Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Harnessing the surface chemistry of methyl ester functionalized polydicyclopentadiene and exploring surface bioactivity†‡
Tong
Li
 ,
Hannah
Shumka
,
Tyler J.
Cuthbert
,
Hannah
Shumka
,
Tyler J.
Cuthbert
 ,
Chang
Liu
and
Jeremy E.
Wulff
,
Chang
Liu
and
Jeremy E.
Wulff
 *
*
Department of Chemistry, University of Victoria, PO Box 3065 STN CSC, Victoria, British Columbia V8W 3V6, Canada. E-mail: wulff@uvic.ca
First published on 24th July 2020
Abstract
In an effort to broaden the utility of the industrially used ROMP polymer polydicyclopentadiene, we introduced an ester-functionalized dicyclopentadiene monomer that can be polymerized and crosslinked under similar conditions to those of the unfunctionalized parent, and for which the resulting functionalized polymer (fPDCPD) maintains the thermal and mechanical properties that make polydicyclopentadiene useful. In the current work, we expand on the utility of fPDCPD by harnessing the embedded functional group to manipulate the surface energy of the material, and to attach a range of biologically relevant functional groups. These include a fluorescent dye used to characterize surface coverage, an RGD peptide used to promote cellular adhesion, and an antibacterial agent (chloramphenicol) used to moderate E. coli growth. Significantly, the chloramphenicol is conjugated to the surface in such a way that it remains inactive until it is cleaved by endogenous bacterial enzymes. This strategy minimizes leaching of the drug into the environment, by effectively relying on colonizing bacteria to dose themselves with surface-available antibiotic.
Introduction
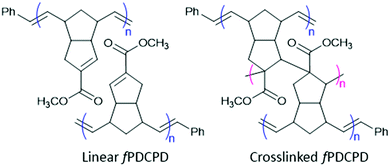
Polydicyclopentadiene (PDCPD), a polymer produced through ring-opening metathesis polymerization (ROMP) of dicyclopentadiene (DCPD), is used industrially to fabricate vehicle body panels and construction equipment.1–5 The extensive crosslinking network within PDCPD contributes to high heat and impact resistance,6–8 making it an attractive material for further applications including composite materials,9–11 aerogels,12,13 and self-healing polymers.14–16 However, PDCPD has a low surface energy when freshly prepared, making it difficult to process or modify, and its lack of chemical functionality – beyond just residual double bonds – limits its broader applications.17To address the low surface energy of PDCPD, and to bestow additional function to the polymer, we installed a C-linked ester group on the dicyclopentadiene monomer.18–20 The ester-functionalized monomer (fDCPD) engages in Ru-catalyzed ROMP similarly to the parent dicyclopentadiene, to afford a linear telechelic polymer that can then be thermally cured to provide a densely crosslinked, functionalized form of polydicyclopentadiene that we call fPDCPD (Fig. 1).21fPDCPD offers a tunable, elevated surface energy, and a higher Tg compared with the parent material.18 Importantly, both the thermal stability of fPDCPD (as assessed by thermogravimetric analysis)18,19 and the storage modulus (measured by dynamic mechanical analysis)20 were shown to be similar to those of the parent polymer. The introduction of new functionality while maintaining durability allows for further modifications, potentially increasing the applications of fPDCPD for industrial and biomedical use.
Through modification of the surface properties of commodity polymers, the response of organisms interacting with the surface can be finely tuned, presenting opportunities to control biological activity.22–24 From promoting cell adhesion and proliferation,25 to inhibition of bacterial growth,26–29 or design of cell patterning surfaces,30,31 bioactive polymers can find applications ranging from antifouling surfaces32 to microfluidics33–35 and tissue engineering.36–38
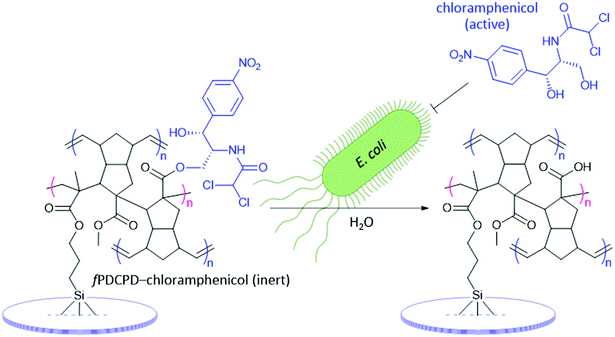
Functionalized surfaces can also be reactive; similar to drug releasing nanoparticles, surfaces may release small molecules or polymers upon reaction or stimulation.39 Releasing antibiotics directly from a surface would result in a high local concentration required for inhibition of bacterial growth or bactericidal effect without the requirement of high systemic dosing typical of broad-spectrum antibiotics. Excess and misuse of antibiotics has in the past led to the emergence of resistant pathogens from the introduction of antibiotics into the environment.40,41 Functionalized surfaces have the potential to mitigate this issue by providing a platform whereby antibiotics can be covalently immobilized to a surface, rendering them biologically inactive, until cleavage occurs when in the presence of the target.42fPDCPD is an attractive polymer substrate for applications of this type because it can be crosslinked into an insoluble polymer appropriate for surface coatings, yet possesses accessible carboxylic acid functionality that can be exploited to attach antibiotics through readily cleaved ester bonds. Ideally, the surface-bound antibiotic will be inactive until it is released from the polymer, resulting in an off-to-on switch similar to a pro-drug strategy.42
As a showcase to illustrate the potential of fPDCPD as a direct-dosing polymer support – and more generally to demonstrate the utility of this novel functionalized polymer – we sought to develop methodology that would permit the attachment of chloramphenicol to the fPDCPD surface (Fig. 2). Chloramphenicol is a broad spectrum antibiotic that inhibits bacterial protein synthesis by binding to the 50S subunit of bacterial ribosomes.43,44 It is a widely prescribed antibiotic with a relatively low dosing cost, and continues to be considered an essential medicine by the World Health Organization.45 Critically, the 1,3-propanediol motif in chloramphenicol is key for its biological activity; if either hydroxyl group is removed or altered, the antibacterial effects of the drug are lost.46 The primary alcohol within this motif is therefore a prime candidate as a point of attachment to the ester functional group of fPDCPD, since immobilization through this group would render chloramphenicol bioinert, while release by promiscuous E. coli enzymes47 would restore antibiotic function.48 In principle, this approach would result in an autonomous stimuli-responsive surface that is capable of dosing the drug only in the presence of bacteria – thereby limiting the release of excess antibiotics to the environment, and minimizing the potential for creating antibiotic-resistant pathogens.
