Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Versatile hybrid acoustic micromixer with demonstration of circulating cell-free DNA extraction from sub-ml plasma samples†
Alvaro J. Conde ab,
Ieva Keraiteab,
Alfredo E. Ongaro
ab,
Ieva Keraiteab,
Alfredo E. Ongaro abc and
Maïwenn Kersaudy-Kerhoas
abc and
Maïwenn Kersaudy-Kerhoas *ab
*ab
aInstitute of Biological Chemistry, Biophysics and Bioengineering, School of Engineering and Physical Science, Heriot-Watt University, Edinburgh, UK. E-mail: m.kersaudy-kerhoas@hw.ac.uk
bInfection Medicine, Edinburgh Medical School, College of Medicine and Veterinary Medicine, The University of Edinburgh, Edinburgh, UK
cDepartment of Civil, Environmental, Aerospace and Materials Engineering (DICAM), University of Palermo, Palermo, Italy
First published on 17th January 2020
Abstract
Acoustic micromixers have attracted considerable attention in the last years since they can deliver high mixing efficiencies without the need for movable components. However, their adoption in the academic and industrial microfluidics community has been limited, possibly due to the reduced flexibility and accessibility of previous designs since most of them are application-specific and fabricated with techniques that are expensive, not widely available and difficult to integrate with other manufacturing technologies. In this work, we describe a simple, yet highly versatile, bubble-based micromixer module fabricated with a combination of low-cost rapid prototyping techniques. The hybrid approach enables the integration of the module into practically any substrate and the individual control of multiple micromixers embedded within the same monolithic chip. The module can operate under static and continuous flow conditions showing enhanced mixing capabilities compared to similar devices. We show that the system is capable of performing cell-free DNA extractions from small volumes of blood plasma (≤500 μl) with up to a ten-fold increase in capture efficiency when compared to control methods.
Introduction
Micromixers are essential components in microfluidic devices which have a direct impact on the efficiency and sensitivity of assays.1 However, mixing in microfluidics is also one of the main challenges since inertial effects are almost irrelevant due to the low Reynolds number in which these systems usually operate. Consequently, researchers endeavour to counterweigh the limits of purely diffusive mixing by introducing structures or active mechanisms that disturb the flow with the ultimate objective of diminishing the striation length and increasing the area across which diffusion takes place, thus enabling a rapid homogenisation of the solution.2Sound field driven, or acoustic, micromixers have attracted considerable attention in the last years since they can deliver high mixing efficiencies without the integration of fragile movable components in opposition to their magnetic counterparts.1,3–5 Surface acoustic wave (SAW) micromixers have shown to be promising, but they require the integration of microelectrodes on piezoelectric substrates which can lead to biocompatibility issues and can drive the fabrication costs up.6 Ultrasonic actuation has been shown to provide efficient mixing; however, significant heat is generated during the operation, carrying the need for bulky active cooling mechanisms.7 Other types of acoustic mixers can be implemented based on the vibration of membranes;8 sharp-edge structures;9 micropillars10 and cilia.11 Nevertheless, all of these devices are fabricated using intricate and laborious techniques which are not widely available and can be cost-prohibitive for many groups.
A surprisingly simple, yet powerful concept, is the acoustic actuation of air bubbles trapped within miniaturised devices. This method has been posed to be the “holy grail” for achieving fast (down to milliseconds) convective mixing at microscale lengths.12 In contrast to mechanical mixers, the microstreaming flow generated by the oscillating bubbles can be driven well beyond the Stokes boundary layer, producing efficient mixing even when dealing with highly viscous fluids.13 Moreover, the actuators for this kind of micromixers are generally economical, low-power and have a reduced footprint.
Researchers have previously developed a variety of highly efficient bubble-based micromixers,13–24 but most of them are fabricated using (SU-8 mould) soft-lithography, a highly manual and laborious technique that requires specialised equipment and is difficult to mass-manufacture. Furthermore, the total cost of microfluidic chips fabricated using this technique is usually underestimated and can be unaffordable for many research groups.25 Other authors have investigated the use of thermoplastics and other fabrication techniques, such as CNC micro-milling,17,18 laser cutting21 and hot embossing,22 however they share a common limitation along with the soft-lithography-based devices: the actuator (piezoelectric transducer) has to be glued using permanent adhesives to the bulk of the chip, impeding the individual control of more than one micromixer per chip and complicating the assembly process.
