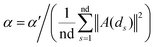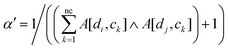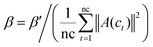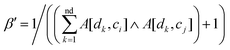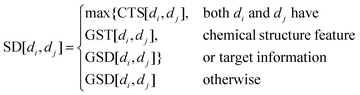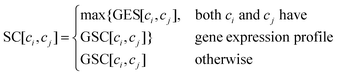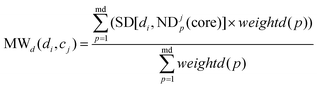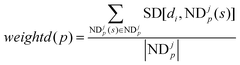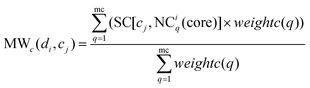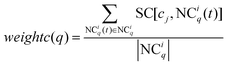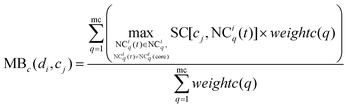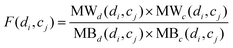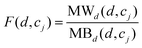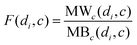DOI:
10.1039/C9MO00162J
(Research Article)
Mol. Omics, 2020,
16, 31-38
Modular within and between score for drug response prediction in cancer cell lines†
Received
3rd November 2019
, Accepted 25th November 2019
First published on 25th November 2019
Abstract
Drug response prediction in cancer cell lines is vital to discover new anticancer drugs. However, it's still a challenging task to accurately predict drug responses in cancer cell lines. In this study, we presented a novel computational approach, named as MSDRP (modular within and between score for drug response prediction), to predict drug responses in cell lines. The method is based on a constructed heterogeneous drug–cell line network with multiple information. Compared with other state-of-the-art methods, MSDRP acquired better predictive performance, and identified potential associations between drugs and cell lines, which have been confirmed by the published literature. The source code of MSDRP is freely available at https://github.com/shimingwang1994/MSDRP.git.
1 Introduction
Drug development often consumes substantial time and cost.1 The rapid growth of various high-throughput biological data such as Cancer Cell Line Encyclopedia (CCLE)2 and Genomics of Drug Sensitivity in Cancer (GDSC),3 makes computational approaches possible and rational to predict the drug response in cancer cell lines. Hence, some methods were proposed to predict drug sensitivity to cell lines using the drug response data. Geeleher et al. predicted clinical drug response using a ridge regression model and obtained good experimental results on before-treatment baseline tumor gene expression data.4 Muhammad et al. employed two types of information (cell line genomic feature and drug chemical properties) to construct the KBMF (kernelized Bayesian matrix factorization) model to predict drug response.5 Recently, several methods were proposed to predict drug response in cancer cell lines through combing more information, including compound chemical and therapeutic properties, baseline gene expression levels in cell lines and the cancer genomic alterations.6–8 These methods usually obtained better predictive ability. Network-based methods are also widely used to predict drug responses. Zhang et al. constructed a dual-layer network (cell line similarity network and drug similarity network) to predict drug responses.9 Stanfield et al. used a random walk algorithm to analyze cell line and drug network.10 GloNetDRP was developed to predict drug response by capturing the global information of drug and cell line in a heterogeneous network.11 HNMDRP12 applied an information flow-based algorithm13 in a heterogeneous network, which consists of the drug–target interaction network, protein–protein interaction(PPI) network, drug–cell line association network, drug similarity network and cell line similarity network, to predict drug response. Machine learning is another effective pattern for predicting drug response.14 For example, a recursive feature selection tool was employed to extract genomic features from the gene expression profile, and then the support vector machine (SVM) was applied to build a predictor in cell lines.15 NLLSS was proposed as a semi-supervised learning method to predict drug response.16 CaDRReS used a recommender system, and reflected its advantages on understanding drug response mechanisms.17 Xu et al. constructed an autoencoder network based on genetic features, such as mutations and RNA expression, and selected important features using the Boruta algorithm and random forest to predict drug response.18
Although many methods have been developed to predict drug response in cancer cell lines, there are still some problems to be solved. For example, predicting drug response to cell lines without any known drug response data still deserves attention. In addition, the topological structure feature of the response network is not taken into account sufficiently. Therefore, we proposed a novel method to solve these problems, called MSDRP (modular within and between score for drug response prediction). The proposed method integrates a drug–cell line response network, a drug chemical structure similarity network, the cell lines’ gene expression similarity network and a Gaussian interaction profile kernel similarity network to predict drug response in cancer cell lines (see Fig. 1).
