Open Access Article
Open Access ArticleCell adhesion and proliferation on common 3D printing materials used in stereolithography of microfluidic devices
Kati
Piironen
 a,
Markus
Haapala
a,
Markus
Haapala
 a,
Virpi
Talman
a,
Virpi
Talman
 b,
Päivi
Järvinen
b,
Päivi
Järvinen
 a and
Tiina
Sikanen
a and
Tiina
Sikanen
 *a
*a
aFaculty of Pharmacy, Drug Research Program, Division of Pharmaceutical Chemistry and Technology, University of Helsinki, Viikinkaari 5E, FI-00014, Finland. E-mail: tiina.sikanen@helsinki.fi; Tel: +358 2941 59173
bFaculty of Pharmacy, Drug Research Program, Division of Pharmacology and Pharmacotherapy, University of Helsinki, Viikinkaari 5E, FI-00014, Finland
First published on 1st June 2020
Abstract
Three-dimensional (3D) printing has recently emerged as a cost-effective alternative for rapid prototyping of microfluidic devices. The feature resolution of stereolithography-based 3D printing is particularly well suited for manufacturing of continuous flow cell culture platforms. Poor cell adhesion or material-induced cell death may, however, limit the introduction of new materials to microfluidic cell culture. In this work, we characterized four commercially available materials commonly used in stereolithography-based 3D printing with respect to long-term (2 month) cell survival on native 3D printed surfaces. Cell proliferation rates, along with material-induced effects on apoptosis and cell survival, were examined in mouse embryonic fibroblasts. Additionally, the feasibility of Dental SG (material with the most favored properties) for culturing of human hepatocytes and human-induced pluripotent stem cells was evaluated. The strength of cell adhesion to Dental SG was further examined over a shear force gradient of 1–89 dyne per cm2 by using a custom-designed microfluidic shear force assay incorporating a 3D printed, tilted and tapered microchannel sealed with a polydimethylsiloxane lid. According to our results, autoclavation of the devices prior to cell seeding played the most important role in facilitating long-term cell survival on the native 3D printed surfaces with the shear force threshold in the range of 3–8 dyne per cm2.
Introduction
Three-dimensional (3D) printing has recently emerged as a cost-effective alternative to rapid prototyping of microfluidic devices.1 The relatively low-cost of commercially available 3D printers of different type (e.g., fused deposition modeling, FDM,2,3 and stereolithography, SLA,2,4), their user-friendly interfaces, and the wide range of materials has facilitated straightforward manufacturing of 3D microstructures. Compared with lithography-based microfabrication (e.g., etching of silicon and glass and SU-8 photolithography) and polymer replication (e.g., soft lithography of polydimethylsiloxane (PDMS), UV and hot embossing, and injection molding), direct 3D printing of microfluidic devices benefits from significantly reduced maintenance costs (no cleanroom requirement) while enabling increased structural complexity, especially in terms of implementation of out-of-the-plane 3D microstructures.5,6 Apart from PDMS adhesive bonding, microchannel sealing is also a challenge for most of the manufacturing materials and methods, which can be resolved by direct 3D printing of enclosed microstructures.5,6 However, the feature resolution is largely dependent of the chosen 3D printing technique and typically, compromises will have to be made between high-resolution vs. printing volume and time. For example, two photon polymerization methods1,7 provide 3D structures with sub-micrometer resolution, but their printing volume is typically below cubic centimeter. Most commercially available benchtop 3D printers employ FDM2,3 or SLA2,4 techniques, which enable fabrication of centimeter-scale structures, but fail to reproduce sub-micrometer features. The achievable feature resolution of SLA is superior to FDM, but depends strongly on the type of the light source. SLA printers based on scanned laser spot routinely reproduce feature resolution in the range of 100 μm, whereas with digital light projection record feature resolution as good as 18 × 20 μm2 (enclosed microchannels) has been reported.8 Overall, SLA techniques are a good match with many microfluidic assays that do not require sub-micrometer structures, such as organ-on-a-chip platforms.Until now 3D printing has been successfully applied to fabrication of several organ-on-a-chip9,10 and organoid-on-a-chip11 assays as well as bacterial cultivation platforms.12 The organ-on-a-chip assays are generally seen as a promising alternative to replace animal experiments in drug screening13 and chemical toxicity testing.14 However, it is critical to distinguish and eliminate the possible material-induced toxic impacts from those of the test chemicals. Prior work on zebrafish embryos15 and early bovine embryos16 indicates that chemical residues leaching from 3D printing materials, including monomers, photoinitiators and plasticizers, may be associated with genotoxicity and developmental abnormalities, but the toxicity depends largely on the chosen manufacturing material. For example, polylactic acid commonly used in FDM is generally known to be biocompatible, whereas SLA and other UV polymerization techniques typically employ resins and photoinitiators that are prominently cytotoxic.17–20 SLA prints were also considerably more toxic to zebrafish embryos compared with FLM prints.15 The biocompatibility of the chosen material and the absence of material-induced adverse effects must therefore be comprehensively established before introducing new materials to organ-on-a-chip development. Some of these effects may not be observed and their impact not understood, if cell viability is only monitored for relatively short-term (24–72 h) as is typical for many standardized biocompatibility tests (e.g., ISO 10993 series). As a result, compliance to these standards gives little or no information on cell proliferation capability when in direct contact with the tested material. Understanding and eliminating the material-induced adverse impacts on cell proliferation is particularly crucial for setting up of 3D organ-on-a-chip cultures when long-term culturing is a prerequisite, as in microfluidic spheroid and organoid models.