In this paper, we describe a novel bubble-based acoustic micromixer module that overcomes all of the aforementioned limitations. The micromixer is fabricated using a hybrid strategy, a combination of rapid prototyping technologies that are low-cost and widely available; it is, for example, possible to order all the necessary parts for their fabrication by mail.25 The hybrid strategy enables the incorporation of the module into practically any substrate and the capability to individually address multiple micromixers integrated within the same monolithic chip. The results show that the micromixer can perform effective homogenisation of two solutions, in continuous and static flow conditions, with enhanced performances compared to similar bubble-based micromixers. Additionally, the mixer can operate over a broad range of volumes (10–800 μl) in static flow conditions, something unique to our system. Finally, we validate the capability of the system to accomplish efficient circulating cell-free DNA (cfDNA) extractions from small volumes of blood plasma samples using a commercial kit based on magnetic beads.
Experimental
Design
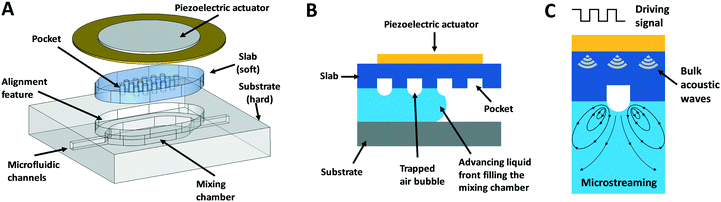
In order to tackle the limitations of previously reported bubble-based micromixers and taking inspiration from a previously reported hybrid microfluidic device,26 we propose the hybrid approach illustrated in Fig. 1A. This hybrid design combines a substrate (hard polymer) with a slab (soft polymer). The substrate contains the mixing chamber and the microfluidic channels, while the slab contains the structures (pockets) necessary to trap the air bubbles (Fig. 1B) that will perform the microstreaming when acoustically stimulated (Fig. 1C).A hermetic seal is formed when the deformable slab is mechanically compressed against the complementary structure on the mixing chamber. This compression force can be given by 1) the piezoelectric actuator when tightened to the chip via a matching frame and four screws (Fig. S1A†), or 2) by an auxiliary chip layer (Fig. S1B†). The first configuration allows for fast design iterations and the reuse of the components, while the second one enables the manufacturing of closed disposable chips. In both configurations, the acoustic energy is concentrated on the slab while much less energy is transferred to the substrate, enabling thus high mixing efficiencies and the individual control of several micromixers per chip as confirmed by the results. All micromixers in this work were fabricated following the first configuration. This solution provides tight sealing up to 100 kPa of pressure (burst test experiments performed as in the ref. 27). An example of a micromixer chip manufactured following the second configuration is shown in Fig. S2.†
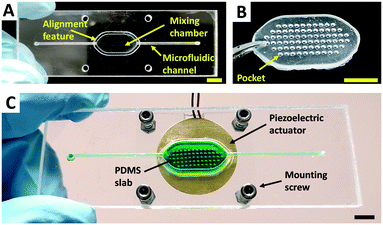
Microfluidic chips fabrication
Microfluidic chips for the mixing characterisation and cfDNA experiments were fabricated from cell-cast polymethyl methacrylate (PMMA) sheets (Clarex, Nitto Jushi Kogyo) using a previously reported rapid prototyping method.27 Briefly, after laser cutting the desired shapes using a CO2 laser cutter (Epilog Mini 18, Epilog, USA), PMMA layers were cleaned with a clean-room tissue soaked in ethanol and blown-dry with compressed air to remove dust. 80 μl of ethanol was spread between each of the layers using a pipette just before bonding. This allows for the partial melting of a superficial layer and the formation of a strong bond between the PMMA elements even if operating below PMMA Tg. Immediately after the ethanol spread, the assembly was placed in custom-made aluminium alignment frames and subsequently positioned between the plates of a heated press (PW100–37, Carver). A load of 2 metric tons was applied for 3 minutes at 70 °C. After the bonding, the chips were blow dry with compressed air and stored until use. A photograph of a fabricated PMMA microfluidic chip is shown in Fig. 2A. More details and photographs of all the chips used for the mixing characterisation experiments are shown in Fig. S3.†To demonstrate compatibility with other manufacturing technologies, 3D-printed chips were manufactured by fused deposition modelling (FDM) and stereolithography (SLA) technologies. FDM chips were fabricated using a commercial FDM printer (Mega i3, Anycubic) using a polylactic acid (PLA) filament (clear 1.75 mm filament, verbatim). The following printing parameters were used: extrusion temperature: 210 °C, bed temperature: 60 °C, printing speed: 30 mm s−1. For visualisation purposes, an optical window at the bottom of the chamber was created by glueing (Super Glue, 3M) a piece of 0.2 mm thick PMMA that was cut using a laser cutter. This piece can also be obtained by manually cutting the thin PMMA sheet with scissors. A photograph of a fabricated and fully assembled FDM micromixer chip is shown in Fig. S4A.† SLA chips were fabricated using a commercial SLA 3D printer (form 2, Formlabs) using a clear resin (Clear V4, Formlabs). After removing the uncured resin by rinsing the parts in pure isopropanol, the parts were cured in a UV crosslinker (CX-2000, UVP/Analytik Jena) for 30 min and subsequently heat-cured in an oven at 60 °C for 1 hour. The UV curing is not mandatory although recommended by the manufacturer. The same strategy followed for the FDM chips was used to create the optical window at the bottom of the mixing chamber. A photograph of a fabricated and fully assembled SLA micromixer chip is shown in Fig. S4B.†