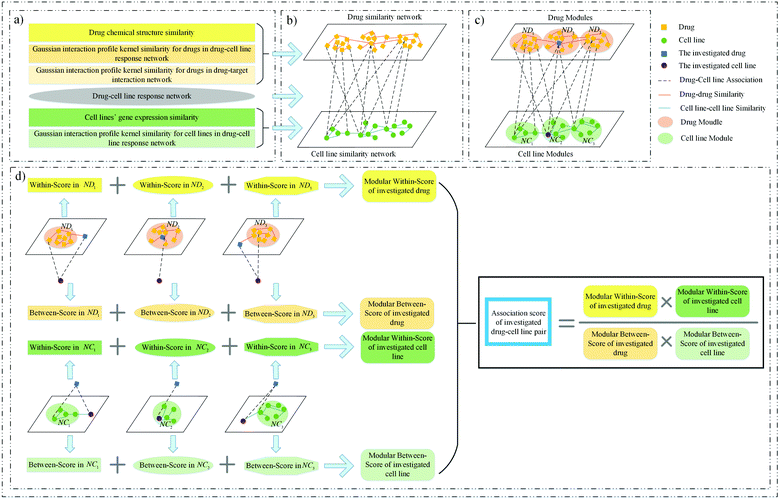 |
| | Fig. 1 Framework of MSDRP. (a) Integrate multiple similarity and response information of drugs and cell lines. (b) Construct a drug similarity network, cell line similarity network and drug–cell line response network. (c) Divide drugs (cell lines) into drug (cell line) modules via the investigated cell line (drug). (d) Calculate the modular within-score and modular between-score of the investigated drug and cell line to acquire the association score. | |
2 Methods
MSDRP includes three parts (see Fig. 1). Firstly, we collect drug's chemical structure information, drug–target interaction information, cell lines’ gene expression information and cell lines’ drug response data to calculate multiple similarities about drugs and cell lines (Fig. 1a). Then, a heterogeneous drug–cell line network is constructed, including the drug similarity network, cell line similarity network and drug–cell line response network (see Fig. 1b). Secondly, drugs (cell lines) are divided into several modules according to cell lines (drugs) associated with them (see Fig. 1c). Finally, the modular within-score and modular between-score of drugs and cell lines are calculated to obtain the association score of drugs and cell lines (see Fig. 1d).
2.1 Construction of the heterogeneous network
2.1.1 Drug–cell line response network.
We download drugs’ log-normalized IC50 values in cancer cell lines19 from GDSC (Downloaded in April 2018, Release 7.0). Staunton et al.'s method20 is used to classify cell lines into two classes: sensitive or resistant, according to their IC50 values with drugs. Then, a drug–cell line response network with nd drugs and nc cell lines is constructed (an edge represents that a cell line is sensitive to a certain drug, and a node represents a drug or a cell line), and its adjacency matrix is A ∈ Rnd×nc.
2.1.2 Drug chemical structure similarity.
The drugs’ chemical structure is downloaded from PubChem,21 and the 1-D and 2-D structure properties are extracted by PaDEL.22 Then, we obtain the chemical structure feature to characterize drugs. Similarly to existing studies,7–9,11,12 the Pearson correlation coefficient23 (Pcc) value of two drugs is calculated to construct the chemical structure similarity matrix CTS.
2.1.3 Cell lines’ gene expression similarity.
We download the gene expression profile for cancer cell lines from GDSC and normalize the data. Similar to previous studies,9,12 the Pcc value of each cell line pair is also calculated to construct the gene expression similarity matrix GES.