In this study, we assessed the biocompatibility of four different commercially available materials, which are commonly used in SLA based 3D printing, with respect to both acute toxicity and long-term cell proliferation for up to 56 days on the native 3D printed surface. For this purpose, we used BALB/c mouse embryonic fibroblasts (3T3), which is the recommended cell line for toxicity testing by the European Reference Laboratory.21 The cell-compatibility of the most feasible material (Dental SG) was also assessed with human hepatoma cells (Huh7) and human induced pluripotent stem cells (hiPSC). In addition, a 3D printed, microfluidic shear force gradient assay was developed to enable distinguishing the impact of surface topography on cell adhesion onto the 3D printed surfaces at different level of shear stress. SLA was chosen as the manufacturing method, because of the affordability and relatively good feature resolution of the commercial benchtop printers, which have substantially increased the use of SLA in rapid prototyping of microfluidic devices in many research laboratories. Depending on the material of choice, SLA also enables manufacturing of optically clear microstructures and has high throughput in terms of printing speed vs. achievable resolution.
Experimental
3D printing and device design
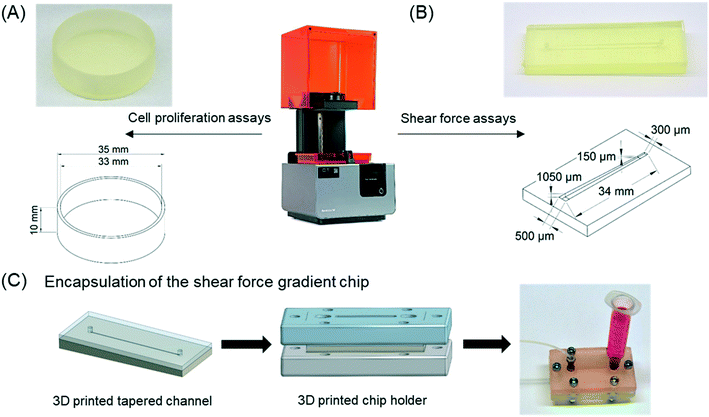
The materials used for 3D printing included four stereolithography resins (Clear, High Temp, Dental SG, and Dental LT Clear) from Formlabs (Somerville, MA). Two of the materials were autoclavable (Dental SG and High Temp), two were biocompatible according to ISO 10993 standard (Dental SG and Dental LT Clear), and one was neither autoclavable nor biocompatible (Clear). All designs were 3D printed on support structures at an angle of 45° using form 2 stereolithography printer and PreForm software (Formlabs). The layer height was 50 μm for Clear, High Temp, and Dental SG resins and 100 μm for Dental LT Clear (in accordance with the printer specifications). After printing, the 3D parts were washed twice with isopropanol (≥99.9%, Sigma Aldrich, Steinheim, Germany), let dry, and post-cured by UV at 60 °C for 15 min, as recommended by the material supplier (Formlabs). The UV cure was performed by using a custom-built dual LED light source (ca. 5 mW cm−2 at 365 nm and 30 mW cm−2 at 405 nm).In this study, two different designs of 3D printed platforms were manufactured, including round dishes (diameter 35 mm, height 10 mm) for cell proliferation experiments under static conditions (Fig. 1A) and 34 mm-long tapered microchannels featuring linearly decreasing width (from 500 to 300 μm) and height (from 1050 to 150 μm) for cell adhesion experiments under through-flow (Fig. 1B). The decreasing microchannel height was realized by printing horizontally aligned channel (bottom) with tilted top surface (Fig. 1B). Prior to use, the 3D printed microchannels were sealed with a PDMS lid. The PDMS lid was made from Sylgard 184 elastomer (Dow Chemical Company, Midland, MI), mixed with the curing agent in a ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and partially cured (ca. 50 min at 55 °C) to achieve a crosslinked PDMS layer with sticky surface so as to facilitate tight sealing of the relatively rough 3D printed surface. To ensure that no oligomers leach out of the PDMS cover layer, further curing was performed at 70 °C for 24 h after bonding, while simultaneously applying an external weight on top of the bonded chip. Before cell seeding, the 3D printed platforms were sterilized by rinsing with 70% (v/v) ethanol for 30 min. Alternatively, the 3D printed platforms made of High Temp and Dental SG were sterilized by autoclavation (saturated water steam, 121 °C, 2 bar, 15 min) using Getinge HS6610 steam sterilizer (Getinge, AB, Gothenburg, Sweden). Clear and Dental LT Clear materials did not tolerate such harsh conditions and became visibly cracked upon autoclavation.
1 and partially cured (ca. 50 min at 55 °C) to achieve a crosslinked PDMS layer with sticky surface so as to facilitate tight sealing of the relatively rough 3D printed surface. To ensure that no oligomers leach out of the PDMS cover layer, further curing was performed at 70 °C for 24 h after bonding, while simultaneously applying an external weight on top of the bonded chip. Before cell seeding, the 3D printed platforms were sterilized by rinsing with 70% (v/v) ethanol for 30 min. Alternatively, the 3D printed platforms made of High Temp and Dental SG were sterilized by autoclavation (saturated water steam, 121 °C, 2 bar, 15 min) using Getinge HS6610 steam sterilizer (Getinge, AB, Gothenburg, Sweden). Clear and Dental LT Clear materials did not tolerate such harsh conditions and became visibly cracked upon autoclavation.
Cell cultures
The cell cultures used in the study included BALB/c mouse embryonic fibroblast (3T3) and human hepatoma (Huh7) cell lines, and human induced pluripotent stem cells (hiPSC).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10.
10.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10.
10.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 Matrigel (Corning, New York, NY) in knockout DMEM (Gibco) as described previously.22 For passaging, hiPSCs were detached using Versene (Life Technologies, Eugene, OR) and replated at 1
50 Matrigel (Corning, New York, NY) in knockout DMEM (Gibco) as described previously.22 For passaging, hiPSCs were detached using Versene (Life Technologies, Eugene, OR) and replated at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 in E8, which was supplemented with 10 μM Rho-associated, coiled-coil containing protein kinase inhibitor (Y-27632) for the first 24 h after seeding.
15 in E8, which was supplemented with 10 μM Rho-associated, coiled-coil containing protein kinase inhibitor (Y-27632) for the first 24 h after seeding.