Results and discussion
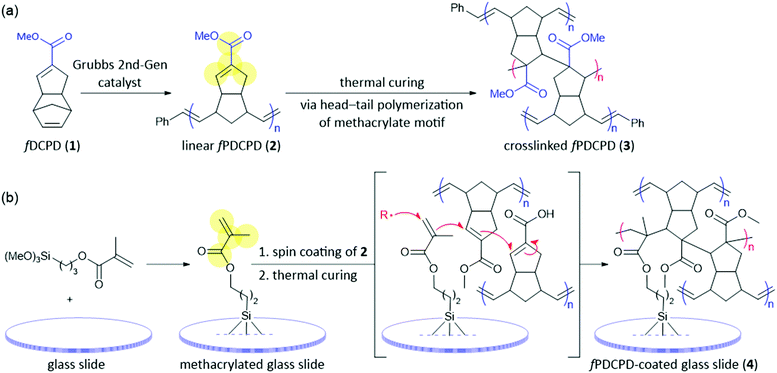
Understanding the chemical properties of a novel material is paramount to discovering its possible future applications. Although some of the unique properties of fPDCPD have been reported previously by our group,18–20 the degree to which the surface can be further functionalized is not yet completely understood. For example, while previous work has shown that partial hydrolysis of the ester functional groups on the surface of fPDCPD can raise the surface energy,18 we have not yet examined the installation of other functionality at this position. With the goal, therefore, of working toward the conjugation of chloramphenicol to fPDCPD, we first needed to gain additional insight into the polymer's ability to accommodate a diverse array of functionality.To establish that a uniformly functionalized surface could be obtained by adding reagents to functionalized polydicyclopentadiene, we began by pursuing the attachment of a fluorophore to the surface of a fPDCPD-coated glass slide. To create the appropriate test-system for this experiment, slides were treated with 3-(trimethoxysilyl)propyl methacrylate and then spin-coated with linear (i.e. uncrosslinked) fPDCPD (2, Scheme 1) prior to thermal curing. One of the advantages of fPDCPD compared with the traditional form of polydicyclopentadiene is that the primary polymerization event (i.e. formation of the linear polymer 2 from monomer 1) is decoupled from the crosslinking step. This difference allows for a facile preparation of thin polymer samples, since standard spin-coating techniques can be used with the uncrosslinked polymer, which is fully soluble in halogenated solvents.
In our previous mechanistic studies,19 we confirmed that the principal crosslinking mechanism for 2 does not take place via secondary metathesis events of the type that are generally considered for unfunctionalized polydicyclopentadiene49–53 but rather through a self-initiated, head–tail radical polymerization event that occurs through the methacrylate motif embedded within 2 (Scheme 1a). In order to achieve robust attachment of fPDCPD to glass slides, we further exploited this mechanism by carrying out the thermal curing event on the 3-(trimethoxysilyl)propyl methacrylate-coated slide, with the goal of engaging the methacrylate groups in the surface-activating reagent directly in the self-initiated radical polymerization event that occurs across the methacrylate groups in linear polymer 2, resulting in the formation of strong covalent bonds that serve to link the fPDCPD polymer to the glass slide. While it is impossible to unambiguously prove the adhesive structure (4) indicated in Scheme 1, this method provided much better adhesion of the polymer to the surface of the glass than analogous experiments where 2 was spin-coated and cured directly onto glass slides: in the absence of the 3-(trimethoxysilyl)propyl methacrylate reagent the polymer exhibited a tendency to delaminate from the slide when it was incubated in culture media.
Having thus affixed fPDCPD firmly to glass slides, we turned to the conjugation of a fluorophore (Scheme 2). To this end, the samples were submerged in methanolic sodium hydroxide to hydrolyze surface-exposed ester groups, affording 5. After washing and drying, the functionalized slides were immersed in a mixture of CH2Cl2, DMF, and SOCl2 to effect conversion to the corresponding acyl chloride surface (6). After further washing and drying, the slides were exposed to a solution of 5-TAMRA-PEO3-amine in DMSO containing triethylamine. The presence of the new amide linkage in the target fPDCPD–TAMRA conjugate (7) was confirmed by FTIR analysis (see Fig. S12–S14 in the ESI‡).
