringe onto the column using Carter 12/8 cassette peristaltic pump at a rate of 0.5 mL min−1. The sample-loaded column was then connected to the Agilent HPLC system and eluted with a gradient of sodium chloride at a rate of 0.25 mL min−1. The method includes the following gradient- 50 mM Tris buffer (pH 8.8) for 5 minutes, followed by a linear increase from 100% 50 mM Tris buffer (pH 8.8) to 100% 50 mM Tris buffer containing 1 M NaCl (pH 8.8) over the course of 40 min and then 50 mM Tris buffer (pH 8.8, 1 M NaCl) for 8 min. Fractions were collected with a Bio-Rad 2110 fraction collector in 2 mL microtubes at a rate of 1 fraction every 4 min, and immediately stored on ice.Anion exchange fractions were concentrated prior to separation by size exclusion chromatography. 10 kDa molecular weight cutoff PES Vivaspin columns were washed with 0.5 mL (×2) Tris pH 8.8 buffer prior to use. Anion exchange fractions were loaded onto the Vivaspin columns and centrifuged at 5000 rpm for 30 min using an Eppendorf MiniSpin. 0.1 mL of each concentrated anion exchange fraction was injected into a Michrom BioResources, Inc. HPLC system containing a Tosoh Bioscience 5 μm particle size TSKgel G3000SWxl size exclusion column with guard column. Samples were eluted using an isocratic method of 50 mM Tris buffer (pH 7.5, 50 mM sodium chloride) at a flow rate of 0.5 mL min−1. Fractions from size exclusion chromatography were collected using a CTC Analytics HTC Pal autosampler system into an acid-washed 1.2 mL 96-well storage plate (Thermo Scientific), covered for storage until analysis with an acid-washed 96 cap sealing mat (Thermo Scientific).
For protein digestion, 90 μL of each sample was diluted with 96 μL 100 mM ammonium bicarbonate solution and 10 μL acetonitrile. Then 15 μL of a freshly-prepared 10 mM dithiothreitol solution was added to the samples and incubated at 70 °C for 30 min with shaking at 400 rpm. The samples were then allowed to cool to room temperature, and 30 μL freshly-prepared 20 mM iodoacetamide was added to each sample and allowed to incubate for 30 min in the dark. Following reduction and alkylation, 10 μL of a 0.03 μg μL−1 trypsin stock (Promega Gold) was added to each sample and then incubated for approximately 18 hours at 37 °C with shaking at 400 rpm. Samples were then analyzed by an LC-MS on a Q-Exactive (Thermo Scientific) after separation on a C18 column following a gradient of 2–95% acetonitrile over a 90 min gradient using a Michrom Paradigm MG4 liquid chromatography system. Samples were ionized by electrospray with 1800 mV potential. Proteomic analyses were used for peptide spectra matching using the SEQUEST algorithm within Proteome Discoverer (v. 2.1) and visualized in Scaffold (v. 4); protein threshold = 99.9%; minimum # peptides = 2; peptide threshold = 99%. All protein identifications were matched by sequence similarity to the specific hit result of BLAST search (Table 1).
| Protein | Exclusive unique peptides observed |
|---|---|
| Bfr 0668 | ILFLEGVPNMTK |
| IGTQNYHQAQMGE | |
| Bfr 0669 | VLANELVGINQYFLHAR |
| ILFLEGLPNLQDLGR | |
| SodA 2621 | LNGLIPGTEFENK |
| AVNNFGSSWTWLVK | |
| KatG 4151 | TGQCPVMHGSNSVISK |
| VNTDWWPNTLNLDILHQHDTK | |
| SNPYGTDFNYAEAFK | |
| AFMTDSQDWWPADWGHYGGLMIR | |
| FSPLNSWPDNASLDK | |
| THGNGK | |
| AEHLGPEPEAADVHEQGLGWNSTK | |
| NTVTSGLEGAWTTHPT | |
| SPAGAHQWEPVNIKEEDK | |
| PVDVEDPSIR | |
| FNDDFEYFSDVFARLAPQK | |
| DWQGNEPERLANVLNALEK | |
| FKAGLAK | |
| PISIADLIVLGGTAAVEQAAK | |
| GDFSVSPEELLLER | |
| SQLMGLTAPEMTVLFGGMR | |
| HGVFTDNVGTLSTDFFVNLTD | |
| AcnB 2248 | IIQSWADAEWFTNK |
| PAVAEK | |
| DGINPEK | |
| PGEVGPISQLEELKAK | |
| FrdB 2645 | YNPDVDNAPR |
| EGVCGSDGVNMNGK | |
| GTIVIRPLPGLPVVR | |
| VKPYLINDQPAAGER | |
| FLADSRDTATEER | |
| LADLDDAFSVFR | |
| QcrA 3485 | SAGAPVEVDISKLEPGQLIR |
| LEPGQLIR | |
| EHEGQLRDPQSQEPQQLESSTNDYR | |
| VEGTDDGFFCPCHGSK | |
| VFQSVPAPLNLEIPPYTFLDDTTILVGEEK | |
| Cyt1 3487 | EQLIFDGSK |
| VGGHILNAITK | |
| WFGAAPPDLTNVAR | |
| GSDWIYTYLK | |
| VVGTETDGKGELSVDEYDQAVR | |
| GELSVDEYDQAVR | |
| DLTNFLEYVGEPTR |
Methodological comparisons were conducted in the same experimental procedure as described, however variation of the method of lysis was explored. For bead-beating lysis, the pellet of cellular material was suspended in 2 mL Tris buffer (pH 8.8) and approximately 100 μL acid-washed glass beads (500 μm, Acros Organics). The pellet was lysed by 3–30 s pulses in an anaerobic environment with a MiniBeadBeater (BioSpec Products), each followed by cooling on ice for 5 min intervals. For freeze-grinding lysis, the cell pellet of was frozen under liquid nitrogen and ground into a fine powder with an acid-washed mortar and pestle. The disrupted cells were thawed under anaerobic conditions. Cytosolic metallome and metalloproteomic data can be accessed via https://www.bco-dmo.org/dataset/808610 and https://www.bco-dmo.org/dataset/808619, respectively.