Assessment of cell survival on 3D printed surfaces
The cell survival on 3D printed surfaces was assessed with respect to short term growth (7 days) and cumulative population doubling level (cPDL) over a period of 56 days using the 3T3 cell line. The cells were passaged every seven days by transferring them onto another new 3D printed dish of the same type (or onto another control dish). In addition, apoptosis and quantitative proliferation rate were assessed with all three cell types (3T3, Huh7, hiPSC). For this purpose, 3T3 and Huh7 were seeded on native, uncoated 3D printed surfaces. Instead the hiPSCs were seeded on 3D printed surfaces coated with diluted Matrigel solution (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 in knockout DMEM) so that a thin protein coating was formed on top of the 3D prints to support hiPSC growth. This is not expected to interfere with examination of the impact of possible leaching monomers and additives on hiPSC culture. Besides the 3D printed surfaces, cell survival tests were conducted on conventional plastic cell culture dishes (as controls).
50 in knockout DMEM) so that a thin protein coating was formed on top of the 3D prints to support hiPSC growth. This is not expected to interfere with examination of the impact of possible leaching monomers and additives on hiPSC culture. Besides the 3D printed surfaces, cell survival tests were conducted on conventional plastic cell culture dishes (as controls).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per dish, seeding density 0.1 × 106 cells per cm2) were seeded in triplicate and the total cell number per dish was calculated every 24 hours for 6 days. For counting the cells, a 10 μL aliquot of the detached cell suspension was stained with Tryptan Blue (Thermo Fisher Scientific) and counted with hemocytometer.
000 cells per dish, seeding density 0.1 × 106 cells per cm2) were seeded in triplicate and the total cell number per dish was calculated every 24 hours for 6 days. For counting the cells, a 10 μL aliquot of the detached cell suspension was stained with Tryptan Blue (Thermo Fisher Scientific) and counted with hemocytometer.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per 3D printed dish, seeding density 0.05 × 106 cells per cm2) were seeded in triplicates and medium was replaced every two to three days. As the doubling time of 3T3 cells is ca. 21 h, the cells reach confluence in a few days. Therefore, the cells were passaged and transferred onto a new culture dish of the same type every 7 days to ensure they maintain the normal growth and space does not become the rate-limiting step. During passaging, the total cell number was counted and the cPDL was estimated using eqn (1):
000 cells per 3D printed dish, seeding density 0.05 × 106 cells per cm2) were seeded in triplicates and medium was replaced every two to three days. As the doubling time of 3T3 cells is ca. 21 h, the cells reach confluence in a few days. Therefore, the cells were passaged and transferred onto a new culture dish of the same type every 7 days to ensure they maintain the normal growth and space does not become the rate-limiting step. During passaging, the total cell number was counted and the cPDL was estimated using eqn (1):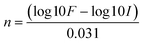 | (1) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per sample Accuri Flow Cytometer, BD Biosciences, Bedford, MA). The data analysis was done with FlowJo software (BD Biosciences).
000 cells per sample Accuri Flow Cytometer, BD Biosciences, Bedford, MA). The data analysis was done with FlowJo software (BD Biosciences).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per sample). The annexinV stain is the apoptosis marker on the cell membrane, whereas the PI stain can only enter the cells with compromised membrane integrity, in late stages of apoptosis. On this basis, annexinV+/PI+ (double positive) cells were concluded to be late apoptotic, annexinV+/PI- cells early apoptotic, and annexinV-/PI+ cells necrotic, whereas total annexinV+ (regardless of PI staining) corresponded to the total apoptotic cell population.
000 cells per sample). The annexinV stain is the apoptosis marker on the cell membrane, whereas the PI stain can only enter the cells with compromised membrane integrity, in late stages of apoptosis. On this basis, annexinV+/PI+ (double positive) cells were concluded to be late apoptotic, annexinV+/PI- cells early apoptotic, and annexinV-/PI+ cells necrotic, whereas total annexinV+ (regardless of PI staining) corresponded to the total apoptotic cell population.
Cell adhesion experiments in 3D printed channels
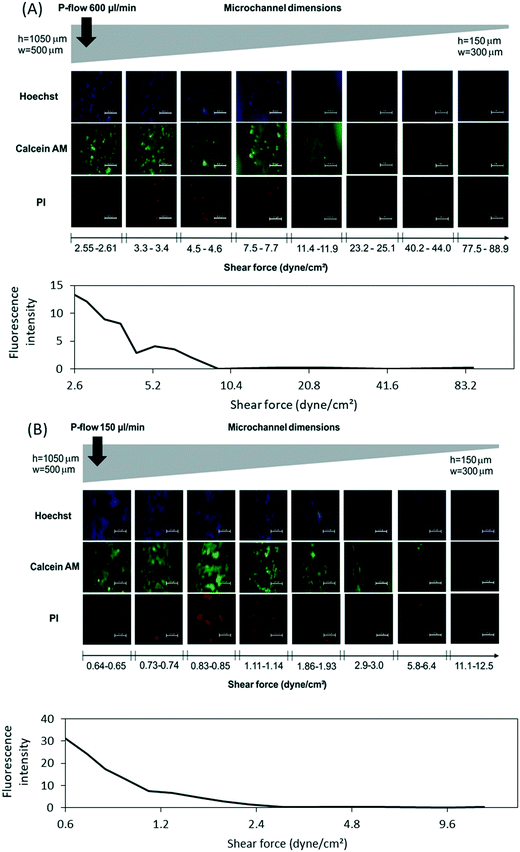
The cell adhesion strength under microfluidic flow was evaluated with 3T3 cells on Dental SG surfaces using tapered microchannels (Fig. 1B) encapsulated in a 3D printed chip holder made from mechanically rigid Clear resin (Fig. 1C). Linearly increasing shear stress was achieved by applying constant flow rate through the tapered (width 500 → 300 μm) and tilted (height 1050 → 150 μm) Dental SG microchannel. The 3T3 cells were seeded into the channel (2 × 106/mL) and allowed to adhere for 4 h prior to application of the flow. The applied volume flow rates were 600 μL min−1 and 150 μL min−1, which corresponded to shear force gradients of 1–89 and 0.22–22 dyne per cm2, respectively. After the experiments, the cells were stained with PI, calcein AM (Thermo Fisher Scientific), and Hoechst 33342 (NucBlue Live, ReadyProbes Reagent, Invitrogen, Thermo Fisher Scientific) to assess the impact of shear stress on 3T3 cell adhesion and viability. The fluorescence intensity of live cells (calcein AM) was quantified from the micrographs taken every 1000 μm along the channel by quantifying the intensity of grey levels using ImageJ.Optical and surface characterization