Slabs fabrication
Slabs were fabricated by casting polydimethylsiloxane (PDMS) onto SLA-3D printed moulds. The moulds contain an array of posts (Fig. S5A†) that serve as negative structures for the formation of the pockets after the casting process. The moulds were fabricated following the same methods as described for the SLA microfluidic chips. A diagram of the casting process is shown in Fig. S5B.† Briefly, a silicone elastomer (Sylgard 184, Dow Chemical Company) was thoroughly mixed in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ratio, degassed in a vacuum chamber and subsequently poured over the clean moulds. Immediately after the casting, a small 0.2 mm thick PMMA piece was placed on top of the moulds to generate a flat and polished surface on the cured PDMS slab. This particular surface is necessary to ensure intimate contact between the slab and the piezoelectric disc and to avoid the use of coupling mediums. The casted moulds were left to cure overnight at room temperature. After this period, the PMMA piece was removed and an additional curing step of 2 hours at 60 °C was performed. Finally, the slabs were carefully peeled from the mould using tweezers and stored until use. A photograph of one of the obtained PDMS slabs is shown in Fig. 2B. More details and photographs of all the PDMS slabs can be found in Fig. S3.†
10 ratio, degassed in a vacuum chamber and subsequently poured over the clean moulds. Immediately after the casting, a small 0.2 mm thick PMMA piece was placed on top of the moulds to generate a flat and polished surface on the cured PDMS slab. This particular surface is necessary to ensure intimate contact between the slab and the piezoelectric disc and to avoid the use of coupling mediums. The casted moulds were left to cure overnight at room temperature. After this period, the PMMA piece was removed and an additional curing step of 2 hours at 60 °C was performed. Finally, the slabs were carefully peeled from the mould using tweezers and stored until use. A photograph of one of the obtained PDMS slabs is shown in Fig. 2B. More details and photographs of all the PDMS slabs can be found in Fig. S3.†
Piezoelectric actuators and driver
Piezoelectric diaphragms with different diameters were used (15 mm 7BB-15-6L0, 20 mm 7BB-20-6 and 27 mm 7BB-27-4, Murata Electronics). Photographs of these devices can be found in Fig. S3.† A function generator (TG215, Thurlby Thandar Instruments) was used as a driver. This generator can work with voltages up to 20 Vp–p. For higher voltages, we used a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 turn ratio audio transformer (NTE10/3, Neutrik) connected with the signal generator.
10 turn ratio audio transformer (NTE10/3, Neutrik) connected with the signal generator.
Image acquisition and analysis
Videos and images for the mixing characterisation experiments were acquired using digital microscopes with white light (AM7013MZT, Dino-Lite) and with fluorescence capability (AM4115T-GFBW, Dino-Lite). Images and videos were captured using the software DinoCapture 2.0 (Dino-Lite) and processed using ImageJ.Plasma samples and cell-free DNA extraction
Blood was drawn from healthy volunteers in EDTA tubes and plasma immediately separated by double centrifugation at low speed (4000g) for 10 min and high speed (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g) for 10 min. The plasma samples were used for cfDNA extraction shortly after separation without any freeze–thaw cycles to avoid any potential degradation of the DNA fragments.
000g) for 10 min. The plasma samples were used for cfDNA extraction shortly after separation without any freeze–thaw cycles to avoid any potential degradation of the DNA fragments.
To extract the cfDNA from plasma, we used the magnetic beads-based MagMAX cfDNA isolation kit (ThermoFisher Scientific) following the manufacturer's recommended protocol for the controls (bench) and a modified protocol for the microfluidic assays as shown in Table S1.†
Special PMMA microfluidic chips with a mixing volume of 255 μl and 1100 μl were fabricated following the same methods as described for the mixing characterisation experiments. As shown in Table S1,† these volumes are necessary for the processing of 100 μl and 500 μl of plasma respectively. A larger PDMS slab was fabricated for the 1100 μl chip. Photographs of the chips and the slabs used for these experiments are shown in Fig. S8.† Before the cfDNA experiments, the chips were coated with a solution of 2% bovine serum albumin (Sigma Aldrich) in 1× PBS (ThermoFisher Scientific) for 5 minutes at room temperature in order to avoid non-specific adsorption of the cfDNA fragments to the chip walls. Immediately after, the chips were washed with nuclease-free water (QIAGEN) and allowed to dry for a couple of minutes before running the experiments.