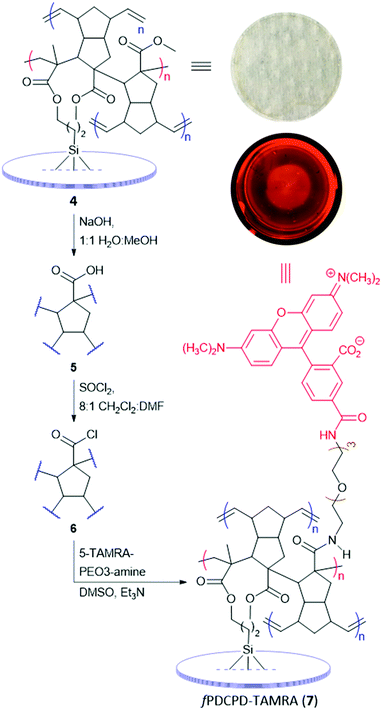 | ||
| Scheme 2 Construction of a fluorescent fPDCPD surface on a glass slide. Inset figures show a visible-light photograph of the initial fPDCPD–ester surface (the mottled surface shown in the photo is from a paper towel that was used as backdrop to image the transparent slide) and a fluorescent image of a fPDCPD–TAMRA surface acquired using a 576–600 nm excitation and a 610–885 nm emission window. The appearance of red fluorescence across the slide confirms an acceptable level of surface coverage to support subsequent experiments. Refer to Fig. S16 (ESI‡) for fluorescent images of negative control slides. | ||
To qualitatively evaluate surface coverage, the TAMRA-coupled slides were imaged using a fluorescent microscope. As shown in the inset to Scheme 2, the entire surface exhibited red fluorescence within the limits of optical resolution, indicating a level of surface coverage that could be harnessed to attach other functionality. Control surfaces (e.g.4 or 5) showed no significant background fluorescence (Fig. S16, ESI‡). The only significant defect appeared to be a series of radial waves in the surface, which result from the spin-coating protocol. Defects on this length scale were not expected to interfere with subsequent experiments. In an attempt to better quantify the density of carboxylate groups available for functionalization, we also treated glass slides covered with 5 with toluidine blue oxide (TBO), a reagent that is known to form a stable complex with surface carboxyl groups.54 This experiment revealed a surface coverage of up to 0.249 ± 0.125 μmol cm−2 (see ESI‡ for details), although we found that the result was highly dependent upon crosslinking time. In retrospect, this is not surprising; we previously showed that crosslinking density in fPDCPD can be controlled with heating time.19 More densely crosslinked samples are presumably less able to swell when exposed to TBO in solvent, which results in a lower uptake of the reagent.
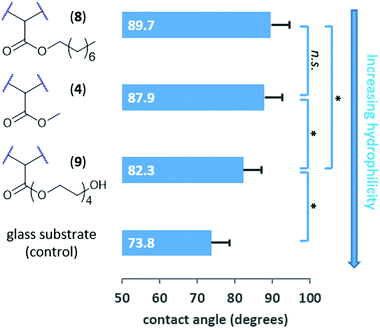
To better evaluate our ability to tune the surface of fPDCPD through rational functional group manipulations, we next prepared an octyl ester derivative and a tetraethylene glycol derivative, and measured the water contact angle on each surface (Fig. 3). As expected, the octyl ester derivative (8) was more hydrophobic than the parent methyl ester surface (4), while the tetraethylene glycol-functionalized surface (9) was more hydrophilic. While the change in contact angle in this experiment was less dramatic than we had previously seen following partial saponification of methyl ester 4,18 we nevertheless observed a statistically significant difference between the 8 and 9 polymer surfaces. Together, these data confirm that chemical changes to the fPDCPD surface can be used to tune the surface energy in more subtle ways than the hydrolysis protocol used in our earlier work.
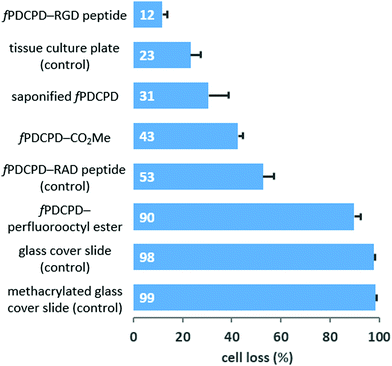
Moving toward more biologically oriented experiments, and mindful of the utility of controlling polymer–cell interactions for microfluidic or cell-patterning applications, we prepared several fPDCPD surfaces containing different functional groups and used these to evaluate the adhesion of tumor cells. Briefly, each polymer surface was prepared on a glass slide that was sized to fit precisely into the bottom of a 24-well tissue culture plate. After inserting the prepared slides into the multiwell plate (using a small amount of epoxy on the underside of each slide to ensure that it adhered to the bottom of the plate), HeLa cells were added in cell culture media. After 48 hours, the surfaces were imaged and then washed with phosphate-buffered saline and then imaged a second time. The populations of adhered cells before and after washing were determined through quantification of the images, and a ratio was taken to determine the amount of cell loss from each surface. As shown in Fig. 4, this experiment revealed an impressive ability to control cellular adhesion through surface functionalization. For example, a perfluorooctyl ester derivative was found to be inhibitory toward cellular attachment (90% loss of cells after washing) relative to the parent methyl ester (43% loss of cells), while saponification to provide carboxylate groups was found to enhance the degree of cellular adhesion (31% loss of cells).
Most significantly, attachment of a cyclic arginine-glycine-aspartate (RGD) tripeptide (which is known to bind to integrin receptors on cells)25 afforded a surface that was very hospitable for cellular attachment, such that only 12% loss of HeLa cells was observed following the washing protocol. This was a more effective attachment than the commercial plasma-treated tissue culture plate used as a positive control, which resulted in 23% loss of HeLa cells after washing. As a negative control for this experiment, we also attached an arginine-alanine-aspartate (RAD) peptide. Despite the fact that the RGD and RAD peptides only differ by a single methyl group (and therefore have similar polarities), the RAD motif has a significantly lower binding affinity for integrin receptors. In the event, use of the RAD-functionalized surface afforded a dramatic reduction in HeLa cell adhesion: 53% of cells were washed away from the RAD–fPDCPD surface, compared with only 12% from the RGD–fPDCPD surface.
Turning at last to the synthesis of the target chloramphenicol conjugate, NMR experiments with the acyl chloride derivative of 1 (as a small-molecule mimic of surface-bound acyl chloride 6) confirmed that the primary alcohol of chloramphenicol underwent condensation with the acyl chloride in either DMSO-d6 or CD3CN, to afford a new ester linkage. We therefore formed the chloramphenicol-functionalized fPDCPD shown in Fig. 1 using a similar protocol to that employed for the generation of fPDCPD–TAMRA in Scheme 2. Freshly prepared glass slides coated with fPDCPD–acyl chloride 6 were submerged in a mixture of triethylamine, chloramphenicol, and DMSO for 48 h. After rinsing (with both acetonitrile and dichloromethane) and drying, FTIR analysis confirmed the formation of the desired ester bond (Fig. S15, ESI‡).