In addition to cell compatibility, both autoclaved and non-autoclaved Dental SG and High Temp materials were characterized for their autofluorescence and surface topography. Autofluorescence was determined from six locations across the plates at three excitation and emission wavelengths (355/455 nm, 488/555 nm, and 530/575 nm) corresponding to the commonly used fluorophores also employed in this study (Hoechst 33342, calcein AM, and PI), by using Varioskan LUX microtiter plate reader (Thermo Fisher Scientific, Waltham, MA). The topography of 3D printed surfaces was analyzed by FEI Quanta FEG scanning electron microscope (FEI, Hillsboro, OR) and by Dektak/XT profilometer (Bruker, Billerica, MA) using a 2 μm stylus tip and 10 mg stylus weight over 3 mm path in 60 s. In addition, the wettability differences between autoclaved and non-autoclaved Dental SG were determined based on water contact angle (WCA) measurement using a CAM200 Optical Contact Angle Meter (KSV instruments Ltd, Helsinki, Finland). In this case, Dental SG was spin coated (3000 rpm, 30 s) on a 3D printed Dental SG sheet and cured under UV-LED (450 nm, 60 s) to be able to create a smooth surface for WCA determination.Statistical analysis
Data are expressed as mean ± standard deviation (SD). Statistical analysis was performed using either one-way ANOVA with Bonferroni type-correction (between three groups) or the Student's t-test (between two groups), considering p < 0.05 as a significant difference.Results and discussion
Cell growth and proliferation on 3D printed surfaces
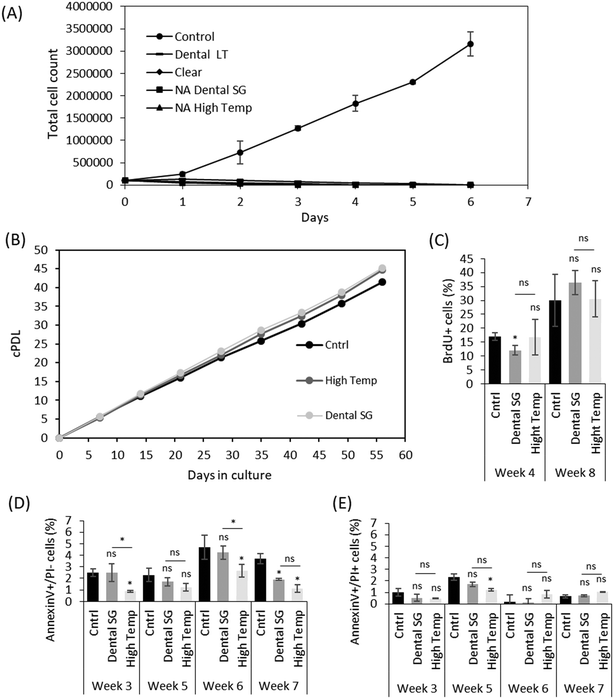
Despite the increasing use of 3D printing in the construction of microfluidic organ-on-a-chip platforms, and the wealth of material characterization data, relatively little is known about cell proliferation and long-term survival on commonly used 3D printing materials. Although material suppliers often provide standardized biocompatibility test results, many of the standards (such as the commonly used ISO 10993 series24,25) do not unequivocally specify the culturing conditions or time, but these depend on the targeted end use (e.g., medical or dental applications). Moreover, biocompatibility may be defined based on various different endpoints, such as cell growth, acute or chronic toxicity or genotoxicity. As a result, the certification does not imply overall biocompatibility and substantial differences in, for instance, cell proliferation may occur between the certified test laboratories and research use. Especially, the long-term material-induced impacts on cell proliferation may not be revealed in simplified, short-term cell viability tests, which was also shown in this study. The better sensitivity of chronic toxicity assays to low concentrations of toxicants has also been shown with doxorubicin, and 3i-1000, an experimental small molecule compound.26 Here, the cell survival was first examined for six days with four different commercial SLA materials from Formlabs, two of which (Dental SG and Dental LT Clear) are certified biocompatible according to EN-ISO 10993-1:2009/AC:2010.24–27,28 Nevertheless, the 3T3 cells did not grow properly on any of these materials (Fig. 2A), unless the culture platforms were autoclaved prior to use (Fig. 2B). Both autoclaved and non-autoclaved (NA) platforms were carefully post-treated by UV, as recommended by the supplier, and sterilized prior to use, suggesting that the autoclavation step is critical to ensuring cell survival in long-term and likely results in favorable, irreversible changes in the material composition. The total cell count from day-to-day is given in Fig. 2A and signifies acute toxicity, whereas the cumulative population doubling level given in Fig. 2B displays suppression of cell growth by, e.g., genotoxic effects and is especially useful when the dose of the toxicant is low. Of the four materials tested, only Dental SG and High Temp materials tolerated the high autoclavation temperatures without any visible damage and thus enabled long-term cell culturing. The cPDL of the 3T3 cells grown on autoclaved Dental SG and autoclaved High Temp materials maintained similar to that of the control (plastic cell culture dish) for at least 56 days (Fig. 2B). Instead, the surfaces made from Clear resin and the biocompatible-certified Dental LT Clear resin were visibly cracked after autoclavation and could only be sterilized with ethanol, but this was proven insufficient to support cell proliferation. However, we did not observe any signs of contamination in the cultures grown on materials sterilized with ethanol only, and therefore the impaired cell growth is not associated with insufficient sterilization. Instead, we hypothesize that despite the UV post-printing treatment, some chemical residues leach out of the bulk polymer and induce cell death on the non-autoclaved platforms, whereas during autoclavation, these residues are likely vaporized out of the bulk polymer which substantially increases the long-term cell survival on autoclaved 3D printed platforms.The proliferation rate of 3T3 cells grown on the autoclaved Dental SG and autoclaved High Temp materials was further quantitated by BrdU staining after 4 and 8 weeks of culturing. Although some variation was observed after 4 weeks of culturing, the 3T3 cells cultured on either of the materials did not have significant differences in their proliferation rate compared with control after 8 weeks (Fig. 2C), suggesting that autoclavation is a sufficient, although critical, post-processing step to ensure long-term cell survival on the 3D printed surfaces made of Dental SG and High Temp. Our results also showed that neither material induced higher apoptosis in the 3T3 cells when compared to the control. The number of early apoptotic cells (Fig. 2D) and late apoptotic cells (Fig. 2E) was similar to or lower than that of the control dishes over the entire culturing period of 7 weeks.
