Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Towards bioproduction of poly-α-olefins from lignocellulose†
Milla
Salmela
 *,
Tapio
Lehtinen
*,
Tapio
Lehtinen
 ,
Elena
Efimova
,
Suvi
Santala
and
Ville
Santala
,
Elena
Efimova
,
Suvi
Santala
and
Ville
Santala
Faculty of Engineering and Natural Sciences, Tampere University, Hervanta Campus, PO Box 527, FI-33014 Tampere, Finland. E-mail: milla.salmela@tuni.fi
First published on 8th July 2020
Abstract
Bioprocesses involving more than one species can alleviate restrictions posed by limited substrate range of single species. Coupled, multistage cultures can be useful when heterogeneous substrates, such as lignocellulosic biomass, are exploited. Here, microbial production of α-olefins (C11) from lignocellulosic substrates, namely cellulose and technical lignin, was investigated. A two-stage culture with cellulose fermentation to organic acids by Clostridium cellulolyticum and subsequent upgrading of the organic acids to 1-undecene by engineered Acinetobacter baylyi ADP1 was established. As a result, A. baylyi ADP1 synthesised 107 μg L−1 of 1-undecene from cellulose. Additionally, ligninolytic effects by A. baylyi ADP1 on softwood were confirmed and downstream processing for continuous 1-undecene collection was introduced. In addition, the synthesis of poly-α-olefin trimers (C33) by the oligomerization of 1-undecene was demonstrated. This study demonstrates the potential of integrated multistage processes in treating challenging substrates.
1. Introduction
Environmentally sustainable production of chemicals and fuels from renewable sources to replace fossil-based products has gained increasing interest in the past years. Especially, lignocellulosic biorefineries that convert biomass (cellulose, hemicellulose and lignin) comprehensively in integrated processes to products present unique opportunities.1,2 Currently, microbial production of biofuels from biomass-derived sugars is well established, although profitable production of advanced biofuels from lignocellulose remains a challenge due to the inherent heterogeneity and recalcitrance of lignocellulose.3,4 Hence, there is an interest in developing robust production hosts with broad substrate and product range, as well as efficient upstream and downstream processing. However, single organisms have limited metabolic flexibility and developing a strain capable of efficient product synthesis from both lignin and cellulose is challenging. For example, difficulties arise from metabolic burden caused by balancing multiple tasks in single organisms and complex synthetic constructs in engineered strains.5Inspired by natural microbial consortia, rationally engineered multispecies systems can overcome some of the challenges related to complex substrates. For example, cellulose solubilisation by bacteria or fungi followed by product synthesis with a second bacterial species have been studied for ethanol and butanol production.6,7 Engineered multispecies systems could also be used for more comprehensive lignin and cellulose conversion to advanced fuels and chemicals. Clostridium cellulolyticum is an example of mesophilic anaerobic bacteria that can solubilise cellulose and ferment glucose to hydrogen and organic acids in consolidated bioprocesses.8,9Acinetobacter baylyi ADP1, on the other hand, is an interesting candidate for upgrading lignocellulosic materials due to its distinctive metabolism, ease of genome engineering, robustness and oleaginous nature.10–14 It is tolerant towards lignocellulose related monomeric compounds, such as phenolic acids, acetate, and ethanol, which typically inhibit microbial growth.15–18 Furthermore, it can utilize monomeric lignin compounds through catabolic β-ketoadipate pathway, which efficiently funnels carbon to biomass and storage compound synthesis.17,19,20 The Acinetobacter genera and A. baylyi ADP1 have also been identified with lignin depolymerizing activities.21,22
Novel biorefineries are expected to produce large quantities of different types of technical lignins as a by-product. For example, hydrolytic pretreatment of lignocellulose results in high molecular weight lignin containing up to 15% of residual cellulose.23 Furthermore, bioprocessing of softwoods is more challenging compared to the processing of hardwoods or agricultural biomass due to higher lignin content (up to 30%), smaller pore size and lower amount of acetylated groups derived of hemicellulose.24,25 However, in the Northern hemisphere softwoods provide a major perennial source for bioprocesses. Currently, the lignin residues are mainly incinerated for heat. On the other hand, upgrading of the material to valuable bioproducts can be considered crucial for future biorefineries.
Medium-chain length linear α-olefins (mcl-LAO) such as 1-alkenes are oleochemicals of particular interest due to their terminal functionality and semi-volatile nature. Applications of mcl-LAO span from “drop-in fuels” to co-monomers in the production of poly-α-olefins (PAO) used as lubricants.26 Recently, the natural biosynthesis of mcl-LAOs has been elucidated in P. aureginosa. It was discovered that a single gene undA catalyses fatty acid (C12) conversion to 1-undecene.27 The heterologous expression of the undA gene enables a biosynthetic pathway for 1-undecene production through fatty acid derived metabolism in oleaginous production hosts such as A. baylyi ADP1.16 Furthermore, the semi-volatile nature of 1-undecene presents opportunities for effective product separation and recovery.
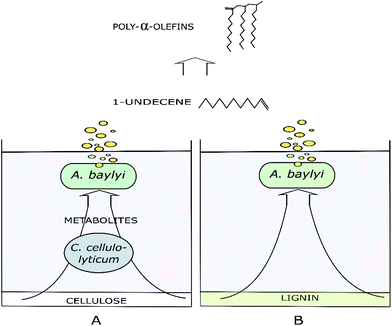
In previous studies, the metabolisms of Acinetobacter baylyi ADP1 and Clostridia butyricum were paired for combined wax ester and hydrogen gas or 1,3-propanediol production from glucose and glycerol.28,29 Here, more challenging substrates were investigated for the production of alkenes. 1-Undecene (C11) biosynthesis from both cellulose and technical lignin was investigated by cultivating C. cellulolyticum and engineered A. baylyi ADP1 in a coupled two-stage system. The metabolic labor was divided between the production of organic acids from cellulose and upgrading of the acids to 1-undecene. Furthermore, ligninolytic capabilities of A. baylyi on softwood lignin were assessed and downstream processes for product separation and oligomerization reactions to PAOs were introduced.