To demonstrate the magnetic beads disaggregation capabilities, two stacked magnets (5 mm diameter, 2 mm thick, N804RS, Eclipse Magnetics) were positioned on top of the piezoelectric disc roughly in the centre of the chamber as shown in Fig. S9A.† For the cfDNA capture experiments, we used a more powerful magnet (5 × 10 × 25 mm, N818RS, Eclipse Magnetics) that was positioned on top of the piezoelectric disc (Fig. S9B†) when required by the protocol to perform the capture of the magnetic beads.
Cell-free DNA quantification and fragment analysis
Real-time quantitative PCR (RT-qPCR) was performed using 2× Power SYBR® Green PCR Master Mix (Thermo Fisher Scientific) to quantify total cfDNA by amplifying LINE-1 (long interspersed nuclear elements) target of 90 bp. The total reaction volume was 12.5 μl with a final concentration of each primer of 200 μM (LINE FW 5′-TGCCGCAATAAACATACGTG-3′, LINE RV 5′-GACCCAGCCATCCCATTAC-3′) and 1 μl cfDNA elution.28 Thermal cycling conditions involved a 10 min cycle at 95 °C followed by 40 cycles with 15 s at 95 °C and 60 s at 60 °C. Samples were amplified in duplicate using Mx3005P qPCR system (Agilent). A melting curve was performed as a control measure for nonspecific amplification. The standard curve for absolute quantification of cfDNA was created with commercially available human genomic DNA (Bioline) with a linear range over 5 orders of magnitude. Linear regression analysis was done in MxPro qPCR software (R2 > 0.98).CfDNA fragment size analysis was performed using the High Sensitivity D5000 ScreenTape assay on the Agilent 4200 TapeStation system. Data were analysed with TapeStation Analysis Software Version A.02.02 (SR1).
Results and discussion
Resonant frequency
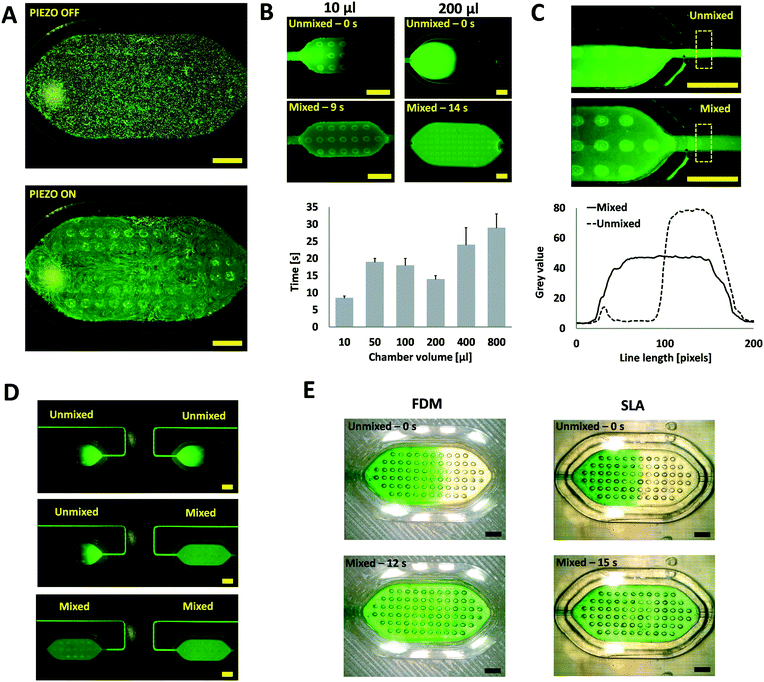
The intensity of the streaming generated by the oscillating bubbles is mainly a function of the applied voltage and frequency. The frequency in which the intensity of the streaming is maximum is called resonant or optimal frequency. This frequency is mainly a function of geometrical bubble parameters and can be theoretically estimated.29 However, the resonant frequency can also be experimentally determined by visual observation due to the limited predictability of the models16 and fabrication tolerances of the 3D printed moulds. Briefly, a frequency sweep is performed using a signal generator, and the microstreaming phenomenon is observed under the digital microscope. To help with this qualitative assessment, fluorescent microparticles (5 μm beads FITC-marked, Sigma Aldrich) were suspended in deionised water (DW) and loaded into the different micromixing chambers. Micrographs of a 100 μl micromixing chamber (Fig. S3C†) with no actuation and actuation at the resonant frequency are shown in Fig. 3A. A video demonstrating this process can be found in the ESI.† The main resonant frequency for all the fabricated micromixers was 4.2 ± 0.5 kHz.Mixing under static-flow conditions
Most of the reported micromixers are designed to operate continuously by disrupting the laminar flow streams to be mixed; however, there are many microfluidic applications that require the mixing of fluids under static-flow conditions. This is especially important in point-of-care (POC) applications where stored reagents must be mixed in confined volume chambers. To demonstrate the capability of the presented system to perform these actions, microfluidic chips integrating mixing chambers with different volumes (10–800 μl) were fabricated. Photographs of these chips are shown in Fig. S3† (A–C, E, I and J). The chips were prefilled with DW and then a solution of fluorescein in DW was injected until approximately half of the chamber was filled. Immediately after filling, the piezoelectric was turned on and the mixing effect observed. The mixing time is calculated by analysing the videos with ImageJ and finding the time in which the mixing is no longer improved through a cut line (averaged width 20 px) placed across the chamber as shown in Fig. S6.† A video of this process for a 200 μl chamber can be found in the ESI.† The assays were done by triplicate and filling from both sides.As shown in Fig. 3B, the hybrid micromixer can effectively mix the samples in less than 35 seconds across all the volume range. These mixing times are comparable or better to similar previous static-flow bubble-based micromixers; however, these devices operate in significantly smaller volumes and have the drawback of the glued piezoelectric disc.16–19 Moreover, the hybrid micromixer can perform efficient mixing over a wide range of volumes, something unique to our system. As a comparison, diffusion only mixing takes approximately 36 hours to homogenise the species in a 200 μl chamber (Fig. S7†).