Additional material considerations
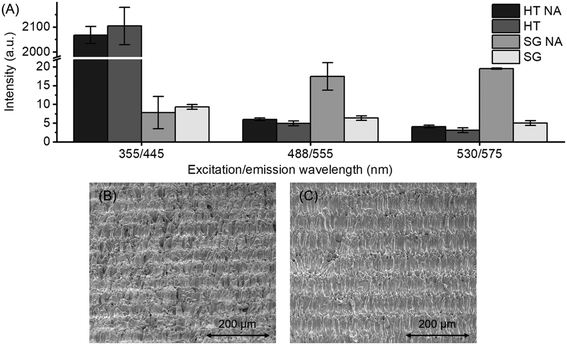
Autofluorescence originating from UV-curable materials, such as (stereo)lithographically defined polymers, has a pronounced impact on the quality of optical detection on microfluidic devices.29 Since high quality in situ monitoring of cells is based on fluorescent stains, the autofluorescence of cell-compatible Dental SG and High Temp materials was further assessed at three excitation and emission wavelengths common to cell stains (Fig. 3A). In the visible range, both materials were shown to induce relatively low autofluorescence at wavelengths equivalent to calcein AM (ex/em 488/555 nm) and PI (ex/em 530/575 nm) stains. Interestingly, autoclavation was shown to further reduce the autofluorescence of Dental SG in the visible range, although its autofluorescence was generally very low at both UV and visible wavelengths. Instead, High Temp was shown to have significantly high autofluorescence in the UV range equivalent to Hoechst 33342 (ex/em 355/455 nm) stain, which was unaffected by the autoclavation. This may impair the detection of blue fluorescent stains on High Temp material, thus favoring the use of Dental SG for fabrication of the 3D printed microfluidic cell culturing assays.Besides autofluorescence, the surface roughness has an impact on the quality of optical detection. SLA-based 3D printing often results in an inherently blurred surface. With the printer settings used in this study, the average roughnesses of the 3D printed surfaces were 3.8 μm for Dental SG and 3.7 μm for High Temp, i.e., somewhat greater than those of photolithographically patterned polymers. These are however only rough estimates and largely impacted by the printer settings (e.g., the printing angle and the layer height) as well as the condition (ageing) of the printer resin tank (the consumable part). SEM analysis also revealed that a typical 3D printed surface has somewhat irregular, undulating topography, which features random small pores and groove-like patterns that result from the layer-by-layer curing principle (Fig. 3B and C). Besides optical quality, these microstructures are likely to impact cell adhesion30 as discussed in the last chapter of Results and discussion.
Compatibility of Dental SG with human hepatoma cells and pluripotent stem cells
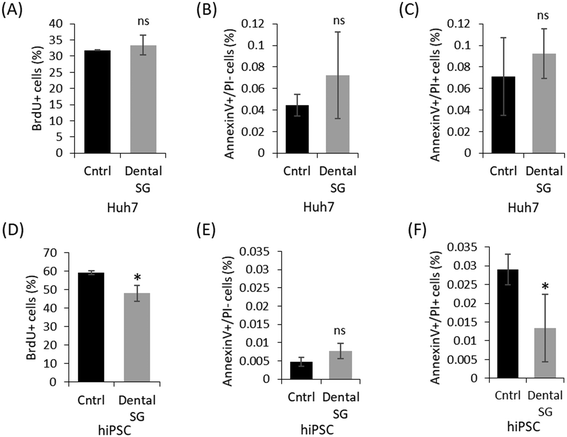
Out of the four commercial SLA resins examined in this study, Dental SG was considered most favorable for 3D printing of microfluidic organ-on-a-chip platforms owing to its better compatibility with the 3T3 cells (the recommended cell line for acute toxicity testing21) over Clear and Dental LT Clear materials and the lower optical background compared with High Temp. The biocompatibility of autoclaved Dental SG was thus further evaluated with the help of Huh7 cells and hiPSCs that are commonly used in, e.g. drug discovery and regenerative medicine, respectively. While Huh7 is a relatively robust cell line, the hiPSCs have been shown to be particularly sensitive to toxicants.19 The biocompatibility of Dental SG with these cells was assessed similar to 3T3 cells, based on BrdU and annexinV/PI staining of cells cultured on Dental SG for total of one week. According to the BrdU staining, the proliferation rate of the Huh7 cells was similar to that of the control (Fig. 4A) and, based on the annexinV/PI staining, the cells did not suffer from material-induced apoptosis either (Fig. 4B and C). For hiPSC culturing, the 3D printed and control dishes were thin-coated with Matrigel, because hiPSCs do not generally attach to artificial surfaces31,32 and thus, surface functionalization is critical for maintaining their phenotype and inherent properties.32 On Matrigel-coated Dental SG, hiPSCs had a slightly lower proliferation rate compared to the control (Fig. 4D), but no indications of material-induced apoptosis was detected compared with similarly coated cell culture dishes (Fig. 4E and F). The number of late apoptotic hiPSCs on Dental SG was even considerably lower than that of the control, although the number of apoptotic cells was very low in both cases. On the basis of these results, the 3D printed Dental SG platforms were concluded well feasible for culturing of human hepatocytes and hiPSCs.Impact of shear force on cell adhesion to Dental SG
Besides material-induced toxicity (e.g., leaching of monomers or other additives), the overall cell compatibility is impacted by surface roughness/topography and wettability, which play critical roles in terms of cell adhesion. The cell adhesion strength for its part greatly affects the biomechanical cues that control cell proliferation and cell cycle progression on planar surfaces.33 The material–cell interactions can be measured by atomic force microscopy,34,35 but the impact of hydrodynamic shear force needs to be determined under through-flow conditions.36,37 The typical surface pattern achieved with the materials and the printer used in this study is given in Fig. 3B and C, which visualize the apparent ‘undulation’ of the 3D printed surfaces that results from layer-by-layer curing principle. While the surface undulation can be to certain extent reduced by adjusting the printing angle (here, 45°), it is inherent to SLA based printers (such as the Form2 used in this study) and its impact on cell adhesion should be accounted for when assessing the overall compatibility of 3D printed platforms for microfluidic cell culturing. For determination of the threshold for applicable shear force on 3D printed Dental SG platforms, we developed a tapered (height 1050 → 150 μm, width 500 → 300 μm) microchannel design to facilitate creation of an on-chip shear force gradient along the microchannel (Fig. 5A). By varying both width and height simultaneously, it was possible to create shear force