2.1.4 Gaussian interaction profile kernel similarity for drugs.
Based on the fact that similar cell lines and similar drugs exhibit similar drug responses,8,9 the topological structure information in the drug–cell line response network can be considered to calculate the Gaussian interaction profile kernel similarity matrix for drugs. Inspired by previous methods,24–27 the similarity between drug di and drug dj is defined as below:| | | GSD[di,dj] = exp(−α‖A(di) − A(dj)‖2) | (1) |
where vector A(di) and A(dj) represent the ith and jth row of matrix A respectively, and α is a parameter to control the kernel bandwidth defined as below:| | 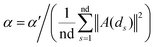 | (2) |
Previous methods24–27 set the bandwidth parameter α′ as 1. Here, we modify α′ by the number of cell lines associated with di and dj simultaneously, as below:
| | 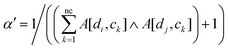 | (3) |
An example demonstrates the rationality of the modified bandwidth parameter visually (see Fig. 2). There is only one cell line associated with d1 and d2 simultaneously, two cell lines associated with d1 and d3, and two cell lines associated with d2 and d3 (see Fig. 2a). So the similarity between d1 and d2 should be lower than the other two drug pairs. However, the similarities calculated by the previous bandwidth parameter are the same, which are 0.4724 (see Fig. 2b). Our modification reflects the difference among the three drug pairs (see Fig. 2c). GSD[d1,d3] and GSD[d2,d3] have the same value of 0.7788, and exceed GSD[d1,d2] of 0.6873, which is more rational.
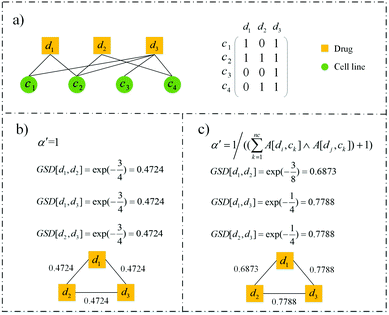 |
| | Fig. 2 An example of the rationality of our modified bandwidth parameter. (a) A drug–cell line response network with three drugs, four cell lines and eight edges. (b) Similarity of drugs calculated by setting the bandwidth parameter as 1. (c) Similarity of drugs calculated with our modified bandwidth parameter. | |
In addition, we collect the drugs’ target information from GDSC and construct the drug–target interaction network. We also consider the target information of drugs, so the steps above are carried out again in this network and get the Gaussian interaction profile kernel similarity matrix GST for drugs.
2.1.5 Gaussian interaction profile kernel similarity for cell lines.
Similar to Section 2.1.4, vector A(ci) and A(cj) are the ith and jth column of matrix A respectively, the Gaussian interaction profile kernel similarity between cell line ci and cell line cj is defined as below:| | | GSC[ci,cj] = exp(−β‖A(ci) − A(cj)‖2) | (4) |
| | 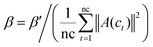 | (5) |
| | 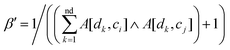 | (6) |
Finally, Gaussian interaction profile kernel similarity matrix GSC can be acquired.
2.1.6 Integrated similarity for drugs and cell lines.
To construct an accurate similarity network for drugs and cell lines, multiple similarity information is integrated by combining the Gaussian interaction profile kernel similarity with drugs’ chemical structure feature similarity and cell lines’ gene expression similarity, respectively. The integrated drug similarity matrix SD and cell line similarity matrix SC is defined as below:| | 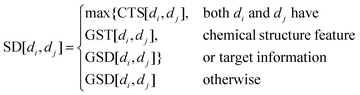 | (7) |
| | 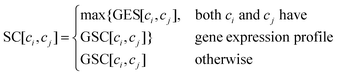 | (8) |
2.2 Module partition of drugs and cell lines
For an investigated pair between drug di and cell line cj, all drugs are divided into drug modules via cj, and all cell lines are divided into cell line modules via di (see Fig. 1c).
Step 1, all drugs are divided into md modules NDj1, NDj2,…,NDjmdvia cj, where md is the number of drugs associated with cj. In order to keep balance among all modules, we control the size of each module not exceeding ⌈nd/md⌉. Firstly, md empty modules are initialized, and each drug associated with cj is assigned into each module and marked as the core of it. Then, for the drug unassigned, the most similar core with it can be found and the drug is added into the core's module. When all drugs are assigned, check the size of all modules. If the size of the module NDjs exceeds ⌈nd/md⌉, the similarity between the core and other drugs in the module will be sorted in descending order, and the last |NDjs| − ⌈nd/md⌉ drugs will be removed from NDjs. Repeat this process until the size of each module is not more than ⌈nd/md⌉.
Step 2, all cell lines are divided into mc modules NCi1, NCi2,…,NCimcvia di in a similar way to step 1.