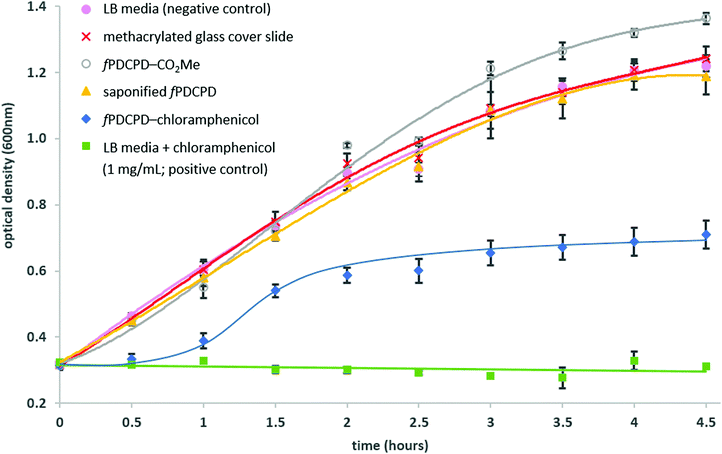
The slides were epoxied to the base of a 24-well tissue culture plate (as described in the HeLa cell experiments above) and 1 mL of E. coli K12 suspension in LB media (at an initial OD600 of 0.3) was added to each well. At regular intervals, aliquots were withdrawn to assay the amount of bacteria present. As shown in Fig. 5, the fPDCPD–chloramphenicol conjugate reduced the growth of the bacteria, relative to controls lacking the chloramphenicol group.
Chloramphenicol is bacteriostatic to E. coli at concentrations lower than 500 μg mL−1, meaning that it slows or inhibits growth but does not induce cell death.55 At concentrations greater than 500 μg mL−1, on the other hand, bacteriocidal effects begin to dominate, resulting in cell lysis. In a positive control experiment (Fig. 5, green squares), 1 mg mL−1 chloramphenicol was shown to fully inhibit the growth of E. coli K12 under identical conditions to those described above (i.e. 1 mL E. coli at 0.3 initial OD600, in the same 24-well plate). The use of the methyl ester-functionalized surface (4; open circles) or the saponified surface (5; orange triangles) resulted in no detectable reduction in bacterial growth relative to the use of either the methacrylated glass slide (red crosses) or the unmodified 24-well plate with no glass slide present (pink circles).
The sigmoidal growth curve obtained for E. coli incubated over the fPDCPD–chloramphenicol surface (data indicated with blue diamonds in Fig. 5) is consistent with the proposed mechanism outlined in Fig. 2. At early time points (<1 h), readily available chloramphenicol (perhaps held on the surface through physical interactions rather than through chemical bonds, or perhaps especially susceptible to secreted bacterial esterases) slows, but does not completely eliminate, the growth of the bacteria. At some stage, however, this modest growth rate produces enough bacteria that the organism is able to overwhelm the small amount of chloramphenicol that is present and grow at similar rates to those observed in the negative controls (1–1.5 h). This increase in bacteria, however, means that more bacterial esterase enzymes are present. These begin to liberate surface-bound chloramphenicol through enzymatic hydrolysis. The liberated chloramphenicol eventually reaches sufficiently high concentrations (>1.5 h) that it can almost fully retard any further growth of the bacteria. Importantly, this reduction in growth (at about an OD600 of 0.6) occurred well below the concentration of bacteria required to limit growth in negative control samples (OD600 ≈ 1.2) due to crowding or nutrient depletion.
While rigorous proof for the mechanism outlined above is beyond the scope of the current work, the fact that the fPDCPD–chloramphenicol conjugate clearly acts to slow the growth of bacteria presents a compelling illustration of the potential utility of rationally installed polydicyclopentadiene surface functionalization.
