Guar gum different from Ganoderma lucidum polysaccharide in alleviating colorectal cancer based on omics analysis
Jianming
Luo†
 ,
Tianxing
Li†
,
Jinli
Xie
,
Hui
Guo
,
Liu
Liu
,
Guangwen
Zhang
,
Tianxing
Li†
,
Jinli
Xie
,
Hui
Guo
,
Liu
Liu
,
Guangwen
Zhang
 * and
Xichun
Peng
* and
Xichun
Peng
 *
*
Department of Food Science and Engineering, Jinan University, Guangzhou 510632, Guangdong, China. E-mail: tzgw@jnu.edu.cn; tpxchun@jnu.edu.cn
First published on 19th December 2019
Abstract
It is unclear if guar gum can alleviate colorectal cancer (CRC). We evaluated the effect of guar gum (unmodified) on the mortality, colon status, serous tumor necrosis factor-alpha (TNF-α) concentration, and gut microbial and colonic epithelial cell gene expression profiles in CRC mice and performed omics analyses to compare these with those of Ganoderma lucidum polysaccharide (GLP), whose main component is β-glucan (>90%). We found that guar gum had a CRC alleviating effect. However, it showed a 20% higher mortality rate, shorter colon length, worse colon status, larger number and size of tumors, higher concentration of serous TNF-α and upregulation of epithelial cell genes (Il10, Cytl1, Igkv7-33, Ighv1-14, Igfbp6 and Foxd3) compared to that of GLP. The higher relative abundance of Akkermansia, the alteration of microbial metabolic pathways, especially those involving chaperones and folding catalysts, fatty acid biosynthesis, glycerophospholipid metabolism, glycolysis/gluconeogenesis, lipid biosynthesis and pyruvate metabolism, and the upregulation of specific genes (Mcpt2, Mcpt9, Des and Sostdc1) were also determined in animals fed a guar gum diet. The results suggested that the alleviating effect of guar gum (an inexpensive polysaccharide) on CRC was inferior to that of GLP (a more expensive polysaccharide). This could potentially be attributed to the increased presence of Akkermansia, the alteration of 10 microbial metabolic pathways and the upregulation of 4 epithelial cell genes.
Introduction
Colorectal cancer (CRC) is a carcinoma with the most estimated new cases among digestive cancers and the estimated cause of the second highest rate of deaths among all cancers in the U.S. in 2019.1 Despite the decreases in the U.S. CRC morbidity and mortality rates, the rapidly rising populations in many developing countries such as China are still suffering from high rates of CRC, possibly due to the adoption of Western diets and lifestyles.2,3 Multiple and extensive investigations and bioinformatics studies have shown that the incidence of CRC is correlated with gut microbiota.4–6 Dietary intervention, which affects intestinal bacteria, will consequently influence the progression of CRC.2 Therefore, intervention with diets with a high content of dietary fiber or nonstarch polysaccharides (NSPs) has become a strategy to prevent or lower the risk of CRC.7,8Dietary fiber or NSPs are defined as food ingredients that can escape digestion in the small intestine and reach the colon.9 Colonic microorganisms ferment NSPs to generate beneficial metabolites and affect colonic epithelial cells, which ultimately prevents CRC.5 Our recent study reported that a diet consisting of Ganoderma lucidum polysaccharide (GLP), of which β-glucan is the main component, with replacement of 10% starch in diet alleviated CRC induced by azoxymethane (AOM) and dextran sulfate sodium (DSS).10 Additionally, polysaccharides from Nostoc commune Vaucher also suppressed CRC.11 It is unclear if all NSPs might have a similar effect on CRC.
As an NSP, guar gum is a complex but easily obtained polysaccharide from the endosperm of Cyamopsis tetragonolobus. It consists of a linear backbone of chains of (1 → 4)-β-D-mannopyranosyl units with branch points with α-D-galactopyranosyl units attached by (1 → 6) linkages.12 In the early 1990s, Heitman et al. found that supplementation with 10% guar gum can suppress colon cancer in rats.13 Nevertheless, McIntyre et al. obtained a contradictory result, indicating that a 10% guar gum diet barely had any effect on rodent colonic tumor suppression.14 With the development of various types of bioinformatics tools, deeper insights into the relationships between food function and gut microbiota became achievable. Therefore, updated research with omics analyses is needed to investigate the exact function of guar gum in CRC by comparing its effects with those of other polysaccharides reported to alleviate CRC, such as GLP.10 This would help to elucidate whether some polysaccharides with lower cost (such as guar gum) could relieve CRC damage and also help in determining the underlying reasons.
In the present research, we investigated the effects of a guar gum diet on the mortality, colon status, tumor numbers and sizes, tumor necrosis factor-alpha (TNF-α) concentration, gut microbial profile and colonic epithelial cell gene expression in CRC mice and sought to explain the differences mainly on the basis of omics analyses by comparing the effects of guar gum with those of GLP.
Materials and methods
Reagents and materials
GLP was provided by Infinitus Co., Ltd (Guangzhou, Guangdong, China) and mainly consisted of β-glucan (β-glucan content >90%). AOM solution was purchased from Sigma-Aldrich Corp. (St Louis, MO, USA), and DSS (MW 36–50 kDa) was obtained from MP Biomedicals LLC (Santa Ana, CA, USA). Unmodified guar gum, paraformaldehyde, hematoxylin solution, eosin solution, phosphate-buffered saline (PBS, pH 7.4) and agarose were obtained from Qiyun Biotechnology Co., Ltd (Guangzhou, Guangdong, China), and the tumor necrosis factor-alpha (TNF-α) enzyme-linked immunosorbent assay (ELISA) kit for mice was obtained from NeoBioscience Corp. (Shenzhen, Guangdong, China). The TIANamp Stool DNA kit was obtained from Tiangen Biotech Co. Ltd (Beijing, China), and FastPfu Polymerase including buffer and dNTPs was obtained from TransGen Biotechnology Co., Ltd (Beijing, China). The AxyPrep DNA gel extraction kit was obtained from Axygen Biosciences (Union City, CA, USA), TRIzol® Reagent was obtained from Invitrogen (Shanghai, China), the TruSeq TM RNA sample prep kit was obtained from Illumina (San Diego, CA, USA), QuantiFluor™-ST was obtained from Promega (Beijing, China), low range ultra agarose was obtained from Bio-Rad Laboratories, Inc. (Shanghai, China) and Phusion DNA polymerase was obtained from NEB (Beijing, China).All animal feeds used in this study were manufactured by the Guangdong Medical Laboratory Animal Center (Foshan, Guangdong, China), which is a research center that has been granted licenses for laboratory animal feed production. The feeds were sterilized by irradiation with Co60 (25 kGy). Formulation no. D12450-B was used as the control diet. A GLP diet and a guar gum diet were prepared by replacing 10% of the corn starch in the diet formulation with equal amounts of GLP and guar gum, respectively (Table 1). The replacement amounts were based on our previous research.10
| Raw material | Control diet | GLP diet | Guar gum diet |
|---|---|---|---|
| Control diet, diet for mice in the N_Con and Ml_Con groups; GLP diet, diet for mice in the M_GLP group; guar gum diet, diet for mice in the M_Guar group. | |||
| Casein | 2500.0 | 2500.0 | 2500.0 |
| L-Cystine | 37.5 | 37.5 | 37.5 |
| Corn starch | 3937.5 | 3543.75 | 3543.75 |
| GLP | 0.0 | 393.75 | 0.0 |
| Guar gum | 0.0 | 0.0 | 393.75 |
| Maltodextrin | 437.5 | 437.5 | 437.5 |
| Sucrose | 4375.0 | 4375.0 | 4375.0 |
| Cellulose | 625.0 | 625.0 | 625.0 |
| Soybean oil | 312.5 | 312.5 | 312.5 |
| Lard | 250.0 | 250.0 | 250.0 |
| Minerals | 437.5 | 437.5 | 437.5 |
| Vitamin | 125.0 | 125.0 | 125.0 |
| Choline | 31.25 | 31.25 | 31.25 |
Animal housing, the CRC model and sample collection
Forty male Balb/c mice (SPF grade, 6–8 weeks old, Guangdong Medical Laboratory Animal Center, Guangdong, China) were housed in polypropylene cages at the Institute of Laboratory Animal Science at Jinan University at a constant temperature (22 ± 2 °C) under a 12 h![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 h light
12 h light![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dark cycle. During the adaption period of ten days, the mice were fed a control diet (no. D12450-B). The experiment lasted for 63 days and was divided into 12 periods. The relationship between the days and periods is as follows: days 1–7 (period 1), days 8–11 (period 2), days 12–17 (period 3), days 18–24 (period 4), days 25–28 (period 5), days 29–32 (period 6), days 33–38 (period 7), days 39–45 (period 8), days 46–49 (period 9), days 50–53 (period 10), days 54–59 (period 11) and days 60–63 (period 12). After adaption, the mice were divided into 4 groups (10 mice per group), which were N_Con, M_Con, M_GLP and M_Guar. Mice in the M_Con, M_GLP and M_Guar groups were administered AOM and DSS solutions to induce the development of colorectal cancer. Briefly, the mice were intraperitoneally injected with AOM (10 mg kg−1) on day 0. On days 8–11 (period 2), 29–32 (period 6) and 50–53 (period 10), the drinking water was replaced by water with 2% DSS,15,16 and on the rest of the days, the mice were supplied with sterilized dH2O (Fig. 1). During the experiment, the mice in groups N_Con and M_Con were offered a control diet, while the mice in groups M_GLP and M_Guar were offered a GLP diet and a guar gum diet, respectively. The detailed treatments used for the groups are listed in Table 2.
dark cycle. During the adaption period of ten days, the mice were fed a control diet (no. D12450-B). The experiment lasted for 63 days and was divided into 12 periods. The relationship between the days and periods is as follows: days 1–7 (period 1), days 8–11 (period 2), days 12–17 (period 3), days 18–24 (period 4), days 25–28 (period 5), days 29–32 (period 6), days 33–38 (period 7), days 39–45 (period 8), days 46–49 (period 9), days 50–53 (period 10), days 54–59 (period 11) and days 60–63 (period 12). After adaption, the mice were divided into 4 groups (10 mice per group), which were N_Con, M_Con, M_GLP and M_Guar. Mice in the M_Con, M_GLP and M_Guar groups were administered AOM and DSS solutions to induce the development of colorectal cancer. Briefly, the mice were intraperitoneally injected with AOM (10 mg kg−1) on day 0. On days 8–11 (period 2), 29–32 (period 6) and 50–53 (period 10), the drinking water was replaced by water with 2% DSS,15,16 and on the rest of the days, the mice were supplied with sterilized dH2O (Fig. 1). During the experiment, the mice in groups N_Con and M_Con were offered a control diet, while the mice in groups M_GLP and M_Guar were offered a GLP diet and a guar gum diet, respectively. The detailed treatments used for the groups are listed in Table 2.
| Treatment | Group | |||
|---|---|---|---|---|
| N_Con | M_Con | M_GLP | M_Guar | |
| The symbol “✗” indicates that the mice were not treated with the corresponding treatment; the symbol “✓” indicates that the mice were treated with the corresponding treatment. | ||||
| AOM, intraperitoneal injection | ✗ | ✓ | ✓ | ✓ |
| DSS, ad libitum (totally 12 days) | ✗ | ✓ | ✓ | ✓ |
| Control diet, ad libitum | ✓ | ✓ | ✗ | ✗ |
| GLP diet, ad libitum | ✗ | ✗ | ✓ | ✗ |
| Guar gum diet, ad libitum | ✗ | ✗ | ✗ | ✓ |
The body weights and feed consumption in each group were recorded each day. On day 63, blood samples were obtained from the orbital sinus and then centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 min. The serum samples were collected and stored at −80 °C until further use. The animals were then killed by cervical dislocation, and the colons were collected. The collected colons were then cut open longitudinally and washed with PBS, and a gross examination was performed to measure the pattern of tumor development, including the tumor numbers and tumor sizes. The tumor sizes were determined by measuring the largest diameter of the identified lesion with an ocular micrometer by microscopy.
000 rpm for 30 min. The serum samples were collected and stored at −80 °C until further use. The animals were then killed by cervical dislocation, and the colons were collected. The collected colons were then cut open longitudinally and washed with PBS, and a gross examination was performed to measure the pattern of tumor development, including the tumor numbers and tumor sizes. The tumor sizes were determined by measuring the largest diameter of the identified lesion with an ocular micrometer by microscopy.
Fecal samples and colonic epithelial cells were also collected after dissection as described previously.17 The colon lengths were measured with a Vernier scale. The serous TNF-α concentration was evaluated with the ELISA detection kit following the manufacturer's protocol.
Colon sections and histological analysis
Parts of the colon were sectioned to an appropriate size and fixed with 4% paraformaldehyde followed by paraffin embedding and hematoxylin–eosin (HE) staining. The thickness of the slides with HE staining was 3 μm. The histological sections were analyzed by light microscopy (using 100× magnification or 200× magnification).18DNA extraction, purification and sequencing
Fecal bacterial DNA from five mice in each group was extracted using the TIANamp Stool DNA kit according to the manufacturer's instructions. Polymerase chain reaction (PCR) was conducted as described in our previous report.19 The PCR products were examined by 2% agarose electrophoresis and purified using the AxyPrep DNA gel extraction kit according to the instructions. Quantitation was performed with QuantiFluor™-ST, and the sequencing of the purified amplicons was performed with an Illumina MiSeq platform based on standard protocols. Approximately 300 samples were included in each sequencing run.Metagenomic analysis and read mapping
Metagenomic analysis with MiSeq techniques and 16S rDNA sequencing of the gut microbiota were performed as described in our previous reports using Quantitative Insights into Microbial Ecology (QIIME) software v1.8.0.20The raw paired-end reads were trimmed and quality-controlled using SeqPrep (https://github.com/jstjohn/SeqPrep) and Sickle (https://github.com/najoshi/sickle) software with the default parameters. Then, the clean reads were separately aligned to the reference genome in the orientation mode using bowtie2 software.21 The mapping criteria used in bowtie2 software were as follows: the sequencing reads should be uniquely matched to the genome by allowing up to 2 mismatches without insertions or deletions. Then, the region of each gene was expanded according to the depth at each site. In addition, the whole genome was split into multiple 15 kbp windows that shared 5 kbp. Newly transcribed regions were defined as those consisting of more than 2 consecutive windows without overlapping regions of genes, in which at least 2 reads were mapped per window in the same orientation.
RNA extraction, library preparation and sequencing
The total RNA of colonic epithelial cells from five rats in each group was extracted using TRIzol® Reagent according to the manufacturer's instructions. The RNA concentration and integrity and the RNA integrity number were determined or confirmed according to the description in our previous research report.10All high-quality RNA samples (concentration >50 ng μL−1, OD260/280 = 1.8–2.2, OD260/230 = 1.8–2.2, RIN > 6.5, 28S:18S ≥ 0.5) were used to construct the sequencing libraries, and the RNA sequencing transcriptome libraries were prepared with the Illumina TruSeq TM RNA sample prep kit using 5 μg of the total RNA. In brief, mRNA was purified according to base pairing rules (polyA-oligo dT) and then fragmented using fragmentation buffer, followed by cDNA synthesis with random hexamers.22 Then, the synthesized cDNA was subjected to end repair. The libraries were size-selected for cDNA target fragments of 200–300 bp with 2% low-range ultra agarose gels followed by PCR amplification using Phusion DNA polymerase for 15 PCR cycles. After quantification using a TBS 380 Fluorometer, the paired-end RNA-seq sequencing library was sequenced with the Illumina HiSeq 4000 (2 × 150 bp read length).
Metabolic function prediction and differential expression analysis
Metabolic function prediction was accomplished using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) software.23 Briefly, the Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used, and a closed reference operational taxonomic unit (OTU) was selected by comparing sampled reads against a Greengenes reference taxonomy (Greengenes 13.5). The 16S copy number was then normalized. After that, the molecular functions were predicted, and the final data were summarized to identify the KEGG pathways.To identify the differentially expressed genes (DEGs) between two different groups, the expression level of each transcript was calculated according to the fragments per kilobase of exon per million mapped reads (FRKM) method. RNA-seq by Expectation–Maximization (RSEM) was used to quantify the gene abundances.24 The R statistical package software Empirical Analysis of Digital Gene Expression in R (EdgeR) was utilized for the differential expression analysis.25
Statistical analysis
The results are expressed as the mean ± the standard deviation (SD). The statistical analysis of the bioinformatics data was performed using R software with the default setting. Other data were analyzed with Student's t-test or one-way analysis of variance (ANOVA) in SPSS (Version 18.0 for Windows, SPSS Inc., Chicago, IL, USA). P values less than 0.05 (p < 0.05) or 0.01 (p < 0.01) were considered statistically significant or highly statistically significant.Results
The effects of the guar gum diet on CRC
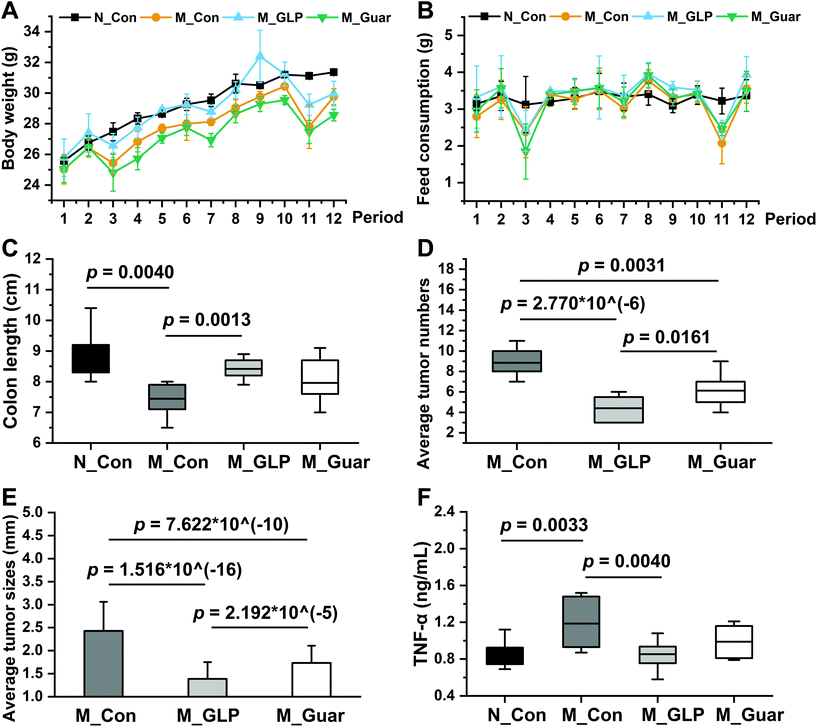
Body weight monitoring of CRC mice (M_Con, M_GLP and M_Guar groups) showed a total of 3 periods of severe weight loss (p < 0.0500) in periods 3 (days 12–17), 7 (days 33–38) and 11 (days 54–59) that were caused by DSS treatment (Fig. 2A). These 3 periods with severe weight loss corresponded to 3 drastic reductions in feed consumption (Fig. 2B). No significant difference was found in the BW of mice between the M_Guar group and the M_GLP group in period 3 (p = 0.0994), but mice in the M_Guar group presented significantly lower BW than those in the M_GLP group in periods 7 and 11 (p = 0.0001 and 0.0134, respectively). As a result, only a 10% reduction in the mortality rate was observed in the M_Guar group, while a 30% mortality rate reduction was observed in the M_GLP group (it was noted that the mortality rate of the M_Con group was 30%). More investigations were performed after the dissection. We found that the colon length of mice in the M_Con group was significantly shortened compared with that in the N_Con group. The guar gum diet (M_Guar group) did not efficiently restrict colon shortening (Fig. 2C). However, it could significantly reduce the tumor numbers and sizes in the colon, although the effect was still significantly weaker than that of the GLP diet (Fig. 2D and E). The guar gum diet also reduced the TNF-α concentration, although this effect did not show statistical significance (Fig. 2F).The HE staining of the histological sections showed that the colon in the M_Con group had a disorganized structure with an unorganized cell arrangement and a looser intercellular space, which was completely different from that in both the N_Con and M_GLP groups. The colonic sections of the M_Guar group showed an improved structure and cell arrangement. However, the colon state was still worse than that in the M_GLP group (Fig. 3).
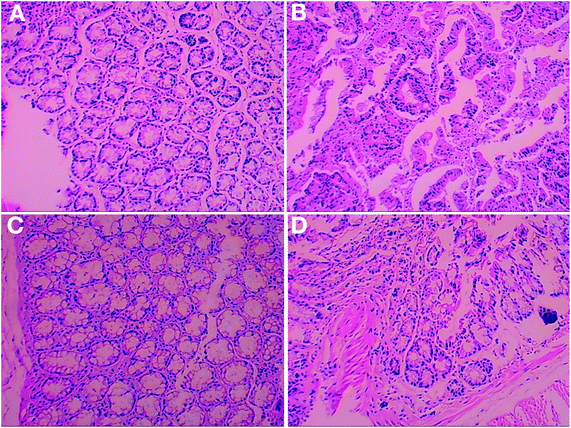 | ||
| Fig. 3 Histological sections of the colon with hematoxylin–eosin staining. (A) N_Con; (B) M_Con; (C) M_GLP; (D) M_Guar. Magnification: 100× for (A) and (C), and 200× for (B) and (D). | ||
Guar gum increased the relative abundance (RA) of Akkermansia
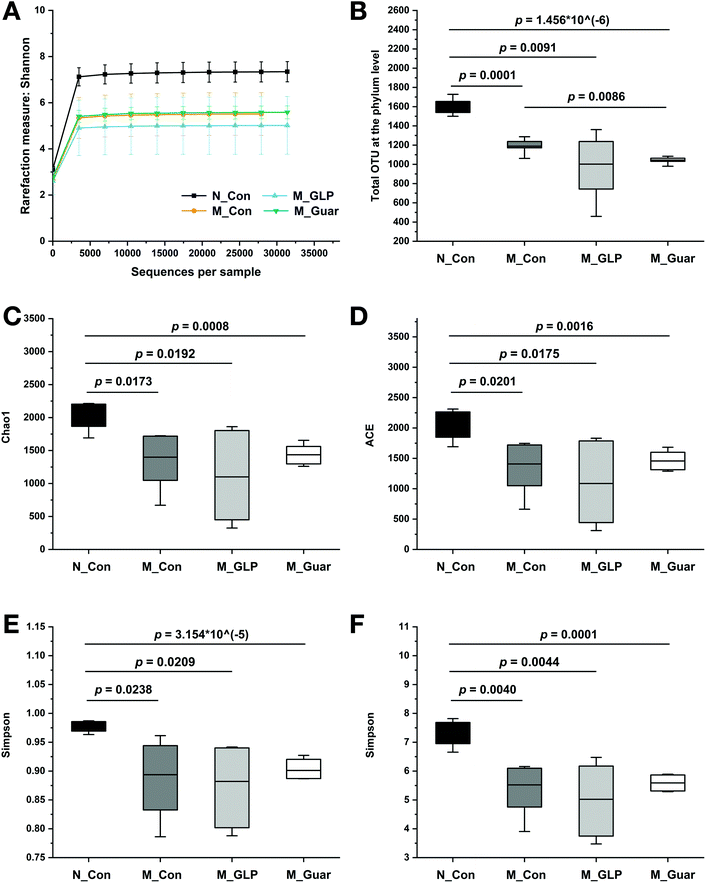
Subsequently, we analyzed the profile of gut microbiota in mice among the four groups. The sequencing depth was sufficient, as indicated by the rarefaction curve in Fig. 4A. Total OTU analysis revealed a highly significant reduction between the N_Con group and the three other groups, but no significant difference was found between mice fed guar gum and GLP (Fig. 4B). The descriptive indices, including Chao1, ACE, Simpson and Shannon, in the M_Guar and M_GLP groups indicated that these two groups had similar α-diversity (Fig. 4C–F).To obtain better insight into the microbial profile, the composition at the phylum and genus level of each group was analyzed. Among the 10 taxa of gut bacteria found at the phylum level, the M_Guar group, together with the M_Con and M_GLP groups, was found to have 9 taxa of bacteria with similar RAs. However, the M_Guar group had a significantly higher RA of Verrucomicrobia than the N_Con and M_GLP groups (p = 4.072 × 10−6 and 0.0012) (Fig. 5A). Therefore, the M_Guar group showed a higher RA of Akkermansia with higher significance than the N_Con and M_GLP groups at the genus level (p = 4.072 × 10−6 and 0.0012) (Fig. 5B). Similar to the other taxa of gut bacteria found at the genus level, such as Oscillospira and an unknown genus of Desulfovibrionaceae, similar RAs were found among the M_Guar, M_Con and M_GLP groups (Fig. 5B).
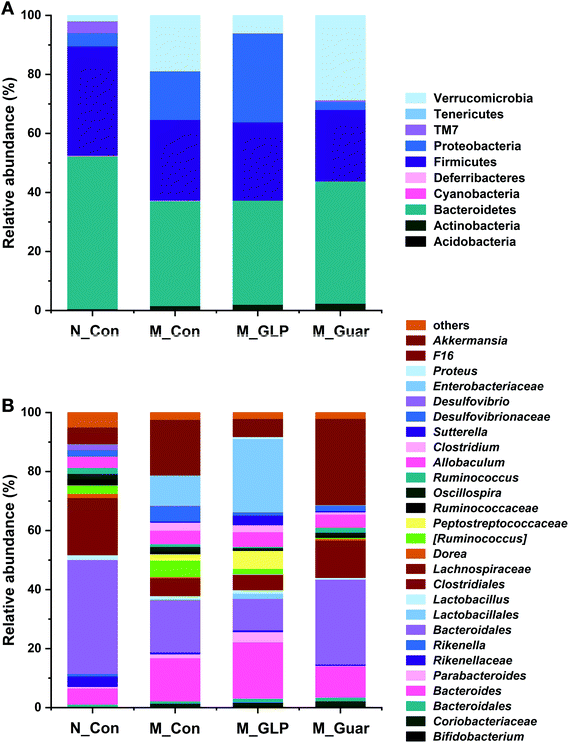 | ||
| Fig. 5 Relative abundances at the phylum and genus level. (A) The phylum level. (B) The genus level. Bacteria with a relative abundance less than 0.1% were classified as “others”. | ||
Guar gum altered the microbial metabolic pathways and colonic epithelial DEGs
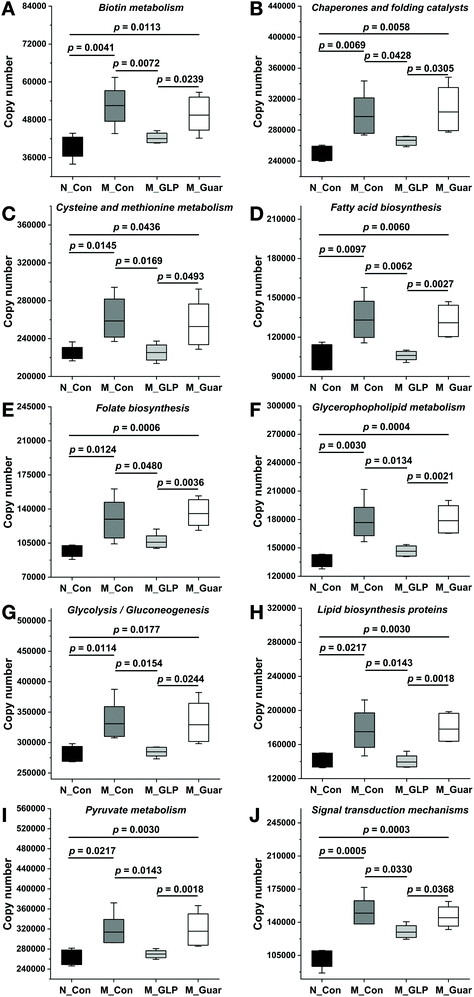
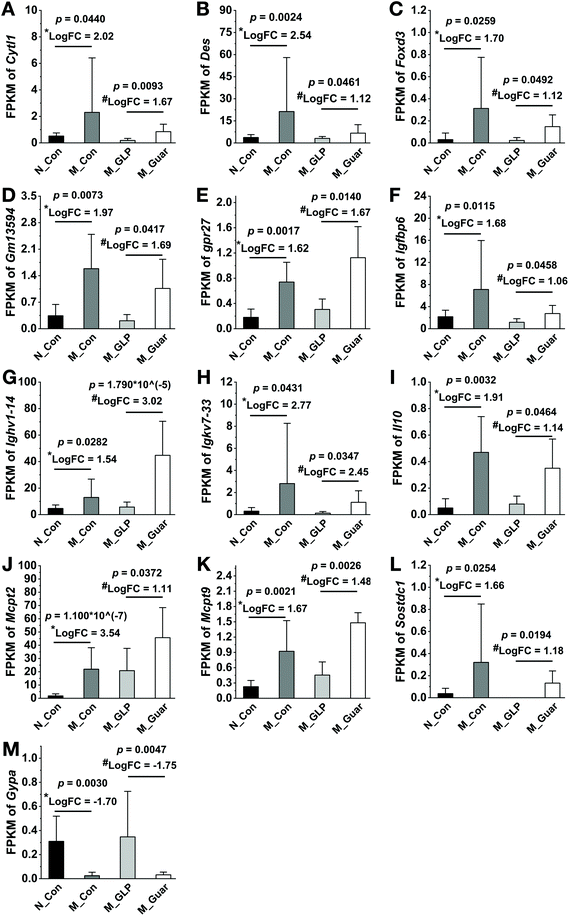
To explain the differences in the CRC alleviating effect among the groups, the metabolic pathways of the gut microbiota were evaluated. Upon the comparison of 328 KEGG pathways at level 3 in the database, we found genes from the colonic microbiota among the groups that were distributed in 273 metabolic pathways. In total, there were 102 metabolic pathways (A) showing significantly higher and 7 showing significantly lower gene copy numbers in the M_Con group compared with those in the N_Con group. Additionally, 122 (130) metabolic pathways (B and C) showed significantly higher copy numbers, while 21 (17) showed significantly lower gene copy numbers in the M_Guar group when compared with those in the M_GLP group (N_Con). Moreover, there were 3 metabolic pathways showing significantly higher and 68 (D) showing significantly lower gene copy numbers in the M_GLP group when compared with those in the M_Con group. We then selected those pathways according to the overlap of (A), (B), (C) and (D) as described above. After that, we excluded those metabolic pathways that were less associated with bacterial metabolism. Finally, guar gum was found to enrich the colonic bacterial genes in 10 microbial metabolic pathways that were involved in CRC (Fig. 6).Since the metabolic pathways of the colonic microbiota were altered, further analysis was applied to evaluate the DEGs in colonic epithelial cells in the host. We found that the M_Con group had 1642 upregulated and 538 downregulated genes (DEGs) compared with N_Con. In addition, the M_Guar group had 389 upregulated and 63 downregulated DEGs compared with N_GLP. Thirteen candidate DEGs (12 upregulated and 1 downregulated) that explained the inferior CRC alleviating effects of the guar gum diet were selected from among the overlapping upregulated and downregulated genes, respectively (Fig. 7).
Discussion
Guar gum has been studied as a functional natural fiber since the 1970s.26 Nevertheless, its CRC alleviating effect is still unclear.13,14 In this report, we offered a guar gum diet to mice during CRC development and studied its CRC alleviating effect by using a GLP diet for comparison. Since the effective alleviation of CRC by the GLP diet has been proven,10 we were able to show that guar gum alleviates CRC, especially in terms of mortality and tumor number and size reduction. However, this effect was inferior to that of GLP, as indicated by the higher mortality rate, shorter colon length, larger number and size of tumors, higher concentration of serous TNF-α, which is a cytokine related to CRC,27 and the worse colon status in histological sections in mice fed guar gum compared to those in mice fed GLP. Another insight obtained from the omics analyses revealed that guar gum, compared with GLP, increased the RA of Akkermansia, but no variation in Oscillospira and an unknown genus of Desulfovibrionaceae was observed; in addition, guar gum altered 10 metabolic pathways and changed the expression of 13 genes in colonic epithelial cells in the host.One of the symptoms of CRC patients is unintentional weight loss, which occurs for multiple reasons. These reasons include progressive tissue depletion caused by altered metabolism, increased resting energy expenditure resulting from tumor-induced changes and decreased nutrient intake.28 It has been reported that long-term weight loss after CRC diagnosis is associated with decreased survival.29 For this reason, the higher mortality rate associated with guar gum intervention should be attributed to the more severe weight loss associated with guar gum than with GLP. This weight loss was the consequence of the altered metabolism or increased resting energy expenditure caused by impaired health due to insufficient protection by guar gum against CRC-induced damage. Since dietary interventions affect the gut microbiota, the colonic microbial profile was investigated here. Previously, we reported that the GLP intervention did not enhance the overall microbial diversity to alleviate CRC.10 In the current study, we found a similar result for the guar gum intervention. Moreover, both guar gum and GLP reduced the abundance of specific microbiota (Oscillospira and an unknown genus of Desulfovibrionaceae) to a similar extent, which was believed to be CRC-associated.10 However, guar gum also increased the abundance of Akkermansia, a mucin-degrading gut bacterium. Akkermansia was found in a high abundance in CRC individuals.30,31 In addition, it was most strongly positively correlated with increased colonic tumor burden.32 We hypothesized that the damaged barrier function caused by CRC would be exacerbated by the mucin degradation caused by Akkermansia. Hence, the increased abundance of Akkermansia probably led to insufficient colonic protection by guar gum against CRC damage.
As we all know, the health status can affect the metabolic pathways of the gut microbiota in hosts. In addition, diet shifts could also change the metabolic networks of the gut microbiota.33 In this study, we found KEGG pathways associated with biotin metabolism, chaperones and folding catalysts, cysteine and methionine metabolism, fatty acid biosynthesis, folate biosynthesis, glycerophospholipid metabolism, glycolysis/gluconeogenesis, lipid biosynthesis proteins, pyruvate metabolism and signal transduction mechanisms that may be linked to the inferior effects of guar gum. The incidence of lipid metabolism abnormalities in patients with CRC was higher than that in healthy individuals.34 Meanwhile, intestinal microorganisms involved in CRC are associated with dyslipidosis.35 The microbial metabolic pathways involved in lipid metabolism (fatty acid biosynthesis, glycerophospholipid metabolism and lipid biosynthesis proteins) likely remain significantly activated, as shown in this study, which might contribute to insufficient protection by guar gum against CRC damage. Moreover, 3 pathways (the glycerophospholipid metabolism, pyruvate metabolism and glycolysis/gluconeogenesis pathways) generate a common metabolite, acetylaldehyde. This played a negative role in CRC alleviation, as it was an agent capable of initiating tumors by forming adducts with proteins and DNA.36 Furthermore, the pathway associated with chaperones and folding catalysts was shown to be activated in CRC patients.35 Therefore, the results related to bacterial metabolic pathways, including those associated with chaperones and folding catalysts, fatty acid biosynthesis, glycerophospholipid metabolism, glycolysis/gluconeogenesis, lipid biosynthesis proteins and pyruvate metabolism, tended to explain why guar gum was inferior to GLP in terms of CRC alleviation.
The alteration of microbial metabolic pathways would ultimately influence the gene expression of host epithelial cells.37 After applying the selection criteria based on the gene regulation patterns among the groups, 13 DEGs were identified. We further divided them into 3 categories according to their annotations and functional descriptions as per previous studies.
Category A: G protein-coupled receptor 27 (Gpr27), Gm13594 and glycophorin A (Gypa). We could not determine the expression of these 3 genes in CRC or other cancers. Further research is needed to verify the actual function of these genes in CRC.
Category B: interleukin 10 (Il10), cytokine-like 1 (Cytl1), immunoglobulin kappa chain variable 7-33 (Igkv7-33), immunoglobulin heavy variable 1-14 (Ighv1-14), insulin-like growth factor binding protein 6 (Igfbp6) and forkhead box D3 (Foxd3). Among these genes, IL10 is a potent anti-inflammatory cytokine.38 Thus, Il10-deficient mice were susceptible to colitis, which could promote tumorigenesis by altering the microbial composition.39 Reports on the direct anti-CRC function of CYTL1 are rare. However, in breast cancer, it was recognized to be a cytokine with tumor-suppressing characteristics that inhibits tumor metastasis.40 FOXD3 also had a positive effect on CRC since the loss of FOXD3 in human colon cancer cells allowed upregulation of the short isoform of doublecortin-like kinase 1 (DCLK1-S), imparting a potent invasive ability to cells.41 In addition, IGFBP-6 could possibly inhibit the invasion and migration of colorectal carcinoma cells via inhibiting the proliferation activity and arresting the cell cycle in vitro.42 Therefore, genes in this category were related to cancer suppression via the rescue of the function of impaired epithelial cells damaged by CRC. It could be inferred that hosts upregulated these genes to combat cancer due to the inferior effect of guar gum on CRC.
Category C: mast cell protease 2 (Mcpt2), mast cell protease 9 (Mcpt9), desmin (Des) and sclerostin domain containing 1 (Sostdc1). Although we could not determine if MCPT2 and MCPT 9 were directly correlated with CRC, another mast cell protease was shown to accumulate in inflammatory cells in colon tissues and support tumor cell growth.43 Serous desmin was significantly elevated in CRC patients compared with healthy volunteers,44 and Sostdc1 was also considered a cancer-related gene.45 In light of these previous reports, we believe that the upregulation of these genes might have caused guar gum to have an inferior effect on CRC alleviation compared to GLP.
This study did not perform an analysis of the metabolites generated by the colonic microbiota or confirm gene expression with quantitative reverse PCR or western blotting. Nevertheless, our study showed the CRC-alleviating effect of guar gum by using multiple methods and interpreted this effect using omics analyses. This study elucidated the CRC-alleviating effect of a polysaccharide that is highly accessible and explored the potential reasons for the differences in its effects compared with those of other polysaccharides. This should be helpful in determining the CRC-alleviating effects of guar gum.
Conclusion
Dietary intake of a guar gum-containing diet had an effect on CRC alleviation. However, this effect was different from that observed in GLP intervention, as indicated by the 20% higher mortality rate, shorter colon length, increased tumor numbers and sizes, higher concentration of serous TNF-α, and worse colon status. In addition, the upregulation of 6 genes in host epithelial cells, including Il10, Cytl1, Igkv7-33, Ighv1-14, Igfbp6 and Foxd3, also reflected the different CRC alleviating effect from that of GLP. Through omics analysis, we also found that guar gum resulted in the increased abundance of the mucin-degrading bacteria Akkermansia, the alteration of 10 microbial metabolic pathways, especially those affecting chaperones and folding catalysts, fatty acid biosynthesis, glycerophospholipid metabolism, glycolysis/gluconeogenesis, lipid biosynthesis proteins and pyruvate metabolism, and the upregulation of 4 epithelial cell genes, including Mcpt2, Mcpt9, Des and Sostdc1. The result suggested that the CRC-alleviating effect of guar gum (an inexpensive polysaccharide) was inferior to that of GLP (a more expensive polysaccharide). This could potentially be attributed to the increased presence of Akkermansia, the alteration of 10 microbial metabolic pathways and the upregulation of 4 epithelial cell genes.Ethics approval and consent to participate
All animal procedures and protocols were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the Committee of Jinan University.Conflicts of interest
The authors declare that they have no conflicts of interest.Acknowledgements
The program was supported by the National Natural Science Funds (No. 31801543) and the funds of the China Postdoctoral Science Foundation (No. 2018M643368). We thank Ruixia Qiu from the Department of Food Science and Engineering, Jinan University, for her valuable contribution to the experiments in this study.References
- SEER Cancer Statistics Review, 1975–2016, https://seer.cancer.gov/csr/1975_2016/results_merged/sect_01_overview.pdf, (accessed September 2019).
- S. J. Bultman, Interplay between diet, gut microbiota, epigenetic events, and colorectal cancer, Mol. Nutr. Food Res., 2017, 61, 1500902 CrossRef PubMed.
- M. Arnold, M. S. Sierra, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, Global patterns and trends in colorectal cancer incidence and mortality, Gut, 2017, 66, 683–691 CrossRef PubMed.
- P. Louis, G. L. Hold and H. J. Flint, The gut microbiota, bacterial metabolites and colorectal cancer, Nat. Rev. Microbiol., 2014, 12, 661–672 CrossRef CAS PubMed.
- S. J. O'keefe, Diet, microorganisms and their metabolites, and colon cancer, Nat. Rev. Gastroenterol. Hepatol., 2016, 13, 691–706 CrossRef PubMed.
- S. Yachida, S. Mizutani, H. Shiroma, S. Shiba, T. Nakajima, T. Sakamoto, H. Watanable, K. Masuda, Y. Nishimoto, M. Kubo, F. Hosoda, H. Rokutan, M. Matsuomto, H. Takamaru, M. Tamada, T. Matsuda, M. Iwasaki, T. Yamaji, T. Yachidia, T. Soga, K. Kurokawa, A. Yoyoda, Y. Ogura, T. Hayashi, M. Hatakeyama, H. Nakagama, Y. Saito, S. Fukuda, T. Shibata and T. Yamada, Metagenomic and metabolomics analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer, Nat. Med., 2019, 25, 968–976 CrossRef CAS PubMed.
- S. J. Bultman, The microbiome and its potential as a cancer preventive intervention, Semin. Oncol., 2016, 43, 97–106 CrossRef CAS PubMed.
- A. T. Kunzmann, H. G. Coleman, W. Y. Huang, C. M. Kitahara, M. M. Cantwell and S. I. Berndt, Dietary fiber intake and risk of colorectal cancer and incident and recurrent adenoma in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial, Am. J. Clin. Nutr., 2015, 102, 881–890 CrossRef CAS PubMed.
- C. A. M. Wegh, M. H. C. Schoterman, E. E. Vaughan, C. Belzer and M. A. Benninga, The effect of fiber and prebiotics on children's gastrointestinal disorders and microbiome, Expert Rev. Gastroenterol. Hepatol., 2017, 11, 1031–1045 CrossRef CAS PubMed.
- J. Luo, C. Zhang, R. Liu, L. Gao, S. Ou, L. Liu and X. Peng, Ganoderma lucidum polysaccharide alleviating colorectal cancer by alteration of special gut bacteria and regulation of gene expression of colonic epithelial cells, J. Funct. Foods, 2018, 47, 127–135 CrossRef CAS.
- M. Guo, Z. Li, Y. Huang and M. Shi, Polysaccharides from Nostoc commune Vaucher activate macrophages via NF-κB and AKT/JNK1/2 pathways to suppress colorectal cancer growth in vivo, Food Funct., 2019, 10, 4269–4279 RSC.
- N. Thombare, U. Jha, S. Mishra and M. Z. Siddiqui, Guar gum as a promising starting material for diverse applications: A review, Int. J. Biol. Macromol., 2016, 88, 361–372 CrossRef CAS PubMed.
- D. W. Heitman, W. E. Hardman and I. L. Cameron, Dietary supplementation with pectin and guar gum on 1,2-dimethylhydrazine-induced colon carcinogenesis in rats, Carcinogenesis, 1992, 13, 815–818 CrossRef CAS PubMed.
- A. McIntyre, P. R. Gibson and G. P. Young, Butyrate production from dietary fibre and protection against large bowel cancer in a rat model, Gut, 1993, 34, 386–391 CrossRef CAS PubMed.
- C. D. Tran, J. M. Ball, S. Sundar, P. Coyle and G. S. Howarth, The role of zinc and metallothionein in the dextran sulfate sodium-induced colitis mouse model, Dig. Dis. Sci., 2007, 52, 2113–2121 CrossRef CAS PubMed.
- J. K. Woo, S. Choi, J. H. Kang, D. E. Kim, B. S. Hurh, J. E. Jeon, S. Y. Kim and S. H. Oh, Fermented barley and soybean (BS) mixture enhances intestinal barrier function in dextran sulfate sodium (DSS)-induced colitis mouse model, BMC Complementary Altern. Med., 2016, 16, 498 CrossRef PubMed.
- J. Luo, L. Han, L. Liu, L. Gao, B. Xue, Y. Wang, S. Ou, M. Miller and X. Peng, Catechin supplemented in a FOS diet induces weight loss by altering cecal microbiota and gene expression of colonic epithelial cells, Food Funct., 2018, 9, 2962–2969 RSC.
- J. Luo, J. Qi, W. Wang, Z. Luo, L. Liu, G. Zhang, Q. Zhou, J. Liu and X. Peng, Antiobesity effect of flaxseed polysaccharide via inducing satiety due to leptin resistance removal and promoting lipid metabolism through the AMP-activated protein kinase (AMPK) signaling pathway, J. Agric. Food Chem., 2019, 87, 7040–7049 CrossRef PubMed.
- J. Luo, Y. Li, Y. Mai, L. Gao, S. Ou, Y. Wang, L. Liu and X. Peng, Flaxseed gum reduces body weight by regulating gut microbiota, J. Funct. Foods, 2018, 47, 136–142 CrossRef CAS.
- C. Zheng, R. Liu, B. Xue, J. Luo, L. Gao, Y. Wang, S. Ou, S. Li and X. Peng, Impact and consequences of polyphenols and fructooligosaccharide interplay on gut microbiota in rats, Food Funct., 2017, 8, 1925–1932 RSC.
- B. Langmead and S. L. Salzberg, Fast gapped-read