2.3 Drug response prediction using the modular within-score and modular between-score
The method WBSMDA26 mentioned the conception of the within-score and between-score. For the investigated drug di and investigated cell line cj, the within-score (between-score) of di is the highest similarity with di among the group of drugs with (without) known association with cj, and the within-score (between-score) of cj is the highest similarity with cj among the group of cell lines with (without) known association with di. MSDRP adds the modular information into within-score and between-score (see Fig. 1d). According to Section 2.2, the core in one drug module is associated with cell line cj, and other drugs in this module are not associated with cj. Hence, we can calculate the within-score and between-score of drug di in each drug module, which is from the local perspective. Then, the within-score and between-score in each drug module can be integrated by the weight of drug modules to obtain the modular within-score and modular between-score of di, which is from the global perspective. It is analogous to calculate the modular within-score and modular between-score of cj. The addition of modular information makes the use of similarity information more comprehensive.
MWd(di,cj) is defined as the modular within-score of di as below:
| | 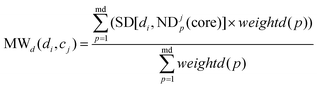 | (9) |
where ND
jp is a drug module and ND
jp(core) is the core of it.
weightd(
p) is the weight of ND
jp as below:
| | 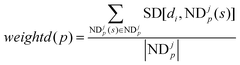 | (10) |
MBd(di,cj) is defined as the modular between-score of di as below:
| | 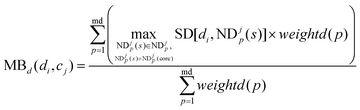 | (11) |
Similarly, MWc(di,cj) is defined as the modular within-score of cj as below:
| | 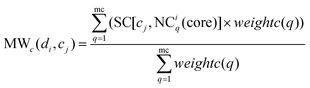 | (12) |
where NC
iq is a cell line module and NC
iq(core) is the core of it.
weightc(
q) is the weight of NC
iq as below:
| | 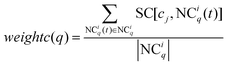 | (13) |
Similarly, MBc(di,cj) is defined as the modular between-score of cj as below:
| | 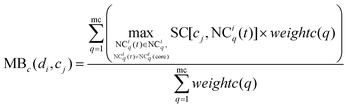 | (14) |
To calculate the association score between di and cj, the modular within-score and modular between-score of di and cj are integrated comprehensively as below:
| | 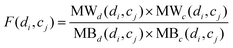 | (15) |
Besides, for a new drug d which does not have any known associations with any cell lines, MSDRP integrates the modular within-score and modular between-score of drugs as below:
| | 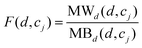 | (16) |
Meanwhile, for a new cell line c which does not have any known associations with any drugs, MSDRP integrates the modular within-score and modular between-score of the cell lines as below:
| | 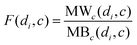 | (17) |
3 Results
3.1 Performance evaluation
To evaluate the predictive performance of MSDRP, Leave-one-out cross validation (LOOCV) is implemented. Each sensitive drug–cell line pair is left out in turn as the test data and the other drug–cell line pairs are used as training data for model learning. The test association with a higher rank represents a greater possibility of correct predictions. The receiver operating characteristic (ROC) curve is drawn and the areas under the ROC curve (AUROC) are computed to show the predictive ability of MSDRP. Meanwhile, because the number of resistant response is much more than sensitive response in the drug–cell line response network (see Table 1), we adopt the precision–recall (PR) curve and the areas under the PR curve (AUPR) to evaluate the predictive performance of our model on this imbalanced data. Hence, the results are evaluated by using the ROC curve (or AUROC) and PR curve (or AUPR) simultaneously in the following sections.
Table 1 Global characteristic of the drug–cell line response network
| Number of drugs |
Dimension of drug chemical structure feature |
Number of cell lines |
Dimension of cell lines’ gene expression profile |
Number of drug–cell line sensitive associations |
Number of resistant drug–cell line pairs |
| 251 |
1444 |
990 |
17737 |
23774 |
178269 |
| Max degree of drugs |
Min degree of drugs |
Average degree of drugs |
Max degree of cell lines |
Min degree of cell lines |
Average degree of cell lines |
| 146 |
34 |
94.72 |
168 |
1 |
24.01 |
3.2 Performance analysis of MSDRP
The global characteristic of the drug–cell line response network is shown in Table 1. The degree of a drug (cell line) is the number of its associations with cell lines (drugs). The drug–target interaction network mentioned in Section 2.1.4 contains 250 drugs, 239 targets and 456 interactions. As a result, MSDRP acquires an AUROC value of 0.8446 and AUPR value of 0.4007 on LOOCV. In addition, we set the bandwidth parameter as 1 to repeat MSDRP and obtain an AUROC value of 0.7529 and AUPR value of 0.3197 (see Fig. 3). These results demonstrate the effectiveness of our modified bandwidth parameters.