From the results shown in Fig. 3B, we have also observed the appearance of a valley in the mixing time for chamber volumes around 100–200 μl. We believe that this phenomenon could be owed to a synergistic interaction between the microstreaming forces and associated secondary flows30 from different bubbles that, as a consequence, generate additional flows that further improve the mixing efficiency for these particular geometrical configurations.
Mixing under continuous-flow conditions
To study the performance of the micromixer under continuous-flow conditions, microfluidic chips with two inlets and a mixing chamber (positioned immediately after the merging point of the two inlet channels) were fabricated (Fig. S3D†). A solution of fluorescein in DW and DW were flown at 100 μl min−1 per channel using two syringe pumps (AL-1000HP, World Precision Instruments). After the laminar flow was established, the piezoelectric was turned on and off, and the mixing process observed and recorded using a fluorescent digital microscope. A video of this experiment can be found in the ESI.† Snapshots extracted from these videos (corresponding to the area marked in yellow at the top in Fig. 3C) were analysed in ImageJ to evaluate mixing efficiency. Briefly, cut lines (averaged width 100 px) were positioned at the outlet of the mixing chamber (dashed yellow rectangle in Fig. 3C, top) and the grey value of the pixels plotted as shown at the bottom in Fig. 3C.The intensity profile showed in Fig. 3C (bottom) indicates that (∼1 second) after the piezoelectric is turned on, a uniform grayscale distribution across the width of the channel is observed, suggesting the complete homogenisation of the species. In the presented configuration, the system can provide efficient mixing from no flow rate (static flow reported earlier) up to a total flow rate of 200 μl min−1 (12 ml h−1), which is significantly higher than results from previous similar devices.14,15,20–23 The system geometrical configuration could be optimised to enable the homogenisation of higher flow rates.
Integration of multiple individually-addressable mixers on the same chip
A common disadvantage of previously reported bubble-based micromixers is that they cannot be controlled individually within the same monolithic chip. To demonstrate that our system overcomes this limitation, we fabricated a PMMA monolithic microfluidic chip integrating two independent micromixers (Fig. S3F†). For running the experiments, the same protocol as for static-flow conditions experiments was used, but the mixers were activated at different times. A video of this experiment can be found in the ESI.†As shown in the sequence of micrographs of Fig. 3D, it is possible to individually address different micromixers integrated within the same monolithic chip. This is a clear advantage over previously reported devices where all the micromixers would be activated simultaneously as they share the same substrate.16–19,22
Integration of the mixer into 3D printed chips
To further demonstrate the versatility and also the accessibility of the micromixer module, we integrated and tested the micromixer in microfluidic chips (Fig. S4†) fabricated with two of the most common and widely available 3D printing technologies; fused deposition modelling (FDM) and stereolithography (SLA). Due to the limited transparency of these 3D printed materials, optical windows were added to better observe the mixing effects. The same approach used in the static-flow condition experiments was followed but using green food dye instead of fluorescein to allow the observation of the 3D printed substrates.As shown in Fig. 3E, the hybrid module can perform efficient mixing operations without noticeable leaking on the devices fabricated with both 3D printing technologies. This capability is enabled by the hybrid nature of the concept: the slab can easily deform and adapt to any planar substrate. An important consequence of this feature is that it allows the fabrication of different micromixers by anyone with access to 3D printers or a small budget to order 3D printed components by mail.25 This accessibility characteristic is unique to our system since all the previously reported acoustic micromixers utilise fabrication technologies that are complex and unaffordable for many.