2. Materials and methods
2.1. Strains, media and components
Escherichia coli XL1-Blue (Stratagene, USA) was used for cloning. A. baylyi wild type (DSM 24193, DSMZ, Germany) was used for strain construction and as a control strain. Clostridium cellulolyticum (ATCC 35319) was used for consolidated saccharification and fermentation of cellulose. The constructed A. baylyi ADP1_UndA strain was used for 1-undecene production. A previously described A. baylyi strain expressing red fluorescent protein (RFP),30 designated here as ADP1_red, was used for growth and depolymerization studies on technical lignin.E. coli XL1 and A. baylyi ADP1 wild type were routinely grown for cloning and transformation purposes on LA plates or LB media with 25 μg ml−1 chloramphenicol and glucose supplementation (0.4–1%). C. cellulolyticum was cultivated in modified minimal CM3 media ((NH2)SO4 1.3 g L−1, KH2PO4 1.5 g L−1, K2HPO4 2.9 g L−1, 5% w/v FeSO4·7H2O solution in 50 mM H2SO4 25 μl L−1, MgCl2·6H2O 0.2 g L−1, CaCl2·2H2O 75 mg L−1, Na–resazurin 5 mg L−1, L-cysteine-HCL 0.5 g L−1) vitamin solution (D-biotin 1 mg L−1, p-amino-benzoic acid 25 mg L−1, nicotinic acid 15 mg L−1, riboflavin 25 mg L−1, pantothenic acid 2.5 mg L−1, thiamin 2.5 mg L−1, cyanocobalamin 10 mg L−1), FeSO4·7H2O 5.00 g L−1, ZnSO4·7H2O, 1.44 g L−1, MnSO4·7H2O 1.12 g L−1, CuSO4·5H2O 0.25 g L−1, Na2B4O7 0.20 g L−1, (Mo)7(NH4)6O24·4H2O 1.00 g L−1, NiCI2 0.04 g L−1, CoCI2 0.02 g L−1, HBO3 0.03 g L−1, Na2SeO3 0.02 g L−1, HCI 0.5 (M).31 The vitamin solution was sterilized by filtration with a 0.2 μm filter and all the other components were autoclaved. Media was made anaerobic by sparging with 100% nitrogen. The media was supplemented either by varying cellulose concentrations (Avicel ∼0.5 μm pore size, Sigma, USA) or by technical hydrolysis lignin (a kind gift from St1 company from their Cellunolix® bioethanol production unit, Kajaani, Finland).
Hydrolysis lignin is a byproduct from steam explosion pre-treatment and enzymatic hydrolysis of softwood originated biomass. The typical lignin content of the hydrolysis lignin is 75–80% of the dry matter and residual cellulose content is between 15–20% (personal communication, Minna Yamamoto, St1). Other components include sugars, acids, phenolic compounds, furanic compounds and proteins. The lignin structure is condensed, and it has low Sulphur and ash content. The technical hydrolysis lignin used in this study was freeze-dried and autoclaved before use.
Studies with A. baylyi were conducted in mineral salts media (MSM) (K2HPO4 3.88 g l−1, NaH2PO4 1.63 g l−1, (NH4)2SO4 2.00 g l−1, MgCl2·6H2O 0.1 g l−1, EDTA 10 mg l−1, ZnSO4·7H2O2 mg l−1, CaCl2·2H2O 1 mg l−1, FeSO4·7H2O 5 mg l−1, Na2MoO4·2H2O 0.2 mg l−1, CuSO4·5H2O 0.2 mg l−1, CoCl2·6H2O 0.4 mg l−1, MnCl2·2H2O 1 mg l−1)32 with appropriate carbon supplementations (glucose, acetate, lactate, the liquid end-products from the C. cellulolyticum fermentation or autoclaved technical lignin). Overnight precultivations were conducted in MSM supplemented with 50 mM of glucose or lactate.
All solvents and reagents were purchased from Sigma-Aldrich (USA) or Merck (USA), except 1-Undecene was purchased from Tokyo Chemical Industry Co (Japan). All cloning reagents including PCR, digestion and ligation were obtained from ThermoScientific (USA) and used according to manufacturer's instructions.
2.2. Strain construction
The construction cassette for genomic integration in A. baylyi wild type contained genes of a leaderless thioesterase from E. coli (‘tesA) and 1-undecene synthesising gene from P. putida (undA) under a T5 promoter. The combination of ‘tesA and undA has been previously shown to enhance 1-undecene production in A. baylyi when compared to undA alone.16 Genomic integration of undA and ‘tesA genes was constructed by homologous recombination using a transformation vector iluxAB_Cmr/pAK400c targeting an integration site ACIAD 3381.33 The luxAB gene was removed from the vector by restriction with NdeI and XhoI and replaced by the genes ‘tesA and undA. The genes encoding undA and ‘tesA were amplified by PCR from pBAV1C-chn-‘tesA-undA16 with primers TAAGCACATATGGCGGACACGTTATTGATTC and TGCTTACTCGAGTTATCAGCCCGCAGCCAACG containing overhangs with NdeI and XhoI restriction sites, and cloned to i/pAK400c with electrocompetent E. coli XL1 blue. The constructed plasmid T5-'tesA-undA/pAK400c was used for natural transformation and homologous recombination in A. baylyi according to a previously described method.34 The genomic integration was verified by primers TGAGAAATCTTTGTCCACGCC targeting upstream of the integration site and TGCTTACTCGAGTTATCAGCCCGCAGCCAACG targeting the undA gene. The constructed strain was designated as ADP1_undA.2.3. Biosynthesis of 1-undecene from defined media
The growth and substrate preferences of A. baylyi ADP1_undA were studied in MSM media with 10 mM of glucose, acetate, and lactate in a 50 ml batch cultivation at 30 °C at 300 rpm for 12 hours. The media was inoculated from an overnight pre-cultivation to an optical density (OD600) of 0.05 measured by a spectrophotometer (Ultrospec 500 pro, Amersham Biosciences, UK) at 600 nm. After a 2-hour incubation, samples were collected hourly for high performance liquid chromatography (HPLC) analysis and OD600 measurements. Experiments were run as duplicates. 1-Undecene production by ADP1_undA was studied in 5 ml of MSM media supplemented with 30 mM of lactate or acetate as the sole carbon source using 20 ml sealable glass vials. The vials were sealed to allow 1-undecene accumulation in the headspace of the vial. Incubation was conducted at 30 °C and 300 rpms for 23 hours and samples collected for HPLC, gas chromatography–mass spectrometry (GC-MS) and OD600 measurements at the end of the cultivation. Samples were run as triplicates and A. baylyi wild type was used as control.2.4. Biosynthesis of 1-undecene from cellulose by a two-stage system
C. cellulolyticum was precultivated with 5 g L−1 of Avicel suspended in minimal CM3 media for 4 days at 34 °C and 240 rpms. From the precultivations, 5%v was inoculated to 50 ml of minimal CM3 media in 120 ml serum bottles. Cellulose concentrations of 0, 5, 10, 20 and 30 g L−1 (Avicel) were used as substrates for end-metabolite formation study. Cultivations were carried out for 11 days at 34 °C and 240 rpm. After 9 days of incubation, 1 ml HPLC samples were collected daily to monitor metabolite formation. The sampling volume was replaced by an equal volume of N2 in the headspace. The cultivation was stopped on day 11 when no more end-metabolites were accumulating in the media. The cultures were centrifuged (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g × 15 min), the supernatants filtered through a 0.2 μm filter and used for subsequent cultivations with ADP1_undA. All experiments were conducted as triplicates except for the cultivation with 30 g L−1 cellulose as a duplicate.
000g × 15 min), the supernatants filtered through a 0.2 μm filter and used for subsequent cultivations with ADP1_undA. All experiments were conducted as triplicates except for the cultivation with 30 g L−1 cellulose as a duplicate.
Overnight precultivations of ADP1_undA were washed with phosphate buffered saline (PBS). MSM components were added to 5 ml of C. cellulolyticum culture supernatants (0 g L−1 and 30 g L−1) and inoculated with the washed ADP1_undA cells to an initial OD of 0.15. Cultivations were carried out in 20 ml sealable glass vials at 30 °C and 300 rpms, and cell growth was measured as OD600. The vials were sealed to allow accumulation of 1-undecene in the headspace of the vial. The cells were incubated for 23 hours at 30 °C and 300 rpms and samples were collected for GC-MS, HPLC and OD600 analyses. Samples were run as triplicates.
2.5. Biosynthesis of 1-undecene from technical lignin
For the multispecies approach on technical lignin, minimal CM3 media was supplemented with 20 g L−1 of technical lignin and inoculated with C. cellulolyticum. Cultivations were carried out anaerobically for 7 days. Then, 5 ml of the culture was transferred aerobically to sealable glass vials and inoculated with ADP1_undA. Identical cultivations without C. cellulolyticum were carried out as control 1. Cultivations without technical lignin were used as control 2. All samples, including controls, were inoculated with ADP1_undA in the second stage and cultivated as triplicates. The experimental procedure was carried out as described in sections 2.4 and 2.3.2.6. Collection of biosynthetically produced 1-undecene
1-Undecene collection system was constructed and integrated to a bioreactor (ESI Fig. 1†). Two separate collection units were attached to the exhaust pipe of the bioreactor with silicon-based gas proof tubing, cooled on ice bath and connected to a condensing unit cooled with circulating cold water (1 °C). The collection vessels were filled with 100 ml heptane to trap 1-undecene. The bioreactor was aerated, which facilitated directing 1-undecene towards the collection vessel.ADP1_undA was cultivated in bioreactor in 1-litre vessel (Sartorius Biostat B plus Twin System, Germany) with a cultivation volume of 500 ml at 30 °C and 350 rpm. The partial oxygen pressure was controlled to 20% of saturation by supply of oxygen/air mixture at 1 vvm. The exhaust pipe of the reactor was connected to the 1-undecene collection system. The cultivation was performed in batch mode, with 500 ml of MSM medium supplemented with 40 mM glucose, 43 mM acetate, and 60 mM. The OD was followed with an online probe (Hamilton Dencytee, Bonaduz, Switzerland), and substrate consumption was followed by HPLC. For 1-undecene measurement, the heptane from the collection system was sampled and subjected directly to analysis by GC-MS. Additionally, at the end of the cultivation, the cells were harvested by centrifugation (25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g × 5 min) and analysed for intracellular 1-undecene. For that, the cellular lipids were extracted by methanol-chloroform extraction as described in product analytics (see section 2.7), and the chloroform phase was used for GC-MS analysis.
000g × 5 min) and analysed for intracellular 1-undecene. For that, the cellular lipids were extracted by methanol-chloroform extraction as described in product analytics (see section 2.7), and the chloroform phase was used for GC-MS analysis.
2.7. Metabolite and product analytics
Acetate, ethanol, glucose and L-lactate concentrations were measured with HPLC (LC-20AD, Shimadzu, Japan) equipped with a Rezex RHM-Monosaccharide H + (8%) 300 × 8 mm column (Phenomenex, USA), refractive index detector (RID, RID-10A) and using 5 mM H2SO4 as mobile phase. The pump (G1211A) flow was adjusted to 0.6 ml min−1, the column temperature to 40 °C, and peaks were identified by comparing the retention times to prepared standards.Intracellular 1-undecene was extracted and analysed from cell pellets by methanol–chloroform extraction and gas chromatography (GC-MS) as previously described.35 Briefly, 3 ml of cell culture was centrifuged (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g × 5 min) and the pellet suspended in 500 μl of methanol. Chloroform (250 μl) was added and the samples were mixed at room temperature for one hour. Chloroform (250 μl) and PBS (250 μl) were added, the samples were mixed for another two hours and centrifuged. A sample from chloroform phase (500 μl) was used in GC-MS analysis (6890N/5975B; Agilent Technologies, Santa Clara, CA). A HP-5MS 30 m × 0.25 mm column (0.25 μm film thickness) was used with 4.7 ml min−1 helium flow rate and 1 μl splitless injection. The following oven program was used: 55 °C hold 5 min, 55–280 °C 20° min−1 ramp and 280 °C hold 3 min. Scanning was set at 50–500 m/z, 1.68 scan per s. 1-Undecene was identified based on the NIST library (Version 2.2/June 2014) and 1-undecene external standards.
000g × 5 min) and the pellet suspended in 500 μl of methanol. Chloroform (250 μl) was added and the samples were mixed at room temperature for one hour. Chloroform (250 μl) and PBS (250 μl) were added, the samples were mixed for another two hours and centrifuged. A sample from chloroform phase (500 μl) was used in GC-MS analysis (6890N/5975B; Agilent Technologies, Santa Clara, CA). A HP-5MS 30 m × 0.25 mm column (0.25 μm film thickness) was used with 4.7 ml min−1 helium flow rate and 1 μl splitless injection. The following oven program was used: 55 °C hold 5 min, 55–280 °C 20° min−1 ramp and 280 °C hold 3 min. Scanning was set at 50–500 m/z, 1.68 scan per s. 1-Undecene was identified based on the NIST library (Version 2.2/June 2014) and 1-undecene external standards.
1-Undecene measurements from culture headspace were conducted according to a previously established method.27 Briefly, 1-undecene was collected with an SPME fibre (df 30 μm, needle size 24 ga, polydimethylsioxane, Supelco, Sigma-Aldrich, USA) from the sealed headspace of Agilent certified 20 ml glass vials used as cultivation vessels. Collection was conducted at 25 °C under constant stirring of the culture media for 12 min. The MS-GC analysis of the samples was performed with Agilent 6890 N GC with 5975B inert XL MSD by desorbing the fibre in a splitless injector for 75 s at 250 °C. Helium was used as carrier gas (1 ml min−1) and the following temperature gradient was used: 50 °C for 3 min, temperature ramped to 130 °C with a rate of 10 °C min−1, then ramped to 300 °C with a rate of 30 °C min−1, 300 °C for 5 min. 1-Undecene was quantified by comparing the peaks to 1-undecene standards. The standards were prepared with 1-undecene mixed with 5 ml of culture media sealed in 20 ml glass vials, collected with the SPME fibre similarly to the samples and analysed with GC-MS.
2.8. Lignin analytics
ADP1_red was cultivated in MSM media supplemented with 20 g L−1 of technical lignin at an initial cell density of OD600 0.2. Separate cultivations with 22 mM of glucose supplementations were carried out as positive controls. Media containing 20 g L−1 of technical lignin without ADP1_undA was used as a negative control. All cultivations were carried out for 7 days as duplicates (30 °C, 300 rpm, 50 ml volume). Cellular growth was monitored daily by measuring fluorescence signal produced by RFP (excitation 560 nm/emission 590 nm, Fluoroskan Ascent plate reader, ThermoLabsystems, Finland). Samples were collected for fluorescent measurements directly from the cultures at a total volume of 200 μL. Relative fluorescence signal was calculated by dividing the fluorescence signal of ADP1_red with the signal from the negative control (sample signal/background signal).After seven days, the cultures were centrifuged (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g × 30 min) and the supernatants were separated from the precipitates. The precipitates were washed with H2O on paper filters (5–13 mesh) and dried in vacuum over sodium hydroxide to obtain samples representing insoluble lignin fraction. Changes in absorption bands in the insoluble lignin fractions were analysed by Fourier-transform infrared spectroscopy (FTIR) spectrometer (PerkinElmer One, USA). The supernatants were filtered through 0.2 μm pore size filters to obtain samples representing water soluble fraction. Changes of the aromatic protons content in the water-soluble fractions were analysed by nuclear magnetic resonance spectroscopy (NMR) (JEOL JNM-ECZ500R spectrometer (500 MHz) equipped with Royal HFX probe. For NMR analysis, samples were concentrated on a rotor-evaporator, then dried in vacuum, re-dissolved in 0.7 mL of D2O, and NMR spectra were measured. The spectra were analyzed with Delta v5.0 program. Absorbance spectra of the water-soluble fractions were also recorded from the centrifuged and filtered supernatants on an ultraviolet–visible spectroscopy (UV-Vis) UV-1800 spectrophotometer (Shimadzu, Japan). For absorbance measurements samples were diluted in water (1
000g × 30 min) and the supernatants were separated from the precipitates. The precipitates were washed with H2O on paper filters (5–13 mesh) and dried in vacuum over sodium hydroxide to obtain samples representing insoluble lignin fraction. Changes in absorption bands in the insoluble lignin fractions were analysed by Fourier-transform infrared spectroscopy (FTIR) spectrometer (PerkinElmer One, USA). The supernatants were filtered through 0.2 μm pore size filters to obtain samples representing water soluble fraction. Changes of the aromatic protons content in the water-soluble fractions were analysed by nuclear magnetic resonance spectroscopy (NMR) (JEOL JNM-ECZ500R spectrometer (500 MHz) equipped with Royal HFX probe. For NMR analysis, samples were concentrated on a rotor-evaporator, then dried in vacuum, re-dissolved in 0.7 mL of D2O, and NMR spectra were measured. The spectra were analyzed with Delta v5.0 program. Absorbance spectra of the water-soluble fractions were also recorded from the centrifuged and filtered supernatants on an ultraviolet–visible spectroscopy (UV-Vis) UV-1800 spectrophotometer (Shimadzu, Japan). For absorbance measurements samples were diluted in water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10).
10).
2.9. Chemical synthesis of poly-α-olefins from 1-undecene
The oligomerization reaction for commercial 1-undecene was carried out under argon atmosphere in anhydrous conditions. All used glassware with magnetic stir bars was heated in gas burner under vacuum (5 mbar), cooled under vacuum and filled up with argon for few times. Toluene was freshly distilled over sodium and additionally dried over molecular sieves (4 Å) and CaH2 and degassed before use. 1-Undecene was dried over CaH2, degassed and stored under argon. Zirconocene dichloride (Cp2ZrCl2) was dried in vacuum over NaOH for few hours before use. MMAO-12 (7% solution in toluene) was taken under argon for the reaction. 1-Undecene (0.5 mL, 2.43 mmol) was introduced into the reaction vial through a membrane. The solution of MMAO-12 (2.9 mL, 3.2 mmol) in toluene (7%) was added to 1-undecene and the mixture was stirred under argon for 15 min. A freshly prepared solution of Cp2ZrCl2 in toluene (0.4 mL, 0.016 mmol) was introduced into the reaction mixture. The reaction mixture was stirred under argon at 50 °C for 5 h and at room temperature overnight. After quenching with HCl (40 mL of a 10% aqueous solution) the resulting mixture was stirred for 30 min. The layers were separated, and the aqueous layer was extracted with diethyl ether (40 mL) twice. The combined organic layers were washed with saturated NaHCO3 solution and dried over Na2SO4. After filtration, all volatiles were removed under vacuum to yield colorless transparent oil (0.22 g, 58.7%).The product was analyzed by NMR and gas chromatograph flame ionization detector (GC-FID). NMR spectra were measured using a Varian Mercury 300 MHz spectrometer (Varian Inc., USA). All chemical shifts are given in ppm relative to tetramethylsilane (TMS) as an internal standard. GC was performed using the instrument Thermo-Finnigan equipped with 100% polydimethyl siloxane 30 m × 0.32 mm × 0.25 μm film column and an FID detector. The inlet temperature was 290 °C, the initial column temperature was 50 °C held at 1 min and the temperature was increased at 25 °C min−1 up to a final temperature 260 °C held for 10 min. Helium flow was 2 mL min−1.
3. Results and discussion
Two of the major components of softwood, cellulose and lignin, are challenging substrates for any currently known bacteria that can be genetically engineered for the synthesis of non-native products. Additional challenges in softwood bioprocessing are caused by the high content of lignin that is more resistant to biological degradation compared to non-wood lignin.36 Although sophisticated genetic tools are available for common industrial hosts, such as E. coli and Saccharomyces cerevisiae, development of a robust cell factory tackling both cellulose and lignin is generally impeded by the limited substrate utilization capabilities of the host, as well as severe metabolic burden caused by complex synthetic metabolic rewiring.5 To alleviate the challenges related to single strain cultures, rationally designed multispecies cultures allow distribution of the metabolic burden and the utilization of wider substrate range. For example, A. baylyi is a microorganism capable of aromatic catabolism19,20 and on the other hand, cellulolytic C. cellulolyticum solubilize and ferment cellulose to end-metabolites acetate, lactate and ethanol.8 In turn, these end-metabolites are applicable substrates for A. baylyi. As a result, by combining the metabolism of two divergent species both cellulose and technical lignin-originated molecules can be funnelled for production. The genetic amenability of A. baylyi allows utilization of synthetic pathways for non-native products. In this case, by heterologous enzyme expressions, the substrates were used to produce semi-volatile 1-undecene. This multispecies biological funnelling approach is illustrated in Fig. 1.3.1. Biosynthesis of 1-undecene
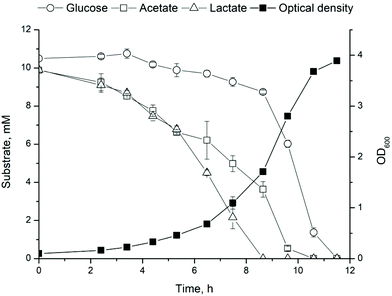
As a first step towards the two-stage production of 1-undecene from cellulose and technical lignins, a strain for 1-undecene production, ADP1_undA, was constructed using genomic integration. Genomic integration of synthetic pathways in bacteria provides advantages over plasmid expression, namely due to improved stability of the construct and avoidance of selection agents such as antibiotics which contribute to significant production costs at industrial level.37,38 Especially, instability caused by nutritional limitations and varying growth conditions becomes a pressing issue when heterologous and seasonably variable feedstocks, such as lignocellulose containing waste streams are used.Growth and product synthesis by ADP1_undA were studied on defined media based on compounds released during cellulose fermentation by C. cellulolyticum, that is glucose, acetate and lactate.9 First, cell growth was studied on 10 mM mixture of glucose, acetate and lactate. The substrates were consumed within 12 hours and the cells reached a final OD of 3.9 (Fig. 2) confirming that the substrate mixture provides an excellent carbon source for rapid growth without significant carbon catabolite repression. Second, the biosynthesis of 1-undecene was verified by supplementing ADP1_undA cultivations with 35 mM of lactate or acetate. Both lactate and acetate served as good substrates for biosynthesis, although lactate seems to be distributed more equally between biomass and product synthesis when compared to acetate (Table 1). Furthermore, the expression of the non-native pathway does not affect cell growth (measured as OD600) (Table 1). Although the titers obtained here are modest, they are comparable to previously obtained results by a plasmid-based expression system in ADP1 using glucose (694 ± 76 μg L−1) or ferulate (72 ± 7.5) as a substrate.16 Similar to observations made by Luo et al.,16 the wild type ADP1 used as a control produced trace amounts of 1-undecene most likely due to a native uncharacterized 1-undecene production activity.
| Strain | Substrate | Substrate consumed, mM | Substrate consumed, g L−1 | 1-Undecene titer, μg L−1 | 1-Undecene yield, μg gsubstrate−1 | OD600 |
|---|---|---|---|---|---|---|
| ADP1 WT | Lactate | 24 | 2.2 | 8 ± 0 | 2 ± 0 | 1.2 ± 0.0 |
| ADP1_undA | Lactate | 24 | 2.2 | 129 ± 11 | 59 ± 3 | 1.4 ± 0.1 |
| ADP1_undA | Acetate | 26 | 1.6 | 128 ± 12 | 82 ± 7 | 1.1 ± 0.0 |
3.2. Biosynthesis of 1-undecene from cellulose by a two-stage system
To evaluate cellulose as a substrate for 1-undecene synthesis, a two-stage cultivation system was investigated where C. cellulolyticum was first employed to enzymatically saccharify cellulose to glucose and simultaneously ferment it to end-metabolites, namely acetate and lactate. The end-metabolites were then used for product synthesis by ADP1_undA.According to preliminary experiments, 30 g L−1 of cellulose (Avicel) was chosen as a substrate due to the highest end-metabolite accumulation of both lactate and acetate (ESI Table 1†). In the first stage of the cultivation, C. cellulolyticum produced 5.2 mM of glucose, 4.9 mM of acetate and 6.8 mM of lactate solely from cellulose. In the second stage, ADP1_undA utilized 80% of the lactate, 16% of the acetate and 3% of the glucose for biomass and 1-undecene synthesis (Table 2). A cellulose control cultivation without Avicel supplementation (i.e. without carbon source) was also conducted. Some 1-undecene was also detected from the control cultivation. As the media was devoid of any carbon sources, and as no growth was observed, it stands to reason that the 1-udecene was produced indirectly from storage compounds accumulated during the precultivation of ADP1_undA. For example, it has previously been demonstrated that wax esters that are produced as storage compounds are degraded during carbon starvation.33,39 Regardless, ADP1_undA produced 1-undecene 2.7-fold compared to the control cultivation verifying that cellulose can indeed be used as sole carbon source for 1-undecene synthesis. These results indicate that the end-metabolites of C. cellulolyticum serve as excellent carbon sources for A. baylyi ADP1, and the 1-undecene titers are comparable to those obtained with defined media. Furthermore, cellulose fermentation by C. cellulolyticum does not produce inhibitors for ADP1_undA as the end-metabolites are readily consumed by ADP1.
| Sample | Strain(s) | Substrate | 1-Undecene titer, μg L−1 | |
|---|---|---|---|---|
| 1-Undecene production from cellulose | Two-stage production | C. cellulolyticum and ADP1_undA | Pure cellulose (Avicel) | 107 ± 8 |
| Cellulose control | C. cellulolyticum and ADP1_undA | No substrate | 39 ± 2 | |
| 1-Undecene production from technical lignin | Two-stage production | C. cellulolyticum and ADP1_undA | Technical lignin | 88 ± 5 |
| C. cellulolyticum control | ADP1_undA | Technical lignin | 56 ± 1 | |
| Technical lignin control | C. cellulolyticum and ADP1_undA | No substrate | 49 ± 1 |
Microbial conversion of lignocellulosic biomass involves a multitude of biological tasks. In multispecies cultures, a biosynthetic pathway can be divided between microorganisms for enhanced production compared to single strain cultures. A typical division of labor for the utilization of lignocellulose is divided between saccharolysis and fermentation to produce ethanol from cellulose.40,41 Here, it was shown for the first time that 1-undecene (C11) can be produced solely from cellulose in a two-stage multispecies approach. Further plug and play configurations in the second stage are also available for wider product range, where ADP1 could be used for native long-chain alkyl ester (C36) production or other non-native fatty-acid derived products, such as alkanes (C17).15,17
3.3. Biosynthesis of 1-undecene from technical lignin
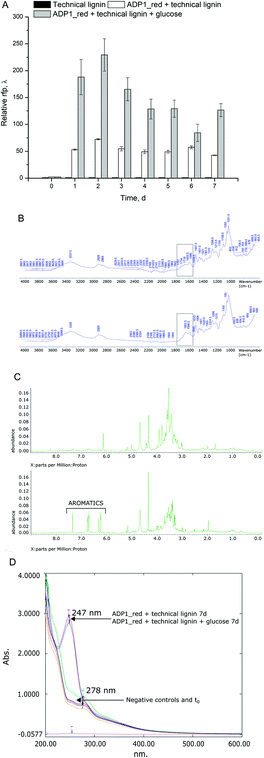
In addition to lignin, technical hydrolysis lignins contain cellulose up to 15–20%, as well as hemicellulose, sugars, phenols, and organic acids, depending on the wood origin and process conditions. Many of these compounds are not optimal substrates for current bioprocesses. The phenols and organic acids are inhibitory to common microbial hosts,42,43 whereas they are reasonably tolerated and consumed by soil bacteria such as A. baylyi ADP1.17,18 Firstly, to investigate the biological conversion potential from softwood-based technical lignin, ADP1 expressing red fluorescent protein (ADP1_red) was employed as a reporter strain and growth and lignin depolymerization were assessed. Secondly, as a proof of principle, ADP1_undA was used to produce 1-undecene from the same cellulose-rich technical lignin by a two-stage cultivation with C. cellulolyticum.Very interestingly, ADP1_red showed signs of lignin depolymerization activities and growth on the technical lignin from soft-wood origins. Most of the APD1_red biomass (measured as RFP) was produced during the first 48 hours (Fig. 3A). As expected, glucose (22 mM) supplementation promoted biomass formation significantly compared to the cultures grown solely on technical lignin. Thus, the lower biomass obtained from technical lignin relates to scarcity of the condense lignin substrate rather than toxic effects on ADP1. After seven-days of cultivation, the effects on lignin by ADP1_red were evaluated by several analytical methods. The FTIR-spectra revealed an increase in the absorption band at 1655 cm−1, which corresponds to vibrations of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds conjugated with aromatic ring indicating of structural changes in the insoluble lignin fraction (Fig. 3B). Furthermore, NMR analysis verified appearance of aromatics in the soluble fraction of the technical lignin (Fig. 3C). Solubilisation of aromatics from lignin was further supported by the changes observed in UV-Vis spectra (Fig. 3D) as a clear shift in the absorption maxima from 278 to 247 nm was detected. This shift is probably due to the leaching of hydroxylated aromatics of lignin into the solution with simultaneous dehydroxylation. The maximum at 278 nm is typical for absorbance spectrum of phenol compounds,44 whereas the maximum at 245 nm is characteristic for dehydroxylated aromatics.45 However, the absorbance-based results should be cautiously interpreted, but taken together with the FTIR and NMR analyses, the results support the hypothesis that A. baylyi ADP1, to at least some extent, can degrade lignin polymer from softwood origins. Previously ADP1 has been described to have ligninolytic activity for non-wood lignin21 and Acinetobacter sp. have been identified with ligninolytic effects on hardwood47 and genes for laccases, which are enzymes capable of oxidation of lignin polymer.22 To our knowledge, this is the first time ADP1 has been observed with ligninolytic activities on softwood lignin, which differs structurally from grassy, herbaceous and hardwood lignins.48 Although softwoods present a vast resource for bioprocessing, it has not been extensively used due to biological resistance.
O bonds conjugated with aromatic ring indicating of structural changes in the insoluble lignin fraction (Fig. 3B). Furthermore, NMR analysis verified appearance of aromatics in the soluble fraction of the technical lignin (Fig. 3C). Solubilisation of aromatics from lignin was further supported by the changes observed in UV-Vis spectra (Fig. 3D) as a clear shift in the absorption maxima from 278 to 247 nm was detected. This shift is probably due to the leaching of hydroxylated aromatics of lignin into the solution with simultaneous dehydroxylation. The maximum at 278 nm is typical for absorbance spectrum of phenol compounds,44 whereas the maximum at 245 nm is characteristic for dehydroxylated aromatics.45 However, the absorbance-based results should be cautiously interpreted, but taken together with the FTIR and NMR analyses, the results support the hypothesis that A. baylyi ADP1, to at least some extent, can degrade lignin polymer from softwood origins. Previously ADP1 has been described to have ligninolytic activity for non-wood lignin21 and Acinetobacter sp. have been identified with ligninolytic effects on hardwood47 and genes for laccases, which are enzymes capable of oxidation of lignin polymer.22 To our knowledge, this is the first time ADP1 has been observed with ligninolytic activities on softwood lignin, which differs structurally from grassy, herbaceous and hardwood lignins.48 Although softwoods present a vast resource for bioprocessing, it has not been extensively used due to biological resistance.
The appearance of the soluble aromatics indicates microbial activities of ADP1 towards lignin polymer. Soluble low molecular weight lignin compounds can also be entwined in the residual holocellulose fraction of the technical lignin. However, as A. baylyi cannot depolymerize cellulose, the most probable origin of the soluble aromatics is from the ligninolytic activities of ADP1. Regardless, this experiment reveals that ADP1 can release soluble aromatics from the compounds present in the technical lignin for further upgrading. Although A. baylyi ADP1 possesses lignin-degrading capabilities, the release of smaller size polymers, oligomers, and monomers for bioconversion needs to be enhanced. In future, improved phenotypes for lignin degradation could be obtained by adaptive laboratory evolution.46
For a proof of concept, the two-stage cultivation by C. cellulolyticum and ADP1_undA was used to produce 1-undecene from the technical lignin which contains, in addition to lignin polymers and oligomers, residual cellulose. In this experimental set-up, C. cellulolyticum was first cultivated with the technical lignin in anaerobic conditions. After the anaerobic phase, the cultivation was transferred to 1-undecene production vessels, inoculated with ADP1_undA and the vessel was sealed for 1-undecene collection. An identical experiment without C. cellulolyticum (blank C. cellulolyticum control) was conducted to compare the effects of cellulose fermentation by C. cellulolyticum on product synthesis by ADP1_undA from the technical lignin. First, the blank control (i.e. technical lignin without C. cellulolyticum) was anaerobically incubated. Then, the control was transferred to 1-undecene production vessels, inoculated with ADP1_undA and the vessel was sealed for 1-undecene collection. As expected, the two-stage system with cellulose fermentation produced 1.5-fold higher titer compared to the C. cellulolyticum blank control (Table 2). To consider the possible 1-undecene production from the storage compounds produced during precultivations, as was seen in the experiments done with pure cellulose, a control cultivation without technical lignin was conducted (technical lignin control, devoid of carbon source). Similarly to the previous experiments, some 1-undecene was also detected from this control. The blank C. cellulolyticum control (i.e. cultivations inoculated only with ADP1_undA) produced slightly more 1-undecene compared to the technical-lignin control (i.e. control without carbon source) indicating that components of the technical lignin can be used for 1-undecene production by ADP1, albeit the differences between the controls were very modest (1.1-fold increase in titer when technical lignin was present). The effects of C. cellulolyticum fermentation on 1-undecene production were, however, notable. Cellulose fermentation produces end-metabolites lactate and acetate, which can be used for the product synthesis by ADP1_undA. Furthermore, the cellulose depolymerization by C. cellulolyticum can release soluble aromatic low-molecular weight lignin-compounds from the entwined holocellulose structure. In turn, these lignin-related soluble aromatic compounds can also be used for biosynthesis by ADP1.16,17 Overall, this experiment demonstrates the potential of multispecies approach for the utilization of heterogenous substrates such as technical lignin for bioproduction purposes.
3.4. Collection of biosynthetically produced 1-undecene
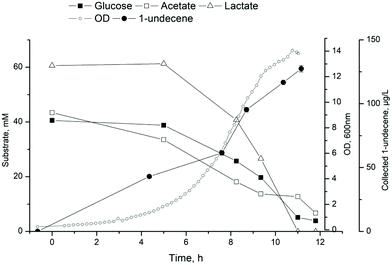
1-Undecene is a semi-volatile carbohydrate (C11) that is partially secreted outside the cells.16,49 This is convenient considering down-stream processes as the product is easily separated from the cultivation broth. Especially in the case of bioreactor with a continuous flow of gas through the system, the 1-undecene produced by the cells can evaporate from the culture vessel. To study continuous recovery of 1-undecane during cultivation, a collection system was constructed and connected to a bioreactor. Initial characterization of the collection system with pure 1-undecene revealed that approximately 12% of 1-undecene was evaporated from the reactor and trapped in heptane in 6 hours in conditions identical to the bacterial cultivations (30 °C, air flow 1 vvm, 350 rpm, 500 ml MSM). To confirm that trapped 1-undecene was not escaping from the system, a separate experiment was carried out by measuring known concentrations of 1-undecene from the trap. The heptane trap was efficient in retaining undecane, and only negligible amounts of undecane evaporated from the trap.ADP1_undA was employed for the production of 1-undecene in a bioreactor setup coupled with the collection system. A synthetic culture media containing 40 mM glucose, 43 mM acetate and 60 mM lactate mimicking the ratios from cellulose fermentation by C. cellulolyticum was used. After 10.5 hours, most of the substrates were consumed and OD of 14 was reached (Fig. 4). 1-Undecene titer of 127.5 ± 2.5 μg L−1 (calculated per cultivation volume) was detected in the collection system, whereas intracellular 1-undecene was detected at levels of 1.5 mg L−1. However, the continuously collected product had higher purity 1-undecene than the extracted intracellular fraction, determined by GS-MS analysis (ESI Fig. 2†). The collection system allows product recovery directly from the culture broth reducing downstream efforts. The purity of the product is important considering further use. Additionally, the collection system would provide means for easy separation and collection from complex and heterogeneous substrates such as technical lignin.
The total titers of 1-undecene obtained here are comparable to other production hosts with undA overexpression. For example, S. cerevisiae and E. coli have been used for heterologous 1-undecene production from glucose or rich media with titers of ∼22 μg L−1 and 6 mg L−1, respectively.27,50 Based on our results, 1-undecene was also accumulated inside the cells. To improve the efficiency of the continuous collection system, engineering of efflux pumps to excrete the 1-undecene outside the cells could facilitate evaporation and collection of the product.51
3.5. Chemical synthesis of poly-α-olefins from 1-undecene
PAOs are examples of branched synthetic hydrocarbons used as industrial lubricants. The uniform distribution of molecular weight of the PAO polymer attributes to high viscosity index (over 130) and excellent low-temperature properties.52 The properties of PAOs obtained from biosynthetically produced 1-undecene would correspond closely with synthetic PAOs and serve as excellent drop-in chemicals. The synthesis of C33 PAOs can be carried out by oligomerization of 1-undecene to trimers. To ensure sufficient amount of 1-undecene for efficient oligomerization reaction, the reactions here were carried out using commercially available 1-undecene.To demonstrate the suitability of 1-undecene for preparation of PAOs, trimerization of 1-undecene using Cp2ZrCl2/MMAO as a catalyst was performed. NMR and gas chromatography analyses confirmed the oligomerization of the starting material (1-undecene) to PAOs (C33) (ESI Fig. 3 and 4†). A 100% conversion of the monomer to reaction products was obtained by using an excess of the co-catalyst/catalyst MMAO (Al/Zr = 200). The NMR proton spectrum of the product corresponded to the theoretical proton ratios of undecene trimer. The presence of the target trimer (C33) was verified with GC. Only minor impurities were present that corresponded to lower molecular weight compounds originating most likely from side reactions. The content of the target trimer in the final product can be estimated as 54%, based on GC-integration. This is reasonably high content for oligomerization reactions. Hence, it was demonstrated that the 1-undecene can be used for the production of trimer PAOs.
4. Conclusions
Here, a platform for the biosynthesis of 1-undecene from technical lignin and cellulose was established. Broad substrate range was achieved by metabolically combining two distinctive bacteria in a two-stage culture setup. Feasible product recovery and possibilities for further downstream processing to PAOs highlight the industrial relevance of the product. However, production metrics including titer, yield, and productivity require resolving before industrial realization. Nonetheless, our study shows the potential of designed multispecies funnelling for enhanced substrate conversion and the power of synthesising tailor-made products to simplify downstream processing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Academy of Finland (grant no. 311986, 310188, 310135, 334822).References
- A. J. Ragauskas, G. T. Beckham, M. J. Biddy, R. Chandra, F. Chen, M. F. Davis, B. H. Davison, R. A. Dixon, P. Gilna, M. Keller, P. Langan, A. K. Naskar, J. N. Saddler, T. J. Tschaplinski, G. A. Tuskan and C. E. Wyman, Science, 2014, 344, 1246843–1246843 CrossRef PubMed.
- G. T. Beckham, C. W. Johnson, E. M. Karp, D. Salvachúa and D. R. Vardon, Curr. Opin. Biotechnol., 2016, 42, 40–53 CrossRef CAS PubMed.
- V. Passoth and M. Sandgren, Appl. Microbiol. Biotechnol., 2019, 103, 5105–5116 CrossRef CAS PubMed.
- O. Rosales-Calderon and V. Arantes, Biotechnol. Biofuels, 2019, 12, 240 CrossRef PubMed.
- G. Wu, Q. Yan, J. A. Jones, Y. J. Tang, S. S. Fong and M. A. G. Koffas, Trends Biotechnol., 2016, 34, 652–664 CrossRef CAS PubMed.
- Q. He, C. L. Hemme, H. Jiang, Z. He and J. Zhou, Bioresour. Technol., 2011, 102, 9586–9592 CrossRef CAS PubMed.
- J. J. Minty, M. E. Singer, S. A. Scholz, C. H. Bae, J. H. Ahn, C. E. Foster, J. C. Liao and X. N. Lin, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 14592–14597 CrossRef CAS PubMed.
- E. Petitdemange, F. Caillet, J. Giallo and C. Gaudin, Int. J. Syst. Bacteriol., 1984, 34, 155–159 CrossRef.
- M. Desvaux, E. Guedon and H. Petitdemange, Appl. Environ. Microbiol., 2000, 66, 2461–2470 CrossRef CAS PubMed.
- V. Barbe, D. Vallenet, N. Fonknechten, A. Kreimeyer, S. Oztas, L. Labarre, S. Cruveiller, C. Robert, S. Duprat, P. Wincker, L. N. Ornston, J. Weissenbach, P. Marlière, G. N. Cohen and C. Médigue, Nucleic Acids Res., 2004, 32, 5766–5779 CrossRef CAS PubMed.
- V. de Berardinis, M. Durot, J. Weissenbach and M. Salanoubat, Curr. Opin. Microbiol., 2009, 12, 568–576 CrossRef PubMed.
- D. Metzgar, J. M. Bacher, V. Pezo, J. Reader, V. Doring, P. Schimmel, P. Marliere and V. de Crecy-Lagard, Nucleic Acids Res., 2004, 32, 5780–5790 CrossRef CAS PubMed.
- R. Kalscheuer and A. Steinbüchel, J. Biol. Chem., 2003, 278, 8075–8082 CrossRef CAS PubMed.
- M. Durot, F. Le Fèvre, V. de Berardinis, A. Kreimeyer, D. Vallenet, C. Combe, S. Smidtas, M. Salanoubat, J. Weissenbach and V. Schachter, BMC Syst. Biol., 2008, 2, 85 CrossRef PubMed.
- T. Lehtinen, E. Efimova, P. L. Tremblay, S. Santala, T. Zhang and V. Santala, Bioresour. Technol., 2017, 243, 30–36 CrossRef CAS PubMed.
- J. Luo, T. Lehtinen, E. Efimova, V. Santala and S. Santala, Microb. Cell Fact., 2019, 18, 48 CrossRef PubMed.
- M. Salmela, T. Lehtinen, E. Efimova, S. Santala and V. Santala, Biotechnol. Bioeng., 2019, 116, 1934–1945 CrossRef CAS PubMed.
- K. Salcedo-Vite, J. C. Sigala, D. Segura, G. Gosset and A. Martinez, Appl. Microbiol. Biotechnol., 2019, 103, 6217–6229 CrossRef CAS PubMed.
- C. S. Harwood and R. E. Parales, Annu. Rev. Microbiol., 1996, 50, 553–590 CrossRef CAS PubMed.
- R. M. Jones, L. S. Collier, E. L. Neidle and P. A. Williams, J. Bacteriol., 1999, 181, 4568–4575 CrossRef CAS PubMed.
- D. Salvachúa, E. M. Karp, C. T. Nimlos, D. R. Vardon and G. T. Beckham, Green Chem., 2015, 17, 4951–4967 RSC.
- L. Ausec, M. Zakrzewski, A. Goesmann, A. Schlüter and I. Mandic-Mulec, PLoS One, 2011, 6, e25724 CrossRef CAS PubMed.
- A. Vishtal and A. Kraslawski, BioResources, 2011, 6, 3547–3568 Search PubMed.
- G. Janusz, A. Pawlik, J. Sulej, U. Świderska-Burek, A. Jarosz-Wilkołazka and A. Paszczyński, FEMS Microbiol. Rev., 2017, 41, 941–962 CrossRef CAS PubMed.
- K. Przybysz Buzała, H. Kalinowska, P. Przybysz and E. Małachowska, Wood Sci. Technol., 2017, 51, 873–885 CrossRef.
- T. Dong, W. Xiong, J. Yu and P. T. Pienkos, RSC Adv., 2018, 8, 34380–34387 RSC.
- Z. Rui, X. Li, X. Zhu, J. Liu, B. Domigan, I. Barr, J. H. D. Cate and W. Zhang, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 18237–18242 CrossRef CAS PubMed.
- M. Salmela, T. Lehtinen, E. Efimova, S. Santala and R. Mangayil, Biotechnol. Biofuels, 2018, 11, 187 CrossRef PubMed.
- R. Mangayil, E. Efimova, J. Konttinen and V. Santala, New Biotechnol., 2019, 53, 81–89 CrossRef CAS PubMed.
- P. S. Losoi, V. P. Santala and S. M. Santala, ACS Synth. Biol., 2019, 8, 2642–2650 CrossRef CAS PubMed.
- E. Guedon, M. Desvaux, S. Payot and H. Petitdemange, Microbiology, 1999, 145, 1831–1838 CrossRef CAS PubMed.
- S. Hartmans, J. P. Smits, M. J. Van Der Werf, F. Volkering and J. A. M. De Bont, Metabolism of Styrene Oxide and 2-Phenylethanol in the Styrene-Degrading Xanthobacter Strain 124X, 1989, vol. 55 Search PubMed.
- S. Santala, E. Efimova, M. Karp and V. Santala, Microb. Cell Fact., 2011, 10, 75 CrossRef CAS PubMed.
- D. Metzgar, Nucleic Acids Res., 2004, 32, 5780–5790 CrossRef CAS PubMed.
- T. Lehtinen, V. Santala and S. Santala, FEMS Microbiol. Lett., 2017, 364 DOI:10.1093/femsle/fnx053.
- J. Y. Zhu and X. J. Pan, Bioresour. Technol., 2010, 101, 4992–5002 CrossRef CAS PubMed.
- M. C. Bassalo, A. D. Garst, A. L. Halweg-Edwards, W. C. Grau, D. W. Domaille, V. K. Mutalik, A. P. Arkin and R. T. Gill, ACS Synth. Biol., 2016, 5, 561–568 CrossRef CAS PubMed.
- C. N. S. Santos and Y. Yoshikuni, Nat. Protoc., 2014, 9, 1320–1336 CrossRef CAS PubMed.
- L. M. Fixter, M. N. Nagi, J. G. Mccormack and C. A. Fewson, Microbiology, 1986, 132, 3147–3157 CAS.
- T. R. Zuroff, S. B. Xiques and W. R. Curtis, Biotechnol. Biofuels, 2013, 6, 59 CrossRef CAS PubMed.
- T. R. Zuroff and W. R. Curtis, Appl. Microbiol. Biotechnol., 2012, 93, 1423–1435 CrossRef CAS PubMed.
- E. Palmqvist and B. Hahn-Hägerdal, Bioresour. Technol., 2000, 74, 25–33 CrossRef CAS.
- L. J. Jönsson and C. Martín, Bioresour. Technol., 2016, 199, 103–112 CrossRef PubMed.
- M. Martynoff, Bull. Soc. Chim. Fr., 1949, 16, 258–261 Search PubMed.
- W. H. Rodebush and I. Feldman, J. Am. Chem. Soc., 1946, 68, 896–899 CrossRef CAS PubMed.
- M. Dragosits and D. Mattanovich, Microb. Cell Fact., 2013, 12, 64 CrossRef PubMed.
- N. Vasudevan and A. Mahadevan, J. Appl. Bacteriol., 1991, 70, 169–176 CrossRef CAS.
- M. Li, C. Foster, S. Kelkar, Y. Pu, D. Holmes, A. Ragauskas, C. M. Saffron and D. B. Hodge, Biotechnol. Biofuels, 2012, 5, 38 CrossRef CAS PubMed.
- J. M. Zechman and J. N. Labows Jr., Can. J. Microbiol., 1985, 31, 232–237 CrossRef CAS PubMed.
- B. Chen, D.-Y. Lee and M. W. Chang, Metab. Eng., 2015, 31, 53–61 CrossRef CAS PubMed.
- B. Chen, H. Ling and M. W. Chang, Biotechnol. Biofuels, 2013, 6, 21 CrossRef CAS PubMed.
- M. M. Wu, S. C. Ho and T. R. Forbus, in Practical Advances in Petroleum Processing, Springer, New York, 2007, pp. 553–577 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0gc01617a |
| This journal is © The Royal Society of Chemistry 2020 |