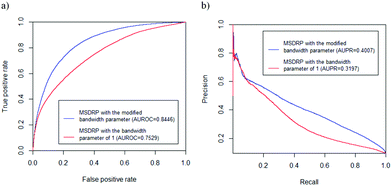 |
| | Fig. 3 ROC curves and PR curves of MSDRP with different bandwidth parameters. (a) ROC curves. (b) PR curves. | |
3.3 Comparison with existing methods
We compare MSDRP with five methods: Xu's method,18 WBSMDA,26 Zhang's method,9 HNMDRP12 and CaDRReS.17 The ROC curves and PR curves of these methods are shown in Fig. 4. Our model has the highest AUROC value and AUPR value. AUROC values of each drug obtained by these methods are calculated and shown in a box-and-whisker plot, where the P-values of paired t-tests between each method and MSDRP are marked (see Fig. 5a). Our model obtains statistically significantly better results than the other five methods according to all P-values not more than 1.2974 × 10−5. For AUPR of all drugs, we also provide a box-and-whisker plot (see Fig. 5b) and five P-values of the paired t-test are not more than 5.2771 × 10−6. These results illustrate that our model possesses excellent ability for predicting drug–cell line associations and potential confidence to detect new associations.
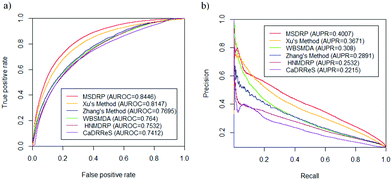 |
| | Fig. 4 ROC curves and PR curves on LOOCV of different methods. (a) ROC curves. (b) PR curves. | |
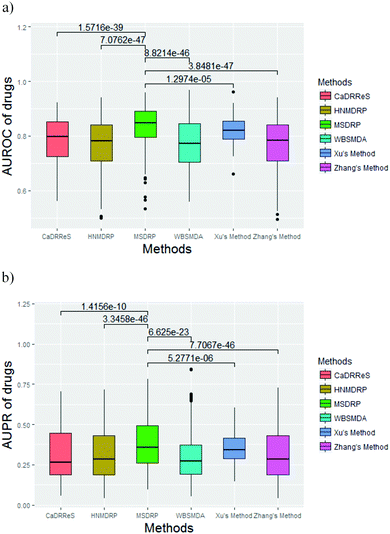 |
| | Fig. 5 The box-and-whisker plots of AUROC and AUPR values of drugs by different methods with the P-value of the paired t-test. (a) AUROC values. (b) AUPR values. | |
3.4 Tissue specific cell line type
Since one cell line belongs to one certain tissue type, we detect differences of drug responses in diverse tissue types. All cell lines are divided into 19 tissue types and the percentage of each tissue type is counted (see Fig. 6a). The major tissue types are lung_NSCLC, urogenital_system, leukemia and aero_dig_tract, which take up 11.21% (111), 10.4% (103), 8.3% (82) and 7.98% (79) of 990 cell lines, respectively. Comparison of AUROC values on the four major tissue types is exhibited (see Fig. 6b). Our model acquires the highest AUROC values no matter in which tissue type, which are 0.829, 0.8314, 0.8035 and 0.8325 from left to right. The AUPR values of the major tissue types are shown in Fig. 6c, where MSDRP gets 0.2587, 0.3286, 0.5677 and 0.2991 from left to right, and still exceeds the other five methods. Meanwhile, predictive performance on the other fifteen tissue types is also detected (see Fig. S1, ESI†) and our model still has an advantage, which illustrates the high efficiency of our model when aiming at a specific tissue type.