Circulating cell-free DNA extraction
The study of circulating cfDNA is gaining interest in various biomedical disciplines, including cancer, metabolic and cardiovascular diseases, prenatal sex determination, sepsis diagnosis and exercise physiology among others. Circulating cfDNA not only provides valuable genetic information: it can also be easily obtained from the body through a minimally invasive blood draw, so-called liquid biopsy. Most of the efforts in the cfDNA field are associated with non-invasive prenatal testing of chromosomal abnormalities, cancer detection, monitoring relapse and drug resistance. In these studies, and due to the very low concentration of the interest analytes, plasma volumes between 1 ml and 5 ml are usually required. However, small volumes of plasma samples (<500 μl) can provide sufficient cfDNA concentrations for their use in different fields, such as sports medicine31 and early fetal sex determination.32Most of the current cfDNA extraction methods are based on silica membranes or silica magnetic microbeads. The latter has been reported to have a higher preference for low molecular weight DNA molecules (up to 300 bp),33 which helps to circumvent cellular genomic DNA contamination in the sample. However, one of the main challenges of this assay is that after the application of a strong magnetic field, the beads tend to “stick” with each other, forming clusters. This issue can compromise the reproducibility and sensitivity of the assays since the cluster population can be quite heterogeneous and less surface area is available for the capture assays. Having less surface area has a significant impact in assays where the target analyte is in very low concentrations, such as cfDNA.
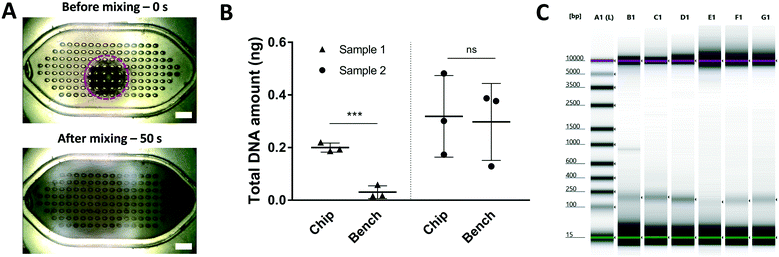
Acoustic microstreaming has been shown to be an efficient mechanism to disaggregate and mix microbeads in microfluidic devices.16 To evaluate the capabilities of our system to perform these operations, we fabricated a microfluidic chip (Fig. S8A†) containing a micromixing chamber of approximately 255 μl. The chamber was loaded with a suspension of magnetic beads in DW, and a stack of neodymium magnets was used to concentrate and aggregate the beads (Fig. S9A†). After the removal of the magnetic field, the piezoelectric was turned on and the mixing effect observed under the digital microscope. As shown in Fig. 4A, the beads can be effectively disaggregated and uniformly distributed across the mixing chamber in less than 50 seconds which is similar to the results obtained in a similar device.16 However, this previous device is fabricated using standard soft-lithography and can only mix 78 nl of sample volume. A video of this disaggregation experiment can be found in the ESI.†
After having this key disaggregation and mixing capability demonstrated, we performed a full cfDNA extraction from 100 μl (sample 1) and 500 μl (sample 2) of blood plasma samples from two different healthy donors using the micromixer and the MagMAX® Cell-Free DNA Isolation Kit. In order to accommodate the required volumes established by the manufacturer's protocol, we used the 255 μl mixing chip (Fig. S8A†) for the 100 μl plasma samples and an 1100 μl mixing chip (Fig. S8B†) for the 500 μl plasma samples. The magnetic capture set-up shown in Fig. S9† was used for these experiments.