Conclusions
The addition of functionality to commodity polymers can facilitate the development of new applications. We harnessed the ester group present on a novel functionalized form of dicyclopentadiene to attach a wide range of functional groups. The installation of polar or lipophilic chains to the ester resulted in predictable changes to surface hydrophobicity, while conjugation of a TAMRA dye afforded a red-fluorescent surface that could be used to confirm the extent of surface coverage. Building upon these results, we attached an RGD peptide known to interact with cellular integrin receptors, and showed that the resulting fPDCPD conjugate led to enhanced adhesion of mammalian cells, relative to a commercial tissue culture plate. Other polymer surfaces – including a control surface incorporating an RAD peptide – led to moderate levels of cell adhesion, while incorporation of a perfluorooctyl ester group almost completely abolished cell attachment. Together, these data indicate an ability to rationally and predictably tune the fPDCPD surface to either enhance or reduce interactions with biological organisms. To further demonstrate the utility of the fPDCPD polymer, we engineered a self-dosing chloramphenicol conjugate that was capable of releasing antibiotic in the presence of bacteria. As designed, this surface limited the growth of E. coli, relative to appropriate controls.Experimental
Materials
Ester-functionalized monomer 1 was prepared as described previously.20 2nd Generation Grubbs catalyst was purchased from Chem-Impex. RGD and RAD cyclic peptides were purchased from Peptides International. 5-TAMRA-PEO3-amine was purchased (as the TFA salt) from ChemoMetec. RPMI1640 media, fetal bovine serum and trypsin–EDTA were purchased from Thermo Fisher. E. coli K12 was purchased from ATCC. Chloramphenicol and all other reagents were sourced from MilliporeSigma. THF was freshly distilled over sodium and benzophenone prior to use; all other reagents were used without further purification. Matsunami round glass cover slides and 24-well plasma-treated cell culture plates were both purchased from VWR.Instrumentation
All NMR spectra were recorded at ambient temperature (298–300 K). 1H NMR spectra were recorded at 300.27 MHz on a Bruker AVANCE 300 spectrometer equipped with a 5 mm PABBO BB-1H/D Z-GRD probe. 1H chemical shifts (δ) are reported in parts-per-million (ppm) relative to tetramethylsilane. NMR data is presented as follows: chemical shift, multiplicity (s = singlet, d = doublet, t = triplet, br = broad, m = multiplet), coupling constants (J, reported in Hz), integration. Infrared spectra were obtained using a PerkinElmer ATR spectrometer. IR wavenumbers (ν) are reported in units of cm−1. Fluorescent imaging and cell counting was accomplished using a Cytation 5 multichannel imaging platereader. Automated agitation of multiwell plates was done using the plate shaker function on a SpectraMax M5 multichannel platereader. Mammalian cell counts were carried out using the standard Cytation 5 Imaging Reader software (v. 3.0.3), using a digital phase contrast of 30 μM, deconvolution of 1 SD, kernal radius of 20 px, cell radius parameters 11 μM minimum to 50 μM maximum, and a threshold setting of 6000. Optical density readings for bacteria were conducted on a SpectraMax M5 platereader. Contact angle measurements were obtained with a Holmarc contact angle meter (HO-IAD-CAM-01) and images were processed using the instrument's standard software package.Generation of polymers and polymer surfaces ring opening metathesis polymerization
Polymer 2 and the linear polymer precursors to 8 and 9 were prepared by adding 1 mol% Grubbs 2nd generation catalyst to 50 mg of the appropriate monomer in 1.0 g dichloromethane. Polymerization was allowed to proceed for 1 h, after which NMR analysis indicated complete consumption of monomer. The reaction was quenched by the addition of 0.5 mL ethyl vinyl ether, and the polymer product was precipitated by the addition of hexanes. Removal of solvent via centrifugation (3750 rpm at 4 °C) followed by drying in vacuo afforded the target linear polymers. Linear polymer 2 (precursor to crosslinked polymer 4): 1H NMR (300 MHz, CDCl3) δ 6.68–6.52 (m, 1H), 5.55–5.27 (m, 2H), 3.75 (br s, 3H), 3.46–3.33 (m, 1H), 3.01–2.84 (m, 2H), 2.76–2.46 (m, 3H), 1.79–1.61 (br, 1H), 1.39–1.20 (br, 1H);20 linear precursor to crosslinked polymer 8: 1H NMR (300 MHz, CDCl3) δ 6.66–6.50 (m, 1H), 5.61–5.24 (m, 2H), 4.20–4.05 (m, 2H), 3.46–3.29 (m, 1H), 3.09–2.83 (m, 2H), 2.76–2.44 (m, 3H), 1.78–1.59 (m, 3H), 1.44–1.16 (m, 11H), 0.88 (br t, 3H); linear precursor to crosslinked polymer 9: 1H NMR (300 MHz, CDCl3) δ 6.70–6.53 (m, 1H), 5.56–5.24 (m, 2H), 4.40–4.19 (m, 2H) 3.81–3.52 (m, 14H), 3.47–3.30 (m, 1H), 3.11–2.77 (m, 3H), 2.77–2.41 (m, 3H), 1.79–1.59 (m, 1H), 1.38–1.17 (m, 1H). For a discussion of fPDCPD molecular weight and polydispersity, see ref. 18. Complete NMR assignments of 1H and 13C shifts for polymer 2 can be found in ref. 20; spectra for the linear precursors to 8 and 9 can be found in the ESI.‡Preparation & methacrylation of glass cover slides
Matsunami 15 mm micro glass cover slides (chosen to fit precisely into the wells of a standard 24-well plate) were sonicated in a solution of Sparkleen detergent and deionized H2O for one hour with occasional stirring. The slides were then thoroughly rinsed with deionized H2O, followed by 95% ethanol and then diethyl ether. Following evaporation of residual solvent at room temperature, the slides were then dried in an oven set to 110 °C. The glass slides were then evenly spread in a glass Petri dish and incubated in a mixture of ethanol, acetic acid and 3-(trimethoxysilyl)propyl methacrylate for three minutes. The incubation medium was prepared by first diluting 1 part glacial acetic acid in 9 parts deionized water, then diluting 3 mL of this dilute acid solution with 96.5 mL of 95% ethanol. To the resulting AcOH/EtOH solution was added 0.5 mL of the methacrylate reagent. After incubation, the slides were then thoroughly rinsed with 95% ethanol and dried in vacuo.fPDCPD spin coating and thermal crosslinking to afford 4, 8 or 9
A 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 solution of toluene:CH2Cl2 containing 4 wt% of linear polymer 2 (or for the experiments in Fig. 3, the linear precursors to 8 or 9; refer to the ESI‡ for details relating to the monomer synthesis, and the section above for polymerization details) was prepared at room temperature in a sintered vial, using sonication to fully dissolve the polymer. 50 μL of the solution was then spin coated onto the methacrylated 15 mm round cover slides using a maximum speed of 3000 rpm, acceleration of 1000 rpm s−1 and a total application time of 30 seconds. The freshly spin-coated slides were then placed on glass Petri dishes and crosslinked at 135 °C for 48 h prior to use. For optimization details of the spin-coating protocol, see Table S1 in the ESI.‡
1 solution of toluene:CH2Cl2 containing 4 wt% of linear polymer 2 (or for the experiments in Fig. 3, the linear precursors to 8 or 9; refer to the ESI‡ for details relating to the monomer synthesis, and the section above for polymerization details) was prepared at room temperature in a sintered vial, using sonication to fully dissolve the polymer. 50 μL of the solution was then spin coated onto the methacrylated 15 mm round cover slides using a maximum speed of 3000 rpm, acceleration of 1000 rpm s−1 and a total application time of 30 seconds. The freshly spin-coated slides were then placed on glass Petri dishes and crosslinked at 135 °C for 48 h prior to use. For optimization details of the spin-coating protocol, see Table S1 in the ESI.‡
Saponification of fPDCPD surfaces to afford 5
Freshly crosslinked methyl ester-functionalized fPDCPD glass cover slides (4) were evenly placed on a glass Petri dish and submerged in a 7 wt% solution of NaOH in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 H2O
1 H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH for 30 min. The surfaces were then washed with water, followed by methanol, and finally dried in vacuo. FTIR analysis confirmed partial hydrolysis of the surface-accessible ester groups (Fig. S12, ESI‡).