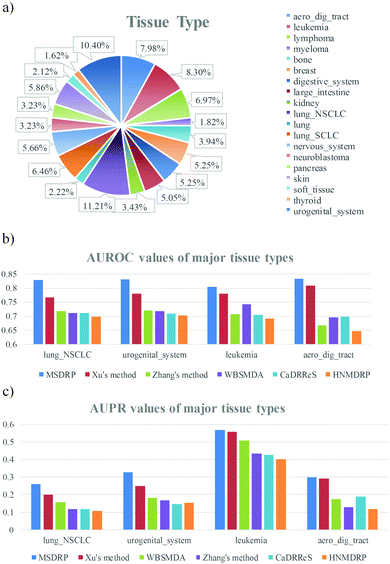 |
| | Fig. 6 Distribution of tissue types and predictive performance in the four major tissue types. (a) Distribution of nineteen tissue types. (b) Comparison of AUROC values between our method and other methods in the four major tissue types. (c) Comparison of AUPR values between our method and other methods in the four major tissue types. | |
3.5 Drug response prediction in the missing data
In the response network consisting of 251 drugs and 990 cell lines, 46447 drug–cell line pairs don’t have log-normalized IC50 values. In other words, 18.69% of 2512990 drug–cell line possible combinations are missing data. Nevertheless, MSDRP has the ability of predicting potential associations in the missing data. Here, we implement drug response prediction in the missing data for the major four tissue types and find literature evidence for the top 20 potential drug–cell line associations in each tissue type manually. When predicting for tissue types of Lung_NSCLC, urogenital_system, leukemia and aero_dig_tract, six, five, four and four association pairs in the top 20 were confirmed sensitive, respectively (see Table S1, ESI†). The drugs involved are used to treat non-small cell lung cancer (NSCLC), endometrium adenocarcinoma (EA), ovarian cancer (OC), endometrial cancer (EC), acute myeloid leukaemia (AML), head-and-neck cancer (HNC) and head and neck squamous cell carcinoma (HNSCC). We also provide the rank of these associations in the other five methods and our method acquires higher rank than them. Here, we introduce the association between drug palbociclib and cell line NCI-H292 in detail. Cell line NCI-H292 belongs to tissue type Lung_NSCLC, which has the highest proportion among all tissue types. We sorted the association scores of all pairs belonging to Lung_NSCLC, and this pair ranked 1st. Gopalan et al. gave palbociclib orally to 19 previously-treated patients with recurrent or metastatic NSCLC at 125 mg daily on days 1–21 of a 28 day cycle.28 Experiments showed that palbociclib therapy alone was well-tolerated, and 50% of evaluable patients kept a stable disease (SD), which suggested the induction of cellular senescence. These results indicate that MSDRP has advantageous performance in predicting unknown drug response and can provide convenience for prediction in networks with missing data.
3.6 Prediction of cell lines without any known drug response data
GDSC (Release 7.0) provides 1001 cell lines with whole exome sequencing, including 990 cell lines with drug response data and 11 cell lines without any drug response data. For the 11 cell lines, which are D-245MG, SNU-283, NCI-H660, U-CH2, MHH-CALL-4, KO52, SC-1, RERF-LC-FM, NCI-H740, NCCIT and BONNA-12, MSDRP can calculate the prediction scores with all drugs and a higher score means a potential sensitive association. After sorting the predictive score of one certain cell line, the top 20 pairs are counted to find literature evidence manually. All evidence with the detailed descriptions is provided (see Table S2, ESI†). The drugs involved are used to treat glioblastoma multiforme (GBM), glioma, glioblastoma, colorectal cancer (CRC), colorectal adenocarcinoma (CRAC), colon cancer (CC), castration-resistant prostate cancers (CRPC), prostate cancer (PC), osteosarcoma, B-Cell Chronic Lymphocytic Leukemia (CLL), AML, diffuse large B-cell lymphoma (DLBCL), lung_small_cell_carcinoma (SCLC), testicular germ cell tumor (TGCT), and hairy cell leukemia (HCL). Here, we introduce the association between drug cetuximab and cell line NCI-H660 in detail. We sorted the association scores between NCI-H660 and all drugs, and cetuximab ranked 1st. NCI-H660 belongs to cell type prostate. The EGF receptor (EGFR) might be a valid therapeutic target of metastatic castration-resistant prostate cancers (mCRPC), because of its overexpression in mCRPC. Cathomas et al. found that cetuximab could improve the outcome of patients with mCRPC by inhibiting EGFR.29 In conclusion, these results demonstrate that MSDRP is an efficient approach to predict the drug response for new cell lines and can provide valuable applications in future research studies.