As shown in Fig. 4B, the micromixer can capture, on average, one order of magnitude more cfDNA than the manual bench protocol (control) for 100 μl of plasma sample. These results indicate that the micromixer enables enhanced capture efficiency due to the superior mixing capabilities when compared to the control methods for this volume range. To the best of our knowledge, this is the first study where a 10-fold improvement in microfluidic-based cfDNA capture efficiency is presented on 100 μl of plasma sample. These results pose the question if the current bench mixing protocols are optimal for such volumes of sample. The difference between the total DNA amounts extracted using the chip and the bench protocol are not statistically significant for the 500 μl plasma sample. We believe that this could be due to the contribution of inertial effects that promote enhanced mixing in the control method for this volume range (>1 ml). The electrophoretic results presented in Fig. 4C confirm that the captured DNA corresponds indeed to cfDNA fragments (∼160 bp). The corresponding electropherograms are shown in Fig. S10.†
In summary, these results demonstrate the capability of the micromixer to perform a sample preparation assay which requires high shear forces (beads disaggregation) and high mixing efficiency (capture of a very low concentrated analyte). The mixer could be potentially integrated into micro and millifluidic cartridges with automated liquid handling operations for its implementation in POC settings. Bringing cfDNA sample preparation assays to the POC would ensure the stability and integrity of these precious molecules since the cfDNA fragments are usually short-lived, with half-lives ranging from minutes up to several hours.34
Conclusions
We have developed a novel bubble-based acoustic micromixer based on hybrid manufacturing. Thanks to the use of an elastomeric slab, less acoustic energy is lost on the substrate, leading to enhanced efficiency compared to previously reported bubble-based acoustic micromixers. Additionally, the micromixer can provide efficient mixing under static and continuous flow conditions with further capabilities such as modularity, augmented static volume range and the ability to individually-address different mixers integrated within a common substrate.The micromixer is fabricated using a combination of rapid prototyping technologies that are low-cost and widely available, enabling quick implementation and broad accessibility. Researchers can reproduce this kind of micromixers with minimal equipment and without the need for trained technicians, expensive equipment and complex facilities as required in the devices fabricated using (SU-8 mould) soft-lithography. The mixer module is highly versatile, it can be integrated into practically any substrate, here we demonstrated implementation with a PMMA layer-by-layer approach and two 3D-printing techniques. The use of a pick-and-place approach can be envisaged to place the elastomeric slab onto moulded chips in a mass-manufacturing setting, following a similar disposable chip concept as shown in Fig. S1.†
We have demonstrated the capability of the hybrid micromixer to significantly outperform the bench protocol for cfDNA extraction in 100 μl plasma samples using a commercial kit based on magnetic beads. For 500 μl plasma samples, the micromixer performed as well as the bench protocol.
We envision that the presented concept will help both industrial and academic developments by catalysing the development of innovative solutions in the lab-on-a-chip field.
Ethical statement
Human blood was obtained from healthy volunteers. Samples were used after obtaining donor consents. Ethical clearance was obtained from the UK National Health Service, East of Scotland Research Ethics Service, reference 19/ES/0056, and Heriot-Watt University EPS Ethics Committee, reference 19/EA/MKK/1.Author contributions
A. J. C. conceived the concept and fabricated the devices. A. J. C. and A. O. and performed the mixing characterisation experiments. A. J. C. and I. K. performed the biological characterisation experiments. M. K. K. acquired funding, supervised the work and provided ethical clearance. A. J. C. and M. K. K. wrote the manuscript. All co-authors edited the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge EPSRC EP/R00398X/1 for providing the funding necessary to complete this research. We are truly thankful to all our blood donors that kindly volunteered for this experiment.References
- G. Cai, L. Xue, H. Zhang and J. Lin, Micromachines, 2017, 8, 274 CrossRef PubMed.
- T. M. Squires and S. R. Quake, Rev. Mod. Phys., 2005, 77, 977–1026 CrossRef CAS.
- L.-H. Lu, K. S. Ryu and C. Liu, J. Microelectromech. Syst., 2002, 11, 462–469 CrossRef CAS.
- A. R. Shields, B. L. Fiser, B. A. Evans, M. R. Falvo, S. Washburn and R. Superfine, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 15670–15675 CrossRef CAS PubMed.
- D. Owen, M. Ballard, A. Alexeev and P. J. Hesketh, Sens. Actuators, A, 2016, 251, 84–91 CrossRef CAS.
- T. D. Luong, V. N. Phan and N. T. Nguyen, Microfluid. Nanofluid., 2011, 10, 619–625 CrossRef CAS.
- I. Iranmanesh, M. Ohlin, H. Ramachandraiah, S. Ye, A. Russom and M. Wiklund, Biomed. Microdevices, 2016, 18, 1–7 CrossRef PubMed.
- H. Van Phan, M. B. Coşkun, M. Şeşen, G. Pandraud, A. Neild and T. Alan, Lab Chip, 2015, 15, 4206–4216 RSC.
- P. H. Huang, Y. Xie, D. Ahmed, J. Rufo, N. Nama, Y. Chen, C. Y. Chan and T. J. Huang, Lab Chip, 2013, 13, 3847–3852 RSC.
- J. van t' Oever, N. Spannenburg, H. Offerhaus, D. van den Ende, J. Herek and F. Mugele, J. Micro/Nanolithogr., MEMS, MOEMS, 2015, 14, 023503 CrossRef.