MeOH for 30 min. The surfaces were then washed with water, followed by methanol, and finally dried in vacuo. FTIR analysis confirmed partial hydrolysis of the surface-accessible ester groups (Fig. S12, ESI‡).
Formation of acyl chloride fPDCPD surfaces (6)
Freshly saponified surfaces (5) were separated into sintered vials sealed with inverted septa and parafilm. Under an atmosphere of argon, 1 mL of a solution containing 80% CH2Cl2, 10% DMF, and 10% SOCl2 was injected into each vial. The reaction mixture was left at room temperature for 2 h, after which the surfaces were washed with CH2Cl2 and dried in vacuo.Synthesis of TAMRA-functionalized fPDCPD (7)
Freshly prepared acyl chloride surfaces (6) were separated into sintered vials sealed with inverted septa and parafilm. Under an atmosphere of argon, surfaces were submerged in 1 mL of a solution containing 0.15 mg mL−1 of 5-TAMRA-PEO3-amine (from 110 μL of a 1.43 mg mL−1 DMSO stock solution), 490 μL DMSO and 400 μL triethylamine. After 48 h at room temperature, the slides were removed from the vials, washed three times with DMSO and placed into a 24-well plate for fluorescent imaging. FTIR analysis confirmed the formation of the target amide (Fig. S14, ESI‡).Synthesis of chloramphenicol-functionalized fPDCPD
Freshly prepared acyl chloride surfaces (6) were separated into sintered vials sealed with inverted septa and parafilm. Under an atmosphere of argon, surfaces were submerged in 1 mL of a solution containing 400 μL of triethylamine, 600 μL of DMSO, and 10 mg of chloramphenicol. After 48 h at room temperature, the slides were removed from the vials, washed with acetonitrile followed by CH2Cl2, and dried in vacuo. FTIR analysis confirmed surface functionalization (Fig. S15, ESI‡).Formation of surfaces for HeLa cell adhesion assays
Methacrylated glass cover slides, fPDCPD–CO2Me-coated cover slides (4) and saponified fPDCPD-coated cover slides (5) were prepared as described above. fPDCPD–perfluorooctyl ester-coated cover slides, as well as fPDCPD–RGD- and fPDCPD–RAD-coated cover slides, were prepared through an analogous procedure to that used for 7 (i.e. acyl chloride surfaces were exposed to either perfluorooctanol or else RGD or RAD amines, in the presence of triethylamine). All surfaces were disinfected with 70% ethanol prior to being brought into a biological safety cabinet, and then were washed with phosphate-buffered saline (PBS) and air-dried for 20 min. Clear LePage Speed Set Epoxy was then applied to the bottom of each well of sterile 24-well tissue culture plates. The prepared glass cover slides were laid over top of the epoxy and light pressure was applied to ensure strong adhesion. The epoxy was left to cure for 30 minutes prior to addition of cells.Measurement of surface properties
Conflicts of interest
The authors declare the following competing financial interest: J. W., T. L., T. J. C. and C. L. are co-inventors on US patent application no. 15/999209, which claims the use of the polymer described herein.Acknowledgements
This work was supported by an ACS PRF award (#57150-ND7) to J. W. The authors thank NSERC of Canada for additional financial support and the Canada Research Chairs program for a salary award to J. W. In addition, the authors thank Rebecca Hof for assistance with the biological and imaging experiments.References
- (a) S. Kovačič and C. Slugovc, Mater. Chem. Front., 2020, 4, 2235–2255 RSC; (b) D. Vervacke, An Introduction to PDCPD, Product Rescue, Waarschoot, 2008 Search PubMed.
- J. C. Mol, J. Mol. Catal. A: Chem., 2004, 213, 39–45 CrossRef CAS.
- (a) L. Grabowski, A. Baier and M. Sobek, IOP Conf. Ser.: Mater. Sci. Eng., 2017, 227, 012051 Search PubMed; (b) M. Placzek, A. Wróbel and A. Baier, IOP Conf. Ser.: Mater. Sci. Eng., 2017, 227, 012095 Search PubMed.
- (a) D. W. Klosiewicz, Method for Making a Dicyclopentadiene Thermoset Polymer, US Pat., 4400340, 1983 Search PubMed; (b) N. R. Newburg, Method for Making Thermoset Poly(dicyclopentadiene) and the Product so Produced, US Pat., 4481344, 1984 Search PubMed.
- B. Autenrieth, H. Jeong, W. P. Forrest, J. C. Axtell, A. Ota, T. Lehr, M. R. Buchmeiser and R. R. Schrock, Macromolecules, 2015, 48, 2480–2492 CrossRef CAS.
- MatWeb overview of material properties for polydicyclopentadiene: http://www.matweb.com/search/DataSheet.aspx? MatGUID = 16d3d6b1e32c4c368fa1ddac6afb2b93.
- S. M. Doughty, G. Recher and Y. S. Yang, Kunstoffe, 1992, 82, 1191–1196 CAS.