4 Conclusions
In this study, we propose a novel method MSDRP to predict drug responses in cancer cell lines and contributions are made at three aspects. Firstly, the innovation on the bandwidth parameter extracts topological information in the drug–cell line response network to the maximum extent, which is beneficial for integrating various similarity networks. Secondly, the addition of modular information into within-score and between-score provides more sufficient similarity information for the investigated drug or cell line than calculating within-score or between-score among the whole response network. Finally, predicting potential drug sensitive associations for new cancer cell lines is a significant development for future biological research. The experiment results confirm these improvements. We hope that MSDRP can provide potential value in future clinical research.
Conflicts of interest
There are no conflicts to declare.
Acknowledgements
This work has been supported by the Natural Science Foundation of Heilongjiang Province (Grant No. F2016016), and National Key Research and Development Program of China (Grant No. 2016YFC0901905).
References
- A. Gottlieb, G. Y. Stein, E. Ruppin and R. Sharan, PREDICT: a method for inferring novel drug indications with application to personalized medicine, Mol. Syst. Biol., 2011, 7, 496 CrossRef PubMed.
- J. Barretina, G. Caponigro, N. Stransky, K. Venkatesan, A. A. Margolin and S. Kim,
et al., The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity (vol 483, pg 603, 2012), Nature, 2012, 492, 290 CrossRef CAS.
- W. J. Yang, J. Soares, P. Greninger, E. J. Edelman, H. Lightfoot and S. Forbes,
et al., Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells, Nucleic Acids Res., 2013, 41, D955–D961 CrossRef CAS PubMed.
- P. Geeleher, N. J. Cox and R. S. Huang, Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines, Genome Biol., 2014, 15, R47 CrossRef PubMed.
- A. U. D. Muhammad, G. Elisabeth, G. N. Mehmet, L. Tuomo, K. Olli and W. Krister,
et al., Integrative and personalized QSAR analysis in cancer by kernelized Bayesian matrix factorization, J. Chem. Inf. Model., 2014, 54, 2347 CrossRef PubMed.
- S. Gupta, K. Chaudhary, R. Kumar, A. Gautam, J. S. Nanda and S. K. Dhanda,
et al., Prioritization of anticancer drugs against a cancer using genomic features of cancer cells: a step towards personalized medicine, Sci. Rep., 2016, 6, 23857 CrossRef CAS PubMed.
- Y. C. Wang, J. W. Fang and S. L. Chen, Inferences of drug responses in cancer cells from cancer genomic features and compound chemical and therapeutic properties, Sci. Rep., 2016, 6, 32679 CrossRef CAS PubMed.
- L. Wang, X. Li, L. Zhang and Q. Gao, Improved anticancer drug response prediction in cell lines using matrix factorization with similarity regularization, BMC Cancer, 2017, 17, 513 CrossRef PubMed.
- N. Q. Zhang, H. Y. Wang, Y. Fang, J. Wang, X. Q. Zheng and X. S. Liu, Predicting Anticancer Drug Responses Using a Dual-Layer Integrated Cell Line-Drug Network Model, PLoS Comput. Biol., 2015, 11, e1004498 CrossRef PubMed.
- Z. Stanfield, M. Coşkun and M. Koyutürk, Drug Response Prediction as a Link Prediction Problem, Sci. Rep., 2017, 7, 40321 CrossRef CAS PubMed.
- D. H. Le and V. H. Pham, Drug Response Prediction by Globally Capturing Drug and Cell Line Information in a Heterogeneous Network, J. Mol. Biol., 2018, 430, 2993–3004 CrossRef CAS PubMed.
- Z. Fei, M. Wang, J. Xi, J. Yang and L. Ao, A novel heterogeneous network-based method for drug response prediction in cancer cell lines, Sci. Rep., 2018, 8, 3355 CrossRef PubMed.
- W. Wenhui, Y. Sen, Z. Xiang and L. Jing, Drug repositioning by integrating target information through a heterogeneous network model, Bioinformatics, 2014, 30, 2923–2930 CrossRef PubMed.
- M. P. Menden, F. Iorio, M. Garnett, U. McDermott, C. H. Benes and P. J. Ballester,
et al., Machine Learning Prediction of Cancer Cell Sensitivity to Drugs Based on Genomic and Chemical Properties, PLoS One, 2013, 8, e61318 CrossRef CAS PubMed.