gradient that covers a range as large as two orders of magnitude. By applying constant flow rate, we were able to define both coarse (e.g., 1–89 dyne per cm2 at 600 μL min−1) and fine (e.g., 0.22–22 dyne per cm2 at 150 μL min−1) shear force gradients on demand in a single experiment. It should be noted that similarly tapered microchannel designs are not easily reproduced by any common (planar) microfabrication technique, which underlines the versatility of 3D printing in rapid prototyping of truly 3D microfluidic assays.The impact of shear force on the adhesion and viability of 3T3 cells seeded (2 × 106/mL, 4 h for adhesion) in 3D printed Dental SG channels was first determined by applying a coarse shear force gradient (1–89 dyne per cm2) using constant flow rate of 600 μL min−1. For this purpose, the 3D printed chip was encapsulated inside a custom-made, 3D-printed chip holder made from Clear resin (Fig. 1C). The holder ensured leakage-free sealing of the 3D printed, tilted channel with the PDMS cover layer even if relatively high pressure was generated inside the channel. Increasing shear stress resulted in detachment of the 3T3 cells with a threshold at ca. τ ≥ 7–8 dyne per cm2 (Fig. 5A). By lowering the flow rate down to 150 μL min−1, a finer shear force gradient (0.22–22 dyne per cm2) could be created, which enabled a more detailed study of cell adhesion in the critical range and suggested loss of adhesion already at lower shear force values with threshold at ca. τ ≥ 3 dyne per cm2 (Fig. 5B). These values are at the low end of the shear force (threshold) values reported for microfluidic devices in the previous literature (varying between 4 and 300 dyne per cm2),36,37 which likely results from the relatively high roughness and irregular topographic pattern of the 3D printed surfaces (Fig. 3B and C). The wettability of Dental SG may also have an impact on the low cell adhesion strength. Namely, the apparent water contact angle of the autoclaved Dental SG was higher (99.5 ± 1.1°) than that of the native Dental SG (75.7 ± 3.5°), which may further reduce the material–cell interactions. Furthermore, PI staining showed substantial amount of dead 3T3 cells even if the shear force values were in the range of 0.8 dyne per cm2 only, i.e., much lower than the threshold value for adhesion loss (Fig. 5B). The quantification of the number of live cells (based on calceim AM stain) also confirmed the adverse impact of increasing shear force on cell viability (Fig. 5A and B). These observations, however, correlate well with earlier studies, in which shear forces as low as τ ≥ 0.65 dyne per cm2 have been shown to cause cellular damage and stress.38
Conclusions
The use of low-cost bench-top 3D printers has substantially increased in rapid prototyping of microfluidic devices along with conventional photolithography-based microfabrication processes. 3D printing of microfluidic cell culturing platforms appears particularly useful in diversifying the experimental settings in cell biology research, but the material–cell interactions are often vaguely examined, which may result in failed experiments without clear understanding of the underlying mechanisms. In this work, we conducted a detailed characterization of cell compatibility of four commercially available 3D printing materials from Formlabs (Clear, High Temp, Dental SG and Dental LT Clear), the provider of affordable SLA printers widely used in research laboratories. According to our results, the certified (ISO) biocompatibility of the materials does not necessarily imply good cell compatibility in organ-on-a-chip applications. Instead, the possibility for autoclavation (i.e., good thermal stability) was concluded to be the most critical factor for ensuring normal cell proliferation and lack of material-induced material induced cell death on the 3D printed surfaces. This applied also to a material that is not certified as biocompatible (High Temp resin). Out of the tested materials, Dental SG was found to be most favorable in terms of microfluidic organ-on-a-chip applications, as it supported long term cell growth and provided low optical background even in the UV range. Besides 3T3 cells, Dental SG was also confirmed suitable for cultivation of human hepatocytes and hiPSCs. However, owing to the inherent high surface roughness and irregularity of the 3D printed surfaces, the cell adhesion strength in 3D printed microchannels was substantially lower (with threshold at ca. τ ≥ 3 dyne per cm2) than that commonly reported for through-flow assays in the literature, which should also be accounted for when designing 3D printed organ-on-a-chip platforms. For rapid determination of the shear force threshold in 3D printed microchannels, we also developed a novel shear force gradient chip design featuring a 3D tapered microchannel.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The work was financially supported by the European Research Council (grant no. 311705/CUMTAS), the Academy of Finland (grants no. 3074641, 308911, 309608, 314303, 325222, and 321564), Sigrid Jusélius Foundation, Instrumentarium Science Foundation, Magnus Ehrnrooth Foundation, and the Finnish Foundation for Cardiovascular Research. The authors thank Annika Korvenpää for technical assistance with hiPSCs. We thank the Electron Microscopy Unit of the Institute of Biotechnology at the University of Helsinki, the DDCB core facility supported by the University of Helsinki and Biocenter Finland, and the Micronova Centre of Micro and Nanotechnology, for providing accesses to the scanning electron microscope, the well-plate reader, and profilometer, respectively.References
- S. Ligon, R. Liska, J. Stampfl, M. Gurr and R. Mülhaupt, Polymers for 3D Printing and Customized Additive Manufacturing, Chem. Rev., 2017, 117, 10212–10290, DOI:10.1021/acs.chemrev.7b00074.