- (a) D. B. Knorr, K. A. Masser, R. M. Elder, T. W. Sirk, M. D. Hindenlang, J. H. Yu, A. D. Richardson, S. E. Boyd, W. A. Spurgeon and J. L. Lenhart, Compos. Sci. Technol., 2015, 114, 17–25 CrossRef CAS; (b) R. M. Elder, D. B. Knorr, J. W. Andzelm, J. L. Lenhart and T. W. Sirk, Soft Matter, 2016, 12, 4418–4434 RSC.
- K. A. M. Vallons, R. Drozdzak, M. Charret, S. V. Lomov and I. Verpoest, Composites, Part A, 2015, 78, 191–200 CrossRef CAS.
- Y. Hu, A. W. Lang, X. Li and S. R. Nutt, Polym. Degrad. Stab., 2014, 110, 464–472 CrossRef CAS.
- Z. He, J. Sun, L. Zhang, Y. Wang, H. Ren and J.-B. Bao, Polymer, 2018, 153, 287–294 CrossRef CAS.
- J. K. Lee and G. L. Gould, J. Sol-Gel Sci. Technol., 2007, 44, 29–40 CrossRef CAS.
- C. Dawedeit, S. H. Kim, T. Braun, M. A. Worsley, S. A. Letts, K. J. Wu, C. C. Walton, A. A. Chernov, J. H. Satcher, A. V. Hamza and J. Biener, Soft Matter, 2012, 8, 3518–3521 RSC.
- S. R. White, N. R. Sottos, P. H. Geubelle, J. S. Moore, M. R. Kessler, S. R. Sriram, E. N. Brown and S. Viswanathan, Nature, 2001, 409, 794–797 CrossRef CAS PubMed.
- E. N. Brown, S. R. White and N. R. Sottos, J. Mater. Sci., 2004, 39, 1703–1710 CrossRef CAS.
- I. M. Radovic, D. B. Stojanovic, A. Kojovic, M. Petrovic, P. S. Uskokovic, V. J. Radojevic and R. R. Aleksic, J. Compos. Mater., 2017, 51, 3003–3016 CrossRef CAS.
- For examples of post-polymerization functionalization of PDCPD through reaction of residual olefins, see: (a) M. Perring and N. B. Bowden, Langmuir, 2008, 24, 10480–10487 CrossRef CAS PubMed; (b) M. Perring, T. R. Long and N. B. Bowden, J. Mater. Chem., 2010, 20, 8679–8685 RSC; (c) S. Kovačič, P. Krajnc and C. Slugovc, Chem. Commun., 2010, 46, 7504–7506 RSC; (d) A.-C. Knall, S. Kovačič, M. Hollauf, D. Reishofer, R. Saf and C. Slugovc, Chem. Commun., 2013, 49, 7325–7327 RSC; (e) C. L. McGann, G. C. Daniels, S. L. Giles, R. B. Balow, J. L. Miranda-Zayas, J. G. Lundin and J. H. Wynne, Macromol. Rapid Commun., 2018, 39, 1800194 CrossRef PubMed.
- J. Chen, F. P. Burns, M. G. Moffitt and J. E. Wulff, ACS Omega, 2016, 1, 532–540 CrossRef CAS PubMed.
- T. J. Cuthbert, T. Li, A. W. H. Speed and J. E. Wulff, Macromolecules, 2018, 51, 2038–2047 CrossRef CAS.
- T. J. Cuthbert, T. Li and J. E. Wulff, ACS Appl. Polym. Mater., 2019, 1, 2460–2471 CrossRef CAS.
- For alternative approaches to functionalized versions of polydicyclopentadiene, see: (a) L. Gong, K. Liu, E. Ou, F. Xu, Y. Lu, Z. Wang, T. Gao, Z. Yang and W. Xu, RSC Adv., 2015, 5, 26185–26188 RSC; (b) S. Saha, Y. Ginzburg, I. Rozenberg, O. Iliashevsky, A. Ben-Asuly and N. G. Lemcoff, Polym. Chem., 2016, 7, 3071–3075 RSC; (c) R. S. Phatake, A. Masarwa, N. G. Lemcoff and O. Reany, Polym. Chem., 2020, 11, 1742–1751 RSC.
- Z. Ma, Z. Mao and C. Gao, Colloids Surf., B, 2007, 60, 137–157 CrossRef CAS.
- N. F. Owens, D. Gingell and P. R. Rutter, J. Cell Sci., 1987, 87, 667–675 CAS.
- J. M. Goddard and J. H. Hotchkiss, Prog. Polym. Sci., 2007, 32, 698–725 CrossRef CAS.
- U. Hersel, C. Dahmen and H. Kessler, Biomaterials, 2003, 24, 4385–4415 CrossRef CAS PubMed.
- W. Tong, X. Yao, S. Duan, B. Yu, X. Ding, X. Ding and F.-J. Xu, Langmuir, 2020, 36, 354–361 CrossRef CAS PubMed.
- T. Çaykara, M. G. Sande, N. Azoia, L. R. Rodrigues and C. J. Silva, Med. Microbiol. Immunol., 2020, 209, 363–372 CrossRef.
- Y. Wang, J. Wu, D. Zhang, F. Chen, P. Fan, M. Zhong, S. Xiao, Y. Chang, X. Gong, J. Yang and J. Zheng, J. Mater. Chem. B, 2019, 7, 5762–5774 RSC.
- A. Asadinezhad, I. Novák, M. Lehocký, V. Sedlařík, A. Vesel, I. Junkar, P. Sáha and I. Chodák, Plasma Processes Polym., 2010, 7, 504–514 CrossRef CAS.
- J. Hou, T. Liu, R. Chen, J. Liu, J. Chen, C. Zhao, L. Yin, C. Li, X. Xu, Q. Shi and J. Yin, Chem. Commun., 2017, 53, 6708–6711 RSC.
- Z. Zhou, P. Yu, H. M. Geller and C. K. Ober, Biomacromolecules, 2013, 14, 529–537 CrossRef CAS PubMed.