Global metaproteomic analysis
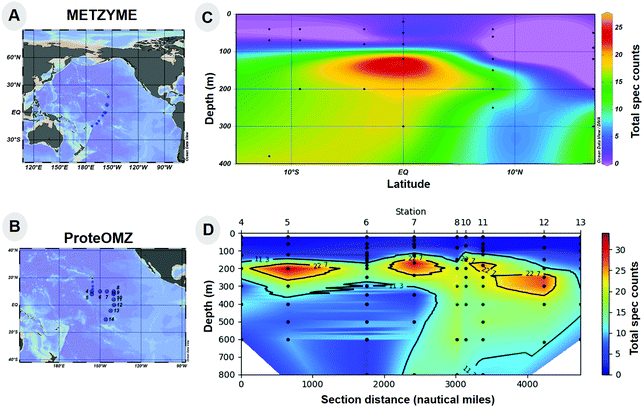
Marine protein samples were collected and analyzed from the METZYME expedition (KM1128) in October 2011 aboard the R/V Kilo Moana and on the ProteOMZ expedition (FK160115) on the R/V Falkor using extraction and analysis methods reported previously.51–54 While cyanobacterial functional proteins, two component regulatory systems and the nitrite oxidoreductase enzyme have been described within these metaproteomic datasets, the Pseudoalteromonas transporter described in this study has not been described. Paired metagenomic analyses were used for peptide spectra matching using the SEQUEST algorithm within Proteome Discoverer.51 Total spectral counts are displayed without normalization approaches.54 Map and protein section were prepared using Ocean Data View (version 5.0.0; Schlitzer, R., Ocean Data View, odv.awi.de, 2018) and the Ocean Protein Portal (https://proteinportal.whoi.edu/). METATRYP software and marine microbial tryptic peptide database (https://metatryp.whoi.edu/)55 was used to query the occurrence of tryptic peptides within the protein coding sequences predicted from genomes and metagenomes.Statistical approaches: linear regressions and singular value decomposition (SVD) analysis
Linear regressions were used to compare metalloproteome results where the metal concentration generated by ICP-MS were compared with proteomics data generated by Orbitrap LC-MS. Because multiple metalloproteins are present in the multi-dimensional dataset, subsets of data along one anion exchange fraction were used for linear regressions (e.g. comparing samples along the 600 mM fraction).An approach for reconstruction of multiple major metalloproteins and the metal distributions across fractions in 2D space was tested using singular value decomposition (SVD). The generalized form of the data matrix can be represented as the following:
Calculation of the singular variable decomposition (SVD), gives the following reduction of the data matrix, A. The pseudoinverse of the protein data matrix was computed through the SVD, and the product of the resultant matrix with that of the Fe data matrix yielded the proportionality constants of each protein within the Fe metalloproteome, effectively the solution of the system of linear equations. U is comprised of columns, composed by the eigenvectors of AAT, D are the singular values, defined as the square roots of eigenvalues from AAT or ATA, the eigenvectors of ATA comprise the columns of V.
| A = UDVT |
where the columns of U and V are orthonormal and the matrix, D, is diagonal with positive real entries.
Results and Discussion
To provide environmental context for the Pseudoalteromonas bacterium in this study, we examined the distribution of a putative TonB Fe acquisition protein attributed to the closely related Alteromonas bacterium in metaproteomic samples from the Central Equatorial Pacific Ocean (Expedition KM1128 METZYME October 2011). This expedition followed a transect from 17°N to 15°S, near the Hawaiian Islands (154.4°W) to the Tonga-Fiji region (173.1°W). A TonB-dependent family protein with the KEGG description Fe complex outermembrane receptor protein (KEGG Orthology, KO2014) was observed in the mesopelagic of this region as depicted in Fig. 2. Taxonomic classification of specific peptides identified by metaproteomics was conducted using the METATRYP least common ancestor analysis (LCA) with all peptides being specific to either the phylum Proteobacteria, and many to the order Alteromonadales or species Alteromonas (Table 2). The chemical structure of Fe complexes used by this oceanic Alteromonas instance of the KO2014 transport system is to our knowledge not yet known, but may be siderophores.57,58 A similarity search of this KO2014 transport protein to the sequence of the ferric enterobactin receptor of BB2-AT2 (BB2AT2_1746) produced specific hits to the CirA protein family, providing additional indication that this TonB system may be responsible for siderophore uptake (ESI†). Metaproteomic data showed these organisms occupy a broad swath of the mesopelagic, ranging from 100–600 m, where the exported biogenic material from the euphotic zone occurs.59,60 A maximum abundance of this Fe complex outermembrane protein was observed at the equator where increased export of organic matter arises from higher productivity associated with equatorial upwelling. These proteins were observed in the free-living bacterial size class (filter size range 0.2–3.0 μm), although it is also likely that these observations also reflect bacteria associated with fragile particles were released during filtration onto the 51 μm prefilter and caught on the downstream smaller 0.2 μm bacterial size during filtration. Together, these results demonstrate Alteromonas is widely distributed in the mesopelagic Pacific Ocean and produces significant quantities of putative TonB Fe complex transporter, implying cellular resources being expended on Fe acquisition.| Peptide | Lowest common ancestor | Genome returned | Metagenome returned |
|---|---|---|---|
| APTPGQANISNISTVADAGGDLYQK | Alteromonadaceae | 0 | 7 |
| GTLAPTNPVSVYYGGK | Alteromonadaceae | 0 | 7 |
| GGVFSTDGGATR | Alteromonadaceae | 0 | 6 |
| LVGDVAEATAGAAR | Alteromonas | 0 | 5 |
| YEDFDTFGSTSNSK | Gammaproteobacteria | 0 | 8 |
| EEQFTVISGNK | Alteromonadaceae | 0 | 6 |
| NVLGGFFFR | Undetermined | 0 | 13 |
| TCPVVPVPASGAGSSTDTALAAIK | Alteromonadaceae | 0 | 6 |
| GAVISFLGSGISDGAQGPDISAIPSIALK | Undetermined | 0 | 20 |
| DGASAQYGSDAIAGIMNFVLR | Alteromonadaceae | 0 | 8 |
| ITQGDDISITAADIALLQSLGVPGAGDLTNFR | Alteromonas | 0 | 4 |
| ADAQGLYDGGNTAIWNYPEPAQIWGSPDVSDDYK | Alteromonadaceae | 0 | 4 |
| DNSEGTQLEIR | Alteromonas | 0 | 4 |
Cellular metal quota and stoichiometry of Pseudoalteromonas (BB2-AT2)
The cellular metal quotas of Pseudoalteromonas BB2-AT2 were quantified by acid digestion of whole cells and ICP-MS analysis (Table 3). Approximately a third of the total metal abundance within this organism's metallome was devoted to Fe (31%), and approximately half was devoted to Zn (48%). Other metal constituents contributed the reminder of the cellular metallome with Mn at 9%, Ni at 6%, Cu at 4%, Mo at 0.6% and Co at 0.4%.| Metal | Cellular metal quota | Cytosolic metalloproteome | Cellular metal![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P (mmol metal P (mmol metal![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol P) mol P) |
Cytosolic metal![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C (mmol metal C (mmol metal![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol C) mol C) |
|---|---|---|---|---|
| Fe | 31.4 ± 0.83% | 39.5 ± 10.7% | 4.27 ± 0.18 | 83.8 ± 0.61 |
| Zn | 47.6 ± 1.40% | 18.6 ± 4.70% | 6.47 ± 0.08 | 126.8 ± 1.52 |
| Mn | 9.58 ± 0.12% | 4.59 ± 1.80% | 1.30 ± 0.03 | 25.5 ± 0.61 |
| Co | 0.43 ± 0.05% | 0.32 ± 0.02% | 0.06 ± 0.01 | 1.16 ± 0.10 |
| Cu | 3.71 ± 0.12% | 1.52 ± 2.95% | 0.50 ± 0.01 | 9.88 ± 0.10 |
| Ni | 6.72 ± 0.54% | 35.0 ± 8.37% | 0.92 ± 0.10 | 18.0 ± 1.87 |
| Mo | 0.57 ± 0.03% | 0.45 ± 0.03% | 0.08 ± 0.02 | 1.51 ± 0.04 |
In contrast to the cellular metal quota, the metalloproteomic separation method releases the soluble metal pools by native extraction, removal of the particulate cell debris by centrifugation, anion exchange chromatography, followed by a 10 kDa molecular weight cutoff, size exclusion chromatography and metal analysis of the resulting supernatants. Combining the sum of these metal fractions can be used to assess the soluble metallome, relative to the total metal quota. The total and soluble metallomes were largely consistent with the exceptions of Zn and Ni (Table 3). The variations between the whole cell and soluble metallome likely reflects the additional contribution of metals associated with the cell surface adsorption, low molecular weight compounds below 10 kDa and membrane proteins. Zn associated with membrane proteins, <10 kDa compounds and extracellular adsorption are likely lost during chromatographic separation The prevalence of Ni within the soluble metalloproteome implies presence of soluble Ni metalloproteins and minimal membrane and extracellular adsorption.
The metal stoichiometry relative to P (also measured by ICPMS) within Pseudoalteromonas was calculated for the whole cellular metallome (Table 3). In the cellular metallome, the Zn![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ratio is the highest amongst all of these trace metal micronutrients compared, followed by Fe, Mn and Ni. Prior studies of coastal Pseudoalteromonas strains (Pseudoalteromonas sp., strain PAtl-P2) measured C
P ratio is the highest amongst all of these trace metal micronutrients compared, followed by Fe, Mn and Ni. Prior studies of coastal Pseudoalteromonas strains (Pseudoalteromonas sp., strain PAtl-P2) measured C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ratios of ∼51
P ratios of ∼51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1,61 significantly below canonical Redfield ratios of 106
1,61 significantly below canonical Redfield ratios of 106![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.62 Using this value as an estimate of the Fe
1.62 Using this value as an estimate of the Fe![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C for Pseudoalteromonas, Fe
C for Pseudoalteromonas, Fe![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C was estimated to be ∼84 μmol
C was estimated to be ∼84 μmol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol Fe
mol Fe![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C, which is ∼11 fold higher than measured in a survey of five marine bacteria (7.5 μmol
C, which is ∼11 fold higher than measured in a survey of five marine bacteria (7.5 μmol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol Fe
mol Fe![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C16,18). Concentrations of P in the cytosolic metalloproteomic fractions were too low to quantitiate relative to the blank, likely reflective of the removal of the phospholipid bilayer, and hence metal: P values for soluble metallome were not calculated. Since this study was conducted in rich organic media compared to prior studies that used low metal concentrations in the media, the stoichiometry presented here also examines the capacity for luxury uptake into cellular storage mechanisms. To our knowledge, there is little metal stoichiometric data to compare for the other metals to for marine heterotrophic bacteria at this time.
C16,18). Concentrations of P in the cytosolic metalloproteomic fractions were too low to quantitiate relative to the blank, likely reflective of the removal of the phospholipid bilayer, and hence metal: P values for soluble metallome were not calculated. Since this study was conducted in rich organic media compared to prior studies that used low metal concentrations in the media, the stoichiometry presented here also examines the capacity for luxury uptake into cellular storage mechanisms. To our knowledge, there is little metal stoichiometric data to compare for the other metals to for marine heterotrophic bacteria at this time.
Cytosolic Fe is localized within bacterioferritin, reactive oxygen stress mitigation and respiration proteins
The Fe metallome of Pseudoalteromonas was comprised of several abundant Fe metalloproteins including the bacterioferritin (Bfr) Fe storage protein, the reactive oxygen stress mitigation proteins, superoxide dismutase (SodA) and catalase (KatG), and the aconitate hydratase (AcnB), the Fe–S cluster subunit of succinate dehydrogenase (FrdB), the ubiquinol-cytochrome c reductase, Fe–S subunit (QcrA) and cytochrome c1 (Cyt1), all involved in respiration (Table 4).| Gene number | Gene | Protein | Anion exchange fraction (mM NaCl) | Molecular weight fraction range | [protein]max location |
|---|---|---|---|---|---|
| BB2AT2_0668 | bfr | Bfr | 450 | 4–8 | 6 |
| BB2AT2_0669 | bfr | Bfr | 450 | 4–8 | 6 |
| BB2AT2_2621 | sodA | SodA | 450 | 9–12 | 10 |
| BB2AT2_4151 | katG | KatG | 450 | 4–13 | 8 |
| BB2AT2_2248 | acnB | AcnB | 450–500 | 4–16 | 10 |
| BB2AT2_2645 | frdB | FrdB | 450 | 4–14 | 11 |
| BB2AT2_3485 | qcrA | QcrA | 300 | 11–14 | 12 |
| BB2AT2_3487 | cyt1 | Cyt1 | 250 | 3–5 | 4 |
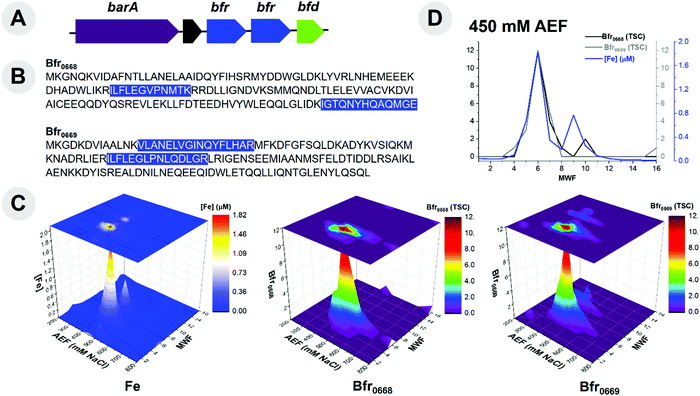
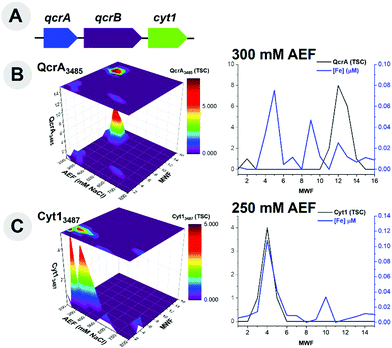
The major reservoir of Fe within this organism was within the Fe storage proteins. Two bacterioferritin genes (bfr) are present within this organism's genome. Notably, the bfr genes have distinct sequences and are localized in one operon (Fig. 3A). Proteomic analysis was able to distinguish between these Bfr protein copies (BB2AT2_0668 and BB2AT2_0669) through tryptic peptides that were unique to each copy (Fig. 3B), indicating that both copies were in active use. The distributions of Fe and Bfr protein along the 450 mM NaCl anion exchange fraction metalloproteome samples (where the majority of this protein eluted) were well correlated (r2 values of 0.87 and 0.91 for Bfr0668 and Bfr0669, respectively; Fig. 3C). These proteins were observed in qualitatively equal abundance and contained the majority of cytosolic Fe, consistent with observations of an elevated Fe cellular quota as described above. Their similar size (18 and 19 kDa) resulted in their elution in similar molecular weight fractions within the 450 mM anion exchange fraction (Fig. 3D). The relatively low total spectral counts of the Bfr proteins implies a high stoichiometry of Fe![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) protein, and future targeted proteomics efforts could determine the precise stoichiometry. Bacterioferritin functions as an Fe cage, encapsulating Fe(OH)3(s) and minimizing the pool of labile Fe within the cell, thereby preventing Fe from forming Fenton ions and their deleterious oxidative reactions to DNA and other biomolecules.63Pseudoalteromonas appear to bear some similarity to the pathogen, Pseudomonas aeruginosa, which also bears two distinct bacterioferritin proteins of comparable molecular weight (∼18 kDa) containing approximately 520 ± 20 Fe atoms per molecule of bacterioferritin.64 The gene neighborhood this pair of bacterioferritins reside in contains other potential proteins related to Fe homeostasis and regulation such as the hybrid signal transduction histidine kinase barA (BB2AT2_0666) and a hypothetical protein (BB2AT2_0667) preceded these bfr genes, but were not observed in the metalloproteome of this organism. Histidine kinases are localized to the membrane and hence were likely not extracted by the native approach used.65 Also proceeding the bfr genes within the genome is a putative bacterioferritin-associated ferredoxin (BB2AT2_0670), bfd, which is likely a motif employed to mobilize Fe from the bacterioferritin to the cellular contents.66,67 This gene cluster and the abundance and duplication of active Bfr proteins demonstrates the importance and capabilities for Fe storage within the Pseudoalteromonas cellular metalloproteome.
protein, and future targeted proteomics efforts could determine the precise stoichiometry. Bacterioferritin functions as an Fe cage, encapsulating Fe(OH)3(s) and minimizing the pool of labile Fe within the cell, thereby preventing Fe from forming Fenton ions and their deleterious oxidative reactions to DNA and other biomolecules.63Pseudoalteromonas appear to bear some similarity to the pathogen, Pseudomonas aeruginosa, which also bears two distinct bacterioferritin proteins of comparable molecular weight (∼18 kDa) containing approximately 520 ± 20 Fe atoms per molecule of bacterioferritin.64 The gene neighborhood this pair of bacterioferritins reside in contains other potential proteins related to Fe homeostasis and regulation such as the hybrid signal transduction histidine kinase barA (BB2AT2_0666) and a hypothetical protein (BB2AT2_0667) preceded these bfr genes, but were not observed in the metalloproteome of this organism. Histidine kinases are localized to the membrane and hence were likely not extracted by the native approach used.65 Also proceeding the bfr genes within the genome is a putative bacterioferritin-associated ferredoxin (BB2AT2_0670), bfd, which is likely a motif employed to mobilize Fe from the bacterioferritin to the cellular contents.66,67 This gene cluster and the abundance and duplication of active Bfr proteins demonstrates the importance and capabilities for Fe storage within the Pseudoalteromonas cellular metalloproteome.
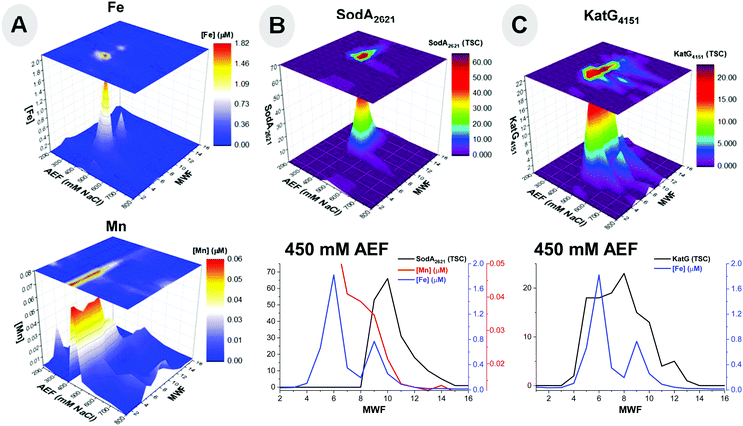
An abundant superoxide dismutase SodA (BB2AT2_2621) from the Fe superoxide dismutase superfamily was observed with over 60 total spectral counts in the metalloproteome. In contrast, SodC (BB2AT2_1013) from the Cu/Zn superoxide dismutase superfamily and SodN (BB2AT2_2930) in the Ni superoxide dismutase superfamily were not observed in this metalloproteomic analysis. Preferential utilization of this Fe-based SodA by BB2-AT2 is noteworthy, particularly in light of this organism's accumulation of Ni within the cytosol (35.0 ± 8.37%; Table 1), and may imply this microbe's preferred use of SodA under replete nutrient conditions as in our media. SodA co-elutes with both Fe and Mn within the cytosol (Fig. 4A and B). Fe and Mn superoxide dismutase isoforms are difficult to distinguish solely by sequence analysis, where the putative binding residues include His 27, His 74, His 162, and a third residue, likely to be Asp.48,68 In BB2-AT2, the SodA has a higher degree of correlation with Fe (r2 = 0.67) than with that of Mn (r2 = 0.38), and Fe is more than 10-fold more abundant than Mn at the SodA maxima (Fig. 4), implying Fe likely dominates its metal use, although the presence of co-eluting Fe-metalloenzyme KatG and AcnB peaks (see below) makes this difficult to be certain of. Additional purification and/or growth of the organism under Fe-depleted, Mn-replete conditions could further confirm the metal use within SodA.
Catalase-peroxidase, KatG, was also present as a defense mechanism against reactive oxygen stress within the metallome of Pseudoalteromonas (BB2AT2_4151). The distributions of catalase-peroxidase and Fe showed an overlap in the elution profiles. Examining the 450 mM anion exchange fraction, where the major elution band of this protein is observed, coelution between Fe and KatG was observed (Fig. 4C). Comparison of the Fe and protein distributions along the 450 mM NaCl anion exchange fraction, where the majority of this protein eluted, reveals a correlation of 0.63 (r2) for KatG. The correlation may have been affected by the broad KatG elution profile (fractions 4–13) and its potential relationship with the smaller of two Fe peaks. Ferrochelatase, hemH, an enzyme tasked with catalyzing the terminal step in the biosynthesis of heme, a critical step in the formation of a functional KatG enzyme, was present in the genome (BB2AT2_3756), however not observed in the proteome assembled from all metalloproteome samples.
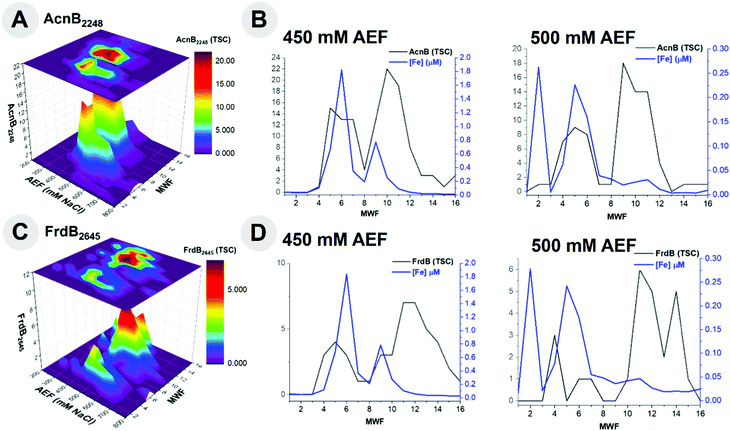
Aconitate hydratase 2 (AcnB, BB2AT2_2248) is a key respiratory protein within heterotrophic bacteria and an additional metalloprotein observed to coelute with Fe in this study. The major distribution of AcnB was observed to be most abundant in the 450 and 500 mM NaCl anion exchange fractions of the metalloproteome of BB2-AT2, consistent with the substantial distribution of Fe (Fig. 5A). Upon further observation of this 450 mM NaCl fraction containing the greatest abundance of AcnB, there is noteworthy overlap between these two distributions (r2 = 0.46, Fig. 5B). Comparison of the 500 mM NaCl anion exchange fraction of AcnB also yields notable overlap between the Fe and protein distributions (centered on fraction 5), and another region of minimal overlap, perhaps indicative of the apo form of this protein (centered on fraction 9–10); due to these multiple AcnB protein peaks the correlation between these metal and protein fractions at 500 mM NaCl yields minimal correlation, r2 = 0.11.
The Fe–S cluster subunit of succinate dehydrogenase (FrdB, BB2AT2_2645) is a respiratory protein complex observed to coelute with Fe (Fig. 5C). Primarily localized along the 450 and 500 mM NaCl anion exchange fractions, this protein separated amongst a variety of molecular weight fractions and coelutes with both the smaller and larger Fe peaks in the 450 mM anion exchange fraction (r2 = 0.56, Fig. 5D). Comparison of the 500 mM NaCl anion exchange fraction of FrdB also yields notable overlap between the Fe and protein distributions (fraction 4, 6–7), and a region of minimal overlap (fractions 11–14), perhaps indicative of either the monomeric or apo form of this protein; the correlation between these metal and protein fractions at 500 mM NaCl yields minimal and negative correlation, r2 = −0.27, likely due to the poorly separated FrdB from the other Fe metalloproteins described above. The two Fe associated FrdB peaks in the 450 and 500 mM fractions appeared to be shouldered under the larger signal associated with the AcnB described above, reflecting their relative contribution to the metallome.
Additional respiratory proteins within the cytochrome family of proteins, were observed within this metalloproteomic analysis; it is apparent that these peripheral membrane proteins can be stripped from the membrane easily with native lysis conditions. Three cytochrome genes (qcrA, qcrB and cyt1) are present within this organism's genome, localized within one operon (Fig. 6A). The ubiquinol-cytochrome c reductase, Fe–sulfur subunit (QcrA, BB2AT2_2645), a component of the electron transport chain, was identified to coelute with the distribution of Fe at the 300 mM NaCl anion exchange fraction (Fig. 6B). Along this 300 mM NaCl anion exchange fraction, QcrA produced an r2 = 0.59. Cytochrome c1 (Cyt1, BB2AT2_3487) was also identified via metalloproteomics to coelute with Fe, with the majority of the abundance of this protein separated along the 250 mM NaCl anion exchange fraction (Fig. 6C), with an r2 = 0.93. These respiratory cytochrome proteins were well detected within the Fe metalloproteome, despite the relatively small Fe demand within each of them due to their good chromatographic separation from other major Fe proteins.
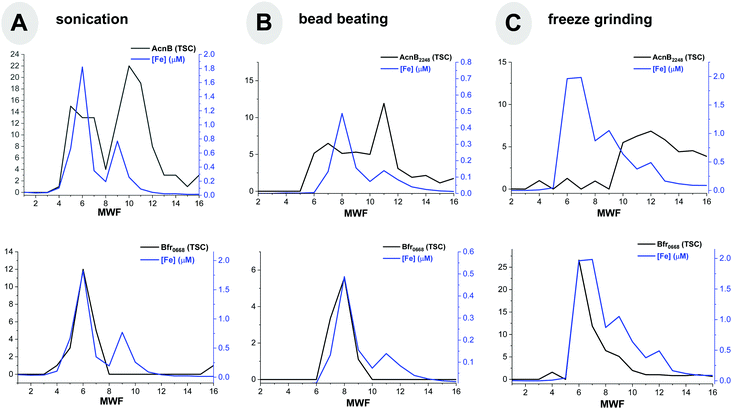
Comparison of cell disruption methods
In order to further optimize the metalloproteome methodology, we compared several methods of cell disruption, summarized in Table 5. A major concern in the analysis of metalloproteins is the denaturation and subsequent loss of metal, which would negate the ability to determine metal–protein relationships. In order to explore the potential for denaturation in future studies, we examined three methods of cell disruption: sonication, bead beating and freeze-grinding lysis. In each case, the use of detergents was avoided in order to prevent metal–protein dissociation. While sonication is performed on ice to minimize denaturation associated with heating, we were concerned about the energy input of sonication, which has the potential to shear DNA and could influence more labile metal–protein interactions. While all three methods were able to identify hundreds of soluble proteins from cultures of Pseudoalteromonas, the total number of proteins (defined here as the sum of all protein total spectral counts obtained from the entire metalloproteomic separation) obtained from freeze-grinding lysis was >30% higher than sonication and bead beating, indicating a more effective extraction of proteins from the cell. Comparison of metalloproteomic chromatographic separation of aconitase and bacterioferritin proteins showed reasonable reproducibility, particularly for the latter although bacterioferritin is known to be heat-stable69 (Fig. 7). It is unclear why the distributions of aconitase appear somewhat different, and could be attributed to the loss of the Fe–S cluster, resulting in the apo form of this protein; the observation of a single, broad distribution of aconitase in the freeze grinding data might indicate minimal demetallation with this methodology. From this comparison, it appears all three methods are capable of producing metalloproteomes, but that freeze-grinding lysis yields a greater number of proteins, and hence could provide a more comprehensive analysis.| Cell disruption method | Σ of metalloproteome proteins identified |
|---|---|
| Sonication | 565 |
| Bead beating | 526 |
| Freeze-grinding lysis | 744 |
Singular variable decomposition analysis of metalloproteomic data
The native separation employed in this metalloproteomic method can result in metal peaks comprised of multiple metalloproteins due to overlap in their elution profiles. Peak shouldering interferes with regression analyses in the less abundant metalloprotein (or with lower metal: protein stoichiometry). As a means of computationally recreating metal profiles from protein distributions, we employed singular variable decomposition (SVD), as described in the methods. To model the relative contributions of metalloproteins to the soluble metal abundances, a low-rank data matrix from selected proteins reconstructed the distribution of Fe. Applied to the metalloproteome, the total metal concentration (in μM), [M], is equal to the sum of the individual protein fractions (P1, P2, P3… Pn), represented in total spectral counts, with a proportionality constant associated with each protein fraction (x1, x2, x3… xn):| [M] = x1[P1] + x2[P2] + x3[P3]… + xn[Pn] | (1) |
These proportionality constants represent the aggregate ionization efficiency of peptides from each protein identified and their metal stoichiometry, and their estimation by SVD. The solution to the values of these proportionality constants is obtained by solving the nonhomogeneous system of linear equations, which can be represented in the general form,
| [M] = [P]x | (2) |
| [Fe] = x1[Bfr0668] + x2[Bfr0669] + x3[SodA2621] + x4[KatG4151] + x5[AcnB2248] + x6[Frd2645] + x7[QcrA3485] + x8[Cyt13487] | (3) |
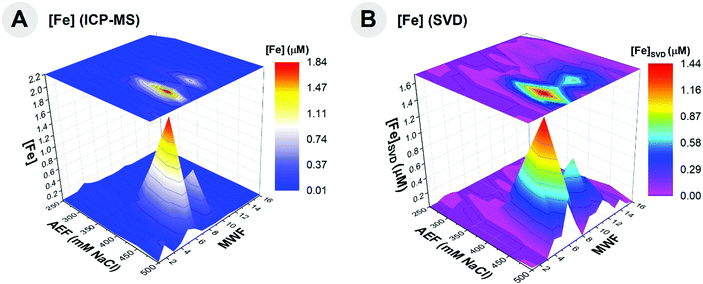
The proportionality constants (x1–x8) were solved for using SVD and eqn (3) for 94 samples where Fe metalloproteins were observed (Table 6). Substituting the protein spectral count data into eqn (3) with the solved constants, a prediction of the Fe metallome from protein data was produced. A comparison of the Fe distribution from ICP-MS distribution and that of the SVD-calculated representation of the Fe distribution indicates good agreement (Fig. 8), with an r2 of 0.84. This method provides a quantitative assessment of the relative influence of metalloproteins to the metalloproteome of the organism, where Bfr0668 and Bfr0669 appear to majorly contribute, based on the magnitude of the singular value (Table 6), and relatively equal contributions of 38.2 and 33.6%, respectively. KatG4151 appears to contribute to the Fe metalloproteome at 18.1%, while SodA2621, AcnB2248, QcrA3485, Cyt13487, and FrdB2645 have minor contributions with 3.5, 3.2, 1.6, 1.6% and a negative value, respectively. SVD appears to be a useful approach to modeling the major features of the metalloproteomic data from the protein total spectral counts. Future applications of SVD on cultures grown on low Fe medium to reduce the dominance of the bacterioferritins would be useful, as well as its application to other metals.70
| Protein | SVD contribution, xn | % contribution |
|---|---|---|
| a FrdB returned a negative constant, likely due to its low abundance and small contribution to the metallome and was not included in the % contribution column. | ||
| Bfr0668 | 0.042 | 38.2 |
| Bfr0669 | 0.037 | 33.6 |
| SodA2621 | 0.004 | 3.5 |
| KatG4151 | 0.020 | 18.1 |
| AcnB2248 | 0.003 | 3.2 |
| FrdB2645 | −0.031a | n.a.a |
| QcrA3485 | 0.0018 | 1.6 |
| Cyt13487 | 0.0018 | 1.6 |
We investigated the cellular quotas and intracellular metal reservoirs of Fe in the abundant marine bacterium Pseudoalteromonas grown under replete culture conditions using metalloproteomic approaches. Despite the scarcity of Fe in marine environments caused by the low solubility of Fe, Pseudoalteromonas demonstrated an impressive ability to accumulate and use Fe within its metalloproteins. These high Fe quotas and Fe metalloprotein used could reflect a dedication to Fe due to its use in the key respiratory enzyme aconitase as well as specialized abilities to acquire it from metal-rich detrital particulate material as described in Fig. 2. Future metalloproteomic studies of cultures grown on low Fe medium to reduce the dominance of the bacterioferritins and potential replacement of FeSOD would be useful to compare to these replete observations. In addition, the study of the Pseudoalteromonas metalloproteome composition with regards to other metals will be valuable.70 This study improves our understanding of the biogeochemistry of marine heterotrophic bacteria by characterizing the contribution of specific metalloproteins to the cellular Fe quota of Pseudoalteromonas and demonstrating the molecular basis for its impressive Fe storage capabilities using bacterioferritin.
Conclusions
Copiotrophs, such as the ubiquitous marine heterotroph, Pseudoalteromonas, appear well-equipped at storing Fe, showcasing an adaptation that could make them competitive for nutrient uptake relative to oligotrophs. Fe![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C in the whole cell metallome was estimated (assuming C
C in the whole cell metallome was estimated (assuming C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P stoichiometry ∼51
P stoichiometry ∼51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to be between ∼83 μmol
1) to be between ∼83 μmol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol Fe
mol Fe![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C, ∼11 fold higher than prior marine bacteria surveys. This substantial Fe quota within Pseudoalteromonas was attributed to luxury uptake, mediated by two active copies of bacterioferritin proteins that together contributed ∼72% of the Fe metallome identified. Catalytic functions have Fe prioritized amongst a small subset of proteins devoted to destruction of reactive oxygen species, such as superoxide dismutase (SodA) and catalase/peroxidase (KatG). Fe was also observed to coelute with AcnB, consistent with its role in catalyzing the isomerization of citrate within the tricarboxylic acid cycle. FrdB, which possesses roles in both respiration and in the oxidation of succinate within the tricarboxylic acid cycle, was also found to coelute with Fe. Respiratory cytochromes (QcrA and Cyt1) were also observed within the Fe metallome Pseudoalteromonas. Three methods of cell disruption were compared for the Fe metalloproteome of BB2-AT2, including sonication, bead beating, and freeze grinding. Elution of Fe-associated proteins were relatively consistent amongst methods and the total number of proteins extracted was greatest from freeze grinding. SVD was used to model the major features of the metalloproteomic data. An Fe complex TonB transporter associated with the closely related Alteromonas bacterium was found to be abundant within the Pacific Ocean mesopelagic environment. This study improves our understanding of the biogeochemistry of marine heterotrophic bacteria by characterizing the contribution of specific metalloproteins to the cellular Fe quota of Pseudoalteromonas and demonstrating the molecular basis for its impressive Fe storage capabilities using bacterioferritin.
C, ∼11 fold higher than prior marine bacteria surveys. This substantial Fe quota within Pseudoalteromonas was attributed to luxury uptake, mediated by two active copies of bacterioferritin proteins that together contributed ∼72% of the Fe metallome identified. Catalytic functions have Fe prioritized amongst a small subset of proteins devoted to destruction of reactive oxygen species, such as superoxide dismutase (SodA) and catalase/peroxidase (KatG). Fe was also observed to coelute with AcnB, consistent with its role in catalyzing the isomerization of citrate within the tricarboxylic acid cycle. FrdB, which possesses roles in both respiration and in the oxidation of succinate within the tricarboxylic acid cycle, was also found to coelute with Fe. Respiratory cytochromes (QcrA and Cyt1) were also observed within the Fe metallome Pseudoalteromonas. Three methods of cell disruption were compared for the Fe metalloproteome of BB2-AT2, including sonication, bead beating, and freeze grinding. Elution of Fe-associated proteins were relatively consistent amongst methods and the total number of proteins extracted was greatest from freeze grinding. SVD was used to model the major features of the metalloproteomic data. An Fe complex TonB transporter associated with the closely related Alteromonas bacterium was found to be abundant within the Pacific Ocean mesopelagic environment. This study improves our understanding of the biogeochemistry of marine heterotrophic bacteria by characterizing the contribution of specific metalloproteins to the cellular Fe quota of Pseudoalteromonas and demonstrating the molecular basis for its impressive Fe storage capabilities using bacterioferritin.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
M. G. M. was supported by the Camille and Henry Dreyfus Foundation Environmental Chemistry Postdoctoral Fellowship. We thank Kay Bidle (Rutgers University) for providing a culture of Pseudoalteromonas (BB2-AT2). We also thank Dawn Moran (WHOI) and Noelle Held (WHOI-MIT) for culturing assistance. We appreciate the Captain and Crew of the R/V Kilo Moana, and the many involved in the METZYME expedition sampling efforts. Discussions with Kevin Waldron (Newcastle University), Alison Butler (University of California, Santa Barbara), Lauren Manck (Scripps Institution of Oceanography), Randie Bundy (University of Washington) and Jake Gebbie (WHOI) were much appreciated. Funding for this research was provided by the Gordon and Betty Moore Foundation (3782), and NSF-OCE 1658030, 1736599, 1657766, 1924554, 1850719, 1924554.References
- K. W. Bruland, K. J. Orians and J. P. Cowen, Geochim. Cosmochim. Acta, 1994, 58, 3171–3182 CrossRef CAS.
- J. H. Martin and S. E. Fitzwater, Nature, 1988, 331, 341–343 CrossRef CAS.
- J. H. Martin, K. H. Coale, K. S. Johnson, S. E. Fitzwater, R. M. Gordon, S. J. Tanner, C. N. Hunter, V. A. Elrod, J. L. Nowicki, T. L. Coley, R. T. Barber, S. Lindley, A. J. Watson, K. Van Scoy, C. S. Law, M. I. Liddicoat, R. Ling, T. Stanton, J. Stockel, C. Collins, A. Anderson, R. Bidigare, M. Ondrusek, M. Latasa, F. J. Millero, K. Lee, W. Yao, J. Z. Zhang, G. Friederich, C. Sakamoto, F. Chavez, K. Buck, Z. Kolber, R. Greene, P. Falkowski, S. W. Chisholm, F. Hoge, R. Swift, J. Yungel, S. Turner, P. Nightingale, A. Hatton, P. Liss and N. W. Tindale, Nature, 1994, 371, 123–129 CrossRef CAS.
- K. H. Coale, K. S. Johnson, S. E. Fitzwater, R. M. Gordon, S. Tanner, F. P. Chavez, L. Ferioli, C. Sakamoto, P. Rogers, F. Millero, P. Steinberg, P. Nightingale, D. Cooper, W. P. Cochlan, M. R. Landry, J. Constantinou, G. Rollwagen, A. Trasvina and R. Kudela, Nature, 1996, 383, 495–501 CrossRef CAS PubMed.
- K. H. Coale, K. S. Johnson, F. P. Chavez, K. O. Buesseler, R. T. Barber, M. A. Brzezinski, W. P. Cochlan, F. J. Millero, P. G. Falkowski, J. E. Bauer, R. H. Wanninkhof, R. M. Kudela, M. A. Altabet, B. E. Hales, T. Takahashi, M. R. Landry, R. R. Bidigare, X. Wang, Z. Chase, P. G. Strutton, G. E. Friederich, M. Y. Gorbunov, V. P. Lance, A. K. Hilting, M. R. Hiscock, M. Demarest, W. T. Hiscock, K. F. Sullivan, S. J. Tanner, R. M. Gordon, C. N. Hunter, V. A. Elrod, S. E. Fitzwater, J. L. Jones, S. Tozzi, M. Koblizek, A. E. Roberts, J. Herndon, J. Brewster, N. Ladizinsky, G. Smith, D. Cooper, D. Timothy, S. L. Brown, K. E. Selph, C. C. Sheridan, B. S. Twining and Z. I. Johnson, Science, 2004, 304, 408–414 CrossRef CAS PubMed.
- D. K. Steinberg, B. A. S. V. Mooy, K. O. Buesseler, P. W. Boyd, T. Kobari and D. M. Karl, Limnol. Oceanogr., 2008, 53, 1327–1338 CrossRef.
- W. G. Sunda, Front. Microbiol., 2012, 3, 204 Search PubMed.
- P. W. Boyd, M. J. Ellwood, A. Tagliabue and B. S. Twining, Nat. Geosci., 2017, 10, 167–173 CrossRef CAS.
- W. G. Sunda, Biol. Oceanogr., 1989, 6, 411–442 Search PubMed.
- F. M. M. Morel and N. M. Price, Science, 2003, 300, 944–947 CrossRef CAS PubMed.
- J. K. Moore, S. C. Doney, D. M. Glover and I. Y. Fung, Deep Sea Res., Part II, 2001, 49, 463–507 CrossRef.
- A. E. Noble, M. A. Saito, D. M. Moran and A. Allen, Front. Chem., 2013, 1, 25 Search PubMed.
- A. Tagliabue, A. R. Bowie, P. W. Boyd, K. N. Buck, K. S. Johnson and M. A. Saito, Nature, 2017, 543, 51–59 CrossRef CAS PubMed.
- K. J. Waldron and N. J. Robinson, Nat. Rev. Microbiol., 2009, 7, 25–35 CrossRef CAS PubMed.
- B. A. Gilston and T. V. O’Halloran, in Encyclopedia of Inorganic and Bioinorganic Chemistry, American Cancer Society, 2013, pp. 1–12 Search PubMed.
- P. D. Tortell, M. T. Maldonado and N. M. Price, Nature, 1996, 383, 330–332 CrossRef CAS.
- M. T. Maldonado and N. M. Price, Deep Sea Res., Part II, 1999, 46, 2447–2473 CrossRef CAS.
- P. D. Tortell, M. T. Maldonado, J. Granger and N. M. Price, FEMS Microbiol. Ecol., 1999, 29, 1–11 CrossRef CAS.
- M. A. Schmidt and D. A. Hutchins, Deep Sea Res., Part II, 1999, 46, 2487–2503 CrossRef CAS.
- P. W. Boyd, K. R. Arrigo, R. Strzepek and G. L. van Dijken, J. Geophys. Res.: Oceans, 2012, 117, C06009 CrossRef.
- R. T. Reid and A. Butler, Limnol. Oceanogr., 1991, 36, 1783–1792 CrossRef CAS.
- R. T. Reid, D. H. Livet, D. J. Faulkner and A. Butler, Nature, 1993, 366, 455–458 CrossRef CAS PubMed.
- J. Granger and N. M. Price, Limnol. Oceanogr., 1999, 44, 541–555 CrossRef CAS.
- K. Barbeau, E. Kujawinski and J. Moffett, Aquat. Microb. Ecol., 2001, 24, 69–81 CrossRef.
- K. Barbeau, E. L. Rue, C. G. Trick, K. W. Bruland and A. Butler, Limnol. Oceanogr., 2003, 48, 1069–1078 CrossRef CAS.
- P. D. Holt, R. R. Reid, B. L. Lewis, G. W. Luther and A. Butler, Inorg. Chem., 2005, 44, 7671–7677 CrossRef CAS PubMed.
- R. M. Bundy, M. Jiang, M. Carter and K. A. Barbeau, Front. Mar. Sci., 2016, 3, 27 Search PubMed.
- B. M. Hopkinson and K. A. Barbeau, Environ. Microbiol., 2012, 14, 114–128 CrossRef CAS PubMed.
- K. L. Roe, S. L. Hogle and K. A. Barbeau, Appl. Environ. Microbiol., 2013, 79, 5753–5762 CrossRef CAS PubMed.
- S. L. Hogle, J. C. Thrash, C. L. Dupont and K. A. Barbeau, Appl. Environ. Microbiol., 2016, 82, 1613–1624 CrossRef CAS PubMed.
- S. L. Hogle, R. M. Bundy, J. M. Blanton, E. E. Allen and K. A. Barbeau, Limnol. Oceanogr. Lett., 2016, 1, 36–43 CrossRef.
- L. Baumann, P. Baumann, M. Mandel and R. D. Allen, J. Bacteriol., 1972, 110, 402–429 CrossRef CAS PubMed.
- J. García-Martínez, S. G. Acinas, R. Massana and F. Rodríguez-Valera, Environ. Microbiol., 2002, 4, 42–50 CrossRef PubMed.
- Y. Shi, J. McCarren and E. F. DeLong, Environ. Microbiol., 2012, 14, 191–206 CrossRef CAS PubMed.
- J. McCarren, J. W. Becker, D. J. Repeta, Y. Shi, C. R. Young, R. R. Malmstrom, S. W. Chisholm and E. F. DeLong, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 16420–16427 CrossRef CAS PubMed.
- S. G. Acinas, J. Antón and F. Rodríguez-Valera, Appl. Environ. Microbiol., 1999, 65, 514–522 CrossRef CAS PubMed.
- E. Ivars-Martinez, A.-B. Martin-Cuadrado, G. D’Auria, A. Mira, S. Ferriera, J. Johnson, R. Friedman and F. Rodriguez-Valera, ISME J., 2008, 2, 1194–1212 CrossRef CAS PubMed.
- S. Hou, M. López-Pérez, U. Pfreundt, N. Belkin, K. Stüber, B. Huettel, R. Reinhardt, I. Berman-Frank, F. Rodriguez-Valera and W. R. Hess, ISME J., 2018, 12, 981–996 CrossRef CAS PubMed.
- A. L. Koch, BioEssays, 2001, 23, 657–661 CrossRef CAS PubMed.
- M. López-Pérez, A. Gonzaga, A.-B. Martin-Cuadrado, O. Onyshchenko, A. Ghavidel, R. Ghai and F. Rodriguez-Valera, Sci. Rep., 2012, 2, 696 CrossRef PubMed.
- K. D. Bidle and F. Azam, Nature, 1999, 397, 508–512 CrossRef CAS.
- K. D. Bidle and F. Azam, Limnol. Oceanogr., 2001, 46, 1606–1623 CrossRef CAS.
- D. C. Smith, M. Simon, A. L. Alldredge and F. Azam, Nature, 1992, 359, 139–142 CrossRef CAS.
- F. Azam and F. Malfatti, Nat. Rev. Microbiol., 2007, 5, 782–791 CrossRef CAS PubMed.
- A. B. Burd, S. Frey, A. Cabre, T. Ito, N. M. Levine, C. Lønborg, M. Long, M. Mauritz, R. Q. Thomas, B. M. Stephens, T. Vanwalleghem and N. Zeng, Glob. Change Biol., 2016, 22, 121–136 CrossRef PubMed.
- S. Tottey, K. Waldron, S. Firbank, B. Reale, C. Bessant, K. Sato, T. Cheek, J. Gray, M. Banfield, C. Dennison and N. Robinson, Nature, 2008, 455, 1138–1142 CrossRef CAS PubMed.
- A. Cvetkovic, A. L. Menon, M. P. Thorgersen, J. W. Scott, F. L. P. Ii, F. E. J. Jr, W. A. Lancaster, J. L. Praissman, S. Shanmukh, B. J. Vaccaro, S. A. Trauger, E. Kalisiak, J. V. Apon, G. Siuzdak, S. M. Yannone, J. A. Tainer and M. W. W. Adams, Nature, 2010, 466, 779–782 CrossRef CAS PubMed.
- J. D. Aguirre, H. M. Clark, M. McIlvin, C. Vazquez, S. L. Palmere, D. J. Grab, J. Seshu, P. J. Hart, M. Saito and V. C. Culotta, J. Biol. Chem., 2013, 288, 8468–8478 CrossRef CAS PubMed.
- M. D. Keller, W. K. Bellows and R. R. L. Guillard, J. Exp. Mar. Biol. Ecol., 1988, 117, 279–283 CrossRef.
- N. M. Price, G. I. Harrison, J. G. Hering, R. J. Hudson, P. M. V. Nirel, B. Palenik and F. M. M. Morel, Biol. Oceanogr., 1989, 6, 443–461 Search PubMed.
- M. A. Saito, M. R. McIlvin, D. M. Moran, T. J. Goepfert, G. R. DiTullio, A. F. Post and C. H. Lamborg, Science, 2014, 345, 1173–1177 CrossRef CAS PubMed.
- M. A. Saito, A. Dorsk, A. F. Post, M. R. McIlvin, M. S. Rappé, G. R. DiTullio and D. M. Moran, Proteomics, 2015, 15, 3521–3531 CrossRef CAS PubMed.
- N. A. Held, M. R. McIlvin, D. M. Moran, M. T. Laub and M. A. Saito, mSystems, 2019, 4, e00317-18 CrossRef PubMed.
- M. A. Saito, M. R. McIlvin, D. M. Moran, A. E. Santoro, C. L. Dupont, P. A. Rafter, J. K. Saunders, D. Kaul, C. H. Lamborg, M. Westley, F. Valois and J. B. Waterbury, Nat. Geosci., submitted Search PubMed.
- M. A. Saito, A. Dorsk, A. F. Post, M. R. McIlvin, M. S. Rappé, G. R. DiTullio and D. M. Moran, Proteomics, 2015, 15, 3521–3531 CrossRef CAS PubMed.
- M. Greenberg, Differential Equations and Linear Algebra, Prentice Hall, Upper Saddle Ridge, NJ, 2001 Search PubMed.
- R. M. Boiteau, D. R. Mende, N. J. Hawco, M. R. McIlvin, J. N. Fitzsimmons, M. A. Saito, P. N. Sedwick, E. F. DeLong and D. J. Repeta, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 14237–14242 CrossRef CAS PubMed.
- R. M. Bundy, R. M. Boiteau, C. McLean, K. A. Turk-Kubo, M. R. McIlvin, M. A. Saito, B. A. S. Van Mooy and D. J. Repeta, Front. Mar. Sci., 2018, 5, 61 CrossRef.
- J. H. Martin, G. A. Knauer, D. M. Karl and W. W. Broenkow, Deep Sea Res., Part I, 1987, 34, 267–285 CrossRef CAS.
- A. E. Santoro, M. A. Saito, T. J. Goepfert, C. H. Lamborg, C. L. Dupont and G. R. DiTullio, Limnol. Oceanogr., 2017, 62, 1984–2003 CrossRef CAS.
- A. M. Posacka, D. M. Semeniuk and M. T. Maldonado, Front. Mar. Sci., 2019, 5, 523 CrossRef.
- A. C. Redfield, B. H. Ketchum and F. A. Richards, The influence of organisms on the composition of sea water, Interscience, New York and London, 1963 Search PubMed.
- R. A. Floyd, Biochem. Biophys. Res. Commun., 1981, 99, 1209–1215 CrossRef CAS.
- G. R. Moore, F. H. A. Kadir, F. K. al-Massad, N. E. L. Brun, A. J. Thomson, C. Greenwood, J. N. Keen and J. B. C. Findlay, Biochem. J., 1994, 304, 493–497 CrossRef CAS PubMed.
- R. Hussain, S. E. Harding, C. S. Hughes, P. Ma, S. G. Patching, S. Edara, G. Siligardi, P. J. F. Henderson and M. K. Phillips-Jones, Biochem. Soc. Trans., 2016, 44, 810–823 CrossRef CAS PubMed.
- R. P. Garg, C. J. Vargo, X. Cui and D. M. Kurtz, Biochemistry, 1996, 35, 6297–6301 CrossRef CAS PubMed.
- Y. Wang, H. Yao, Y. Cheng, S. Lovell, K. P. Battaile, C. R. Midaugh and M. Rivera, Biochemistry, 2015, 54, 6162–6175 CrossRef CAS PubMed.
- Y. M. Garcia, A. Barwinska-Sendra, E. Tarrant, E. P. Skaar, K. J. Waldron and T. E. Kehl-Fie, PLoS Pathog., 2017, 13, e1006125 CrossRef PubMed.
- S. C. Andrews, N. E. L. Brun, V. Barynin, A. J. Thomson, G. R. Moore, J. R. Guest and P. M. Harrison, J. Biol. Chem., 1995, 270, 23268–23274 CrossRef CAS PubMed.
- M. G. Mazzotta, M. R. McIlvin, D. M. Moran and M. A. Saito, in preparation.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0mt00034e |
| This journal is © The Royal Society of Chemistry 2020 |