- G. Salentijn, P. Oomen, M. Grajewski and E. Verpoorte, Fused Deposition Modeling 3D Printing for (Bio)analytical Device Fabrication: Procedures, Materials, and Applications, Anal. Chem., 2017, 89(13), 7053–7061, DOI:10.1021/acs.analchem.7b00828.
- S. Waheed, J. M. Cabot, N. P. Macdonald, T. Lewis, R. M. Guijt, B. Paullab and M. C. Breadmore, 3D printed microfluidic devices: enablers and barriers, Lab Chip, 2016, 16, 1993–2013, 10.1039/C6LC00284F.
- A. Au, N. Bhattacharjee, L. Horowitz, C. Chang and A. Folch, 3D-printed microfluidic automation, Lab Chip , 2015, 15, 1934–1941, 10.1039/C5LC00126A.
- R. Gajasinghe, S. Senveli, S. Rawal, A. Williams, A. Zheng, R. Datar, R. Cote and O. Tigli, Experimental study of PDMS bonding to various substrates for monolithic microfluidic applications, J. Micromech. Microeng., 2014, 24, 075010, DOI:10.1088/0960-1317/24/7/075010.
- N. Bhattacharjee, A. Urrios, S. Kang and A. Folch, The upcoming 3D-printing revolution in microfluidics, Lab Chip, 2016, 16, 1720–1742, 10.1039/C6LC00163G.
- M. Farsari and B. Chichkov, Two-photon fabrication, Nat. Photonics, 2009, 3, 450–452, DOI:10.1038/nphoton.2009.131.
- H. Gong, B. Bickham, A. Woolley and G. Nordin, Custom 3D printer and resin for 18 μm × 20 μm microfluidic flow channels, Lab Chip, 2017, 17, 2899–2909, 10.1039/c71c00644f.
- M. Matsusaki, K. Sakaue, K. Kadowaki and M. Akashi, Three-dimensional human tissue chips fabricated by rapid and automatic inkjet cell printing, Adv. Healthcare Mater., 2013, 2, 534–539, DOI:10.1002/adhm.201200299.
- S. Knowlton, C. H. Yu, F. Ersoy, S. Emadi, A. Khademhosseini and S. Tasoglu, 3D-printed microfluidic chips with patterned, cell-laden hydrogel constructs, Biofabrication, 2016, 8(2), 025019, DOI:10.1088/1758-5090/8/2/025019.
- T. Tao, Y. Wang, W. Chen, Z. Li, W. Su, Y. Guo, P. Dengad and J. Qin, Engineering human islet organoids from iPSCs using an organ-on-chip platform, Lab Chip, 2019, 19, 948–958, 10.1039/C8LC01298A.
- C. D. Robinson, J. M. Auchtung, J. Collins and R. A. Britton, Epidemic Clostridium difficile strains demonstrate increased competitive fitness compared to nonepidemic isolates, Infect. Immun., 2014, 82(7), 2815–2825, DOI:10.1128/IAI.01524-14.
- Y. Jodat, M. Kang, K. Kiaee, G. Kim, A. Martinez, A. Rosenkranz, H. Bae and S. Shin, Human-Derived Organ-on-a-Chip for Personalized Drug Development, Curr. Pharm. Des., 2018, 24, 5471–5486, DOI:10.2174/1381612825666190308150055.
- A. O. Erhirhie, C. P. Ihekwereme and E. E. Ilodigwe, Advances in acute toxicity testing: strengths, weaknesses and regulatory acceptance, Interdiscip. Toxicol., 2018, 11(1), 5–12, DOI:10.2478/intox-2018-0001.
- S. M. Oskui, G. Diamante, C. Liao, W. Shi, J. Gan, D. Schlenk and W. H. Grover, Assessing and Reducing the Toxicity of 3D-Printed Parts, Environ. Sci. Technol. Lett., 2016, 3, 1–6, DOI:10.1021/acs.estlett.5b00249.
- M. Ferraz, H. Henning, P. da Costa, M. Malda, S. Le Gac, F. Bray, M. Duursen, J. Brouwers, C. van de Lest, I. Bertijn, L. Kraneburg, P. Vos, T. Stout and B. Gadella, Potential Health and Environmental Risks of Three-Dimensional Engineered Polymers, Environ. Sci. Technol. Lett., 2018, 5, 80–85, DOI:10.1021/acs.estlett.7b00495.
- F. Ejserholm, J. Stegmayr, P. Bauer, F. Johansson, L. Wallman, M. Bengtsson and S. Oredsson, Biocompatibility of a polymer based on Off-Stoichiometry Thiol-Enes + Epoxy (OSTE+) for neural implants, Biomater. Res., 2015, 19, 19, DOI:10.1186/s40824-015-0041-3.
- R. Bail, A. Patel, H. Yang, C. M. Rogers, F. R. Rose, J. I. Segal and S. M. Ratchev, The effect of a type I photoinitiator on cure kinetics and cell toxicity in projection-microstereolithography, Procedia CIRP., 2013, 5, 222–225, DOI:10.1016/j.procir.2013.01.044.
- C. Lin, Y. Lin, Y. Lai and S. Lee, Mechanical properties, accuracy, and cytotoxicity of UV-polymerized 3D printing resins composed of Bis-EMA, UDMA, and TEGDMA, J. Prosthet. Dent., 2020, 123, 349–354, DOI:10.1016/j.prosdent.2019.05.002.
- T. Puškar, B. Trifković, D. Koprivica, V. Kojić, A. Jevremović, S. Mirković and D. Eggbeer, In vitro cytotoxicity assessment of the 3D printed polymer based epoxy resin intended for use in dentistry, Vojnosanit. Pregl., 2019, 76, 502–509, DOI:10.2298/VSP170721127P.
- EURL ECVAM recommendation on the 3T3 Neutral Red Uptake cytotoxicity assay for acute oral toxicity testing, European Union, 2013, DOI:10.2788/88799.
- S. T. Karhu, M. J. Välimäki, M. Jumppanen, S. M. Kinnunen, L. Pohjolainen, R. S. Leigh, S. Auno, G. Földes, G. Boije af Gennäs, J. Yli-Kauhaluoma, H. Ruskoaho and V. Talman, Stem cells are the most sensitive screening tool to identify toxicity of GATA4-targeted novel small-molecule compounds, Arch. Toxicol., 2018, 92, 2897–2911, DOI:10.1007/s00204-018-2257-1.
- B. Lener, R. Rafał Kozieł, H. Pircher, E. Hutter, R. Greussing, D. Herndler-Brandstetter, M. Hermann, H. Unterluggauer and P. Jansen-Durr, The NADPH oxidase Nox4 restricts the replicative lifespan of human endothelial cells, Biochem. J., 2009, 423, 363–374, DOI:10.1042/BJ20090666.
- ISO 10993-3:2014, Biological evaluation of medical devices - Part 3: Tests for genotoxicity, carcinogenicity and reproductive toxicity.
- ISO 10993-1:2018, Biological evaluation of medical devices - Part 1: Evaluation and testing within a risk management process.
- S. Karhu, S. Kinnunen, M. Tölli, M. Välimäki, Z. Szabó, V. Talman and H. Ruskoaho, GATA4-targeted compound exhibits cardioprotective actions against doxorubicin-induced toxicity in vitro and in vivo: establishment of a chronic cardiotoxicity model using human iPSC-derived cardiomyocytes, Arch. Toxicol., 2020 DOI:10.1007/s00204-020-02711-8.
- https://formlabs.com/media/upload/DentalLTClear-DataSheet-EN.pdf .
- https://formlabs.com/media/upload/DentalSG-DataSheet.pdf .
- A. Piruska, I. Nikcevic, S. H. Lee, C. Ahn, W. R. Heineman, P. A. Limbach and C. J. Seliskar, The autofluorescence of plastic materials and chips measured under laser irradiation, Lab Chip, 2005, 5(12), 1348–1354, 10.1039/B508288A.
- E. Martinez, E. Engel, J. A. Planell and J. Samitier, Effects of artificial micro- and nano-structured surfaces on cell behaviour, Ann. Anat., 2009, 191, 126–135, DOI:10.1016/j.aanat.2008.05.006.
- C. Xu, M. S. Inokuma, J. Denham, K. Golds, P. Kundu, J. D. Gold and M. K. Carpenter, Feeder-free growth of undifferentiated human embryonic stem cells, Nat. Biotechnol., 2001, 19(10), 971–974, DOI:10.1038/nbt1001-971.
- M. T. Lam and M. T. Longaker, Comparison of several attachment methods for human iPS, embryonic and adipose-derived stem cells for tissue engineering, J. Tissue Eng. Regener. Med., 2012, 6(03), 80–86, DOI:10.1002/term.1499.
- S. Huang and D. E. Ingber, The structural and mechanical complexity of cell-growth control, Nat. Cell Biol., 1999, 1, E131 CrossRef CAS PubMed.
- G. Sagvolden, I. Giaever, E. O. Pettersen and J. Feder, Cell adhesion force microscopy, Proc. Natl. Acad. Sci. U. S. A., 1999, 96(2), 471–476, DOI:10.1073/pnas.96.2.471.
- Y. Shen, M. Nakajima, S. Kojima, M. Homma, M. Kojima and T. Fukuda, Single cell adhesion force measurement for cell viability identification using an AFM cantilever-based micro putter, Meas. Sci. Technol., 2011, 22, 115802, DOI:10.1088/0957-0233/22/11/115802.
- H. Lu, L. Y. Koo, W. M. Wang, D. A. Lauffenburger, L. G. Griffith and K. F. Jensen, Microfluidic Shear Devices for Quantitative Analysis of Cell Adhesion, Anal. Chem., 2004, 76, 5257–5264, DOI:10.1021/ac049837t.
- P. Rupprecht, L. Golé, J. P. Rieu, C. Vézy, R. Ferrigno, H. C. Mertani and C. Rivière, A tapered channel microfluidic device for comprehensive cell adhesion analysis, using measurements of bdetachment kinetics and shear stress-dependent motion, Biomicrofluidics, 2012, 6(1), 14107–1410712, DOI:10.1063/1.3673802.
- N. A. Stathopoulos and J. D. Hellums, Shear stress effects on human embryonic kidney cells in vitro, Biotechnol. Bioeng., 1985, 27, 1021, DOI:10.1002/bit.260270713.
| This journal is © The Royal Society of Chemistry 2020 |