- E. K. Riga, M. Vöhringer, V. T. Widyaya and K. Lienkamp, Macromol. Rapid Commun., 2017, 38, 1700216 CrossRef PubMed.
- T. Rohr, D. F. Ogletree, F. Svec and J. M. J. Fréchet, Adv. Funct. Mater., 2003, 13, 264–270 CrossRef CAS.
- J. Lahann, M. Balcells, H. Lu, T. Rodon, K. F. Jensen and R. Langer, Anal. Chem., 2003, 75, 2117–2122 CrossRef CAS PubMed.
- Y. Cheng, Y. Yu, F. Fu, J. Wang, L. Shang, Z. Gu and Y. Zhao, ACS Appl. Mater. Interfaces, 2016, 8, 1080–1086 CrossRef CAS.
- P. Y. W. Dankers, M. C. Harmsen, L. A. Brouwer, M. J. A. Van Luyn and E. W. Meijer, Nat. Mater., 2005, 4, 568–574 CrossRef CAS.
- M. Wang, L. J. Chen, J. Ni, J. Weng and C. Y. Yue, J. Mater. Sci.: Mater. Med., 2001, 12, 855–860 CrossRef CAS.
- S. Stratton, N. B. Shelke, K. Hoshino, S. Rudraiah and S. G. Kumbar, Bioact. Mater., 2016, 1, 93–108 CrossRef PubMed.
- P. T. Wong and S. K. Choi, Chem. Rev., 2015, 115, 3388–3432 CrossRef CAS PubMed.
- E. M. H. Wellington, A. B. A. Boxall, P. Cross, E. J. Feil, W. H. Gaze, P. M. Hawkey, A. S. Johnson-Rollings, D. L. Jones, N. M. Lee, W. Otten, C. M. Thomas and A. P. Williams, Lancet Infect. Dis., 2013, 13, 155–165 CrossRef CAS.
- E. Wistrand-Yuen, M. Knopp, K. Hjort, S. Koskiniemi, O. G. Berg and D. I. Andersson, Nat. Commun., 2018, 9, 1599 CrossRef.
- E. M. Larsen and R. J. Johnson, Drug Dev. Res., 2019, 80, 33–47 CrossRef CAS.
- M. J. Kasten, Mayo Clin. Proc., 1999, 74, 825–833 CrossRef CAS PubMed.
- F. E. Hahn, Chloramphenicol, in Modes and Mechanisms of Microbial Growth Inhibitors, ed. F. E. Hahn, Antibiotics, Springer, Heidelberg, 1983, vol. 6 Search PubMed.
- World Health Organization Model List of Essential Medicines, 21st List, Geneva, 2019.
- V. S. Malik, Adv. Appl. Microbiol., 1972, 15, 297–336 CAS.
- U. T. Bornscheuer, FEMS Microbiol. Rev., 2002, 26, 73–81 CrossRef CAS PubMed.
- For an alternative strategy to prepare an antimicrobial form of PDCPD, see: M. Abbas, A. Leitgeb, J. Kienberger and C. Slugovc, J. Chem. Soc. Pak., 2013, 35, 359–362 Search PubMed.
- Wagener showed that metathesis crosslinks are generally not important for unfunctionalized polydicyclopentadiene that is polymerized using early transition metal catalysts; see ref. 50. However, Ru catalysts can support the metathesis of cyclopentenes (see ref. 51), which makes metathesis crosslinks relevant for polydicyclopentadiene prepared using Grubbs-type catalysts (see ref. 52). For the most compelling spectroscopic evidence of metathesis crosslinks in Ru-polymerized PDCPD, see ref. 53.
- (a) T. A. Davidson and K. B. Wagener, J. Mol. Catal. A: Chem., 1998, 133, 67–74 CrossRef CAS; (b) T. A. Davidson, K. B. Wagener and D. B. Priddy, Macromolecules, 1996, 29, 786–788 CrossRef CAS.
- (a) C. Mugemana, K. V. Bukhryakov, O. Bertrand, K. B. Vu, J.-F. Gohy, N. Hadjichristidis and V. O. Rodionov, Polym. Chem., 2016, 7, 2923–2928 RSC; (b) K. J. Ivin and J. C. Mol, Monocyclic Alkenes and Polyenes in Olefin Metathesis and Metathesis Polymerization, Academic Press, Cambridge, 1997, pp. 260–287 Search PubMed; (c) R. Tuba and R. H. Grubbs, Polym. Chem., 2013, 4, 3959–3962 RSC.
- (a) N. D. Steese, D. Barvaliya, X. D. Poole, D. E. McLemore, J. C. DiCesare and H.-J. Schanz, J. Polym. Sci., Part A: Polym. Chem., 2018, 56, 359–364 CrossRef CAS; (b) S. Y. Lee, J. Lyu, S. Kang, S. J. Lu and C. W. Bielawski, J. Phys. Chem. C, 2018, 122, 11855–11861 CrossRef CAS.
- P. Shieh, W. Zhang, K. E. L. Husted, S. L. Kristufek, B. Xiong, D. J. Lundberg, J. Lem, D. Veysset, Y. Sun, K. A. Nelson, D. L. Plata and J. A. Johnson, Nature, 2020, 583, 542–547 CrossRef CAS PubMed.
- Z. Ma, M. Kotaki and S. Ramakrishna, J. Membr. Sci., 2006, 272, 179–187 CrossRef CAS.
- J. J. Rahal and M. S. Simberkoff, Antimicrob. Agents Chemother., 1979, 16, 13–18 CrossRef CAS PubMed.
Footnotes |
| † We dedicate this work to the memory of Dr Chang Liu. |
| ‡ Electronic supplementary information (ESI) available: Detailed experimental procedures and additional characterization data. See DOI: 10.1039/d0ma00480d |
| This journal is © The Royal Society of Chemistry 2020 |