- S. Orbay, A. Ozcelik, H. Bachman and T. J. Huang, J. Micromech. Microeng., 2018, 28, 025012 CrossRef PubMed.
- A. Hashmi, G. Yu, M. Reilly-Collette, G. Heiman and J. Xu, Lab Chip, 2012, 12, 4216–4227 RSC.
- S. Orbay, A. Ozcelik, J. Lata, M. Kaynak, M. Wu and T. J. Huang, J. Micromech. Microeng., 2017, 27, 1–7 CrossRef.
- D. Ahmed, X. Mao, B. K. Juluri and T. J. Huang, Microfluid. Nanofluid., 2009, 7, 727–731 CrossRef CAS.
- A. Ozcelik, D. Ahmed, Y. Xie, N. Nama, Z. Qu, A. A. Nawaz and T. J. Huang, Anal. Chem., 2014, 86, 5083–5088 CrossRef CAS PubMed.
- H. Chen, Y. Gao, K. Petkovic, S. Yan, M. Best, Y. Du and Y. Zhu, Microfluid. Nanofluid., 2017, 21, 30 CrossRef.
- R. H. Liu, R. Lenigk, R. L. Druyor-Sanchez, J. Yang and P. Grodzinski, Anal. Chem., 2003, 75, 1911–1917 CrossRef CAS PubMed.
- R. H. Liu, J. Yang, M. Z. Pindera, M. Athavale and P. Grodzinski, Lab Chip, 2002, 2, 151–157 RSC.
- Y. Okabe, Y. Chen, R. Purohit, R. M. Corn and A. P. Lee, Biosens. Bioelectron., 2012, 35, 37–43 CrossRef CAS PubMed.
- D. Ahmed, X. Mao, J. Shi, B. K. Juluri and T. J. Huang, Lab Chip, 2009, 9, 2738–2741 RSC.
- S. S. Wang, Z. J. Jiao, X. Y. Huang, C. Yang and N. T. Nguyen, Microfluid. Nanofluid., 2009, 6, 847–852 CrossRef CAS.
- A. P. Lee, M. V. Patel, A. R. Tovar and Y. Okabe, JALA, 2010, 15, 449–454 CAS.
- M. R. Rasouli and M. Tabrizian, Lab Chip, 2019, 19, 3316–3325 RSC.
- H. Chen, C. Chen, S. Bai, Y. Gao, G. Metcalfe, W. Cheng and Y. Zhu, Nanoscale, 2018, 10, 20196–20206 RSC.
- A. K. Au, W. Lee and A. Folch, Lab Chip, 2014, 14, 1294–1301 RSC.
- A. J. Conde, A. Bianchetti, F. E. Veiras, A. Federico, J. M. Cabaleiro, M. Dufva, R. E. Madrid and L. Fraigi, RSC Adv., 2015, 5, 49996–50000 RSC.
- A. Liga, J. A. S. Morton and M. Kersaudy-Kerhoas, Microfluid. Nanofluid., 2016, 20, 1–12 CrossRef CAS.
- S. Breitbach, S. Tug, S. Helmig, D. Zahn, T. Kubiak, M. Michal, T. Gori, T. Ehlert, T. Beiter and P. Simon, PLoS One, 2014, 9, e87838 CrossRef PubMed.
- C. Chindam, N. Nama, M. Ian Lapsley, F. Costanzo and T. Jun Huang, J. Appl. Phys., 2013, 114, 194503 CrossRef PubMed.
- A. Marin, M. Rossi, B. Rallabandi, C. Wang, S. Hilgenfeldt and C. J. Kähler, Phys. Rev. Appl., 2015, 3, 41001 CrossRef.
- S. Breitbach, S. Tug, P. Simon, R. Primacio, H. Milot, C. Jacob, R. Gyanchandani, E. Kvam, R. Heller, E. Finehout, N. Smith, K. Kota, J. R. Nelson, W. Griffin, S. Puhalla, A. M. Brufsky, N. E. Davidson and A. V. Lee, J. Preg. Child Health, 2017, 8, 4–7 Search PubMed.
- R. Primacio, H. Milot and C. Jacob, J. Preg. Child Health, 2017, 4, 4–7 Search PubMed.
- C. Stemmer, M. Beau-Faller, E. Pencreac'h, E. Guerin, A. Schneider, D. Jaqmin, E. Quoix, M.-P. Gaub and P. Oudet, Clin. Chem., 2003, 49, 1953–1955 CrossRef CAS PubMed.
- S. Khier and L. Lohan, Future Sci. OA, 2018, 4, FSO295 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9lc01130g |
| This journal is © The Royal Society of Chemistry 2020 |