- Z. Dong, N. Zhang, C. Li, H. Wang, Y. Fang and J. Wang,
et al., Anticancer drug sensitivity prediction in cell lines from baseline gene expression through recursive feature selection, BMC Cancer, 2015, 15, 489 CrossRef PubMed.
- X. Chen, B. Ren, M. Chen, Q. Wang, L. Zhang and G. Yan, NLLSS: Predicting Synergistic Drug Combinations Based on Semi-supervised Learning, PLoS Comput. Biol., 2016, 12, e1004975 CrossRef PubMed.
- S. Chayaporn, B. Denis and N. Niranjan, Predicting Cancer Drug Response Using a Recommender System, Bioinformatics, 2018, 34(22), 3907–3914 CrossRef PubMed.
- X. Lu, H. Gu, Y. Wang, J. Wang and P. Qin, Autoencoder Based Feature Selection Method for Classification of Anticancer Drug Response, Front. Genet., 2019, 10, 233 CrossRef PubMed.
- J. L. Sebaugh, Guidelines for accurate EC50/IC50 estimation, Pharm. Stat., 2011, 10, 128–134 CrossRef CAS PubMed.
- J. E. Staunton, D. K. Slonim, H. A. Coller, P. Tamayo, M. J. Angelo and J. Park,
et al., Chemosensitivity prediction by transcriptional profiling, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 10787–10792 CrossRef CAS PubMed.
-
E. E. Bolton, Y. Wang, P. A. Thiessen and S. H. Bryant, Chapter 12 – PubChem: Integrated Platform of Small Molecules and Biological Activities, Elsevier Science & Technology, 2008 Search PubMed.
- Y. Chun Wei, PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints, J. Comput. Chem., 2011, 32, 1466–1474 CrossRef PubMed.
- P. Ahlgren, J. Bo and R. Rousseau, Requirements for a cocitation similarity measure, with special reference to Pearson's correlation coefficient, J. Assoc. Inf. Sci. Technol., 2014, 54, 550–560 CrossRef.
- X. Chen, Z. C. Jiang, D. Xie, D. S. Huang, Q. Zhao and G. Y. Yan,
et al., A novel computational model based on super-disease and miRNA for potential miRNA-disease association prediction, Mol. BioSyst., 2017, 13, 1202 RSC.
- X. Chen, Y. A. Huang, Z. H. You, G. Y. Yan and X. S. Wang, A novel approach based on KATZ measure to predict associations of human microbiota with non-infectious diseases, Bioinformatics, 2016, 33, 733–739 Search PubMed.
- X. Chen, C. C. Yan and X. Zhang, WBSMDA Within and Between Score for MiRNA-Disease Association prediction, Sci. Rep., 2016, 6, 21106 CrossRef CAS PubMed.
- X. Chen, D. Xie, L. Wang, Q. Zhao and H. Liu, BNPMDA: Bipartite network projection for MiRNA–Disease association prediction, Bioinformatics, 2018, 34, 3178–3186 CrossRef CAS PubMed.
-
P. K. Gopalan, M. C. Pinder, A. Chiappori, A. M. Ivey, A. Gordillo Villegas and F. J. Kaye, A phase II clinical trial of the CDK 4/6 inhibitor palbociclib (PD 0332991) in previously treated, advanced non-small cell lung cancer (NSCLC) patients with inactivated CDKN2A, American Society of Clinical Oncology, 2014 Search PubMed.
- R. Cathomas, C. Rothermundt, D. Klingbiel, L. Bubendorf, R. Jaggi and D. C. Betticher,
et al., Efficacy of cetuximab in metastatic castration-resistant prostate cancer might depend on EGFR and PTEN expression: results from a phase II trial (SAKK 08/07), Clin. Cancer Res., 2012, 18, 6049–6057 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1. Predictive performance in the other fifteen tissue types. (a) Comparison of AUROC values between our method and other methods in the other fifteen tissue types. (b) Comparison of AUPR values between our method and other methods in the other fifteen tissue types. Table S1. Literature evidence of the top 20 potential drug-cell line responses for major tissue types in the missing data. Table S2. Literature evidence of potential drug responses for new cell lines in the top 20. See DOI: 10.1039/c9mo00162j |
|
| This journal is © The Royal Society of Chemistry 2020 |
Click here to see how this site uses Cookies. View our privacy policy here.  and
Jie
Li
and
Jie
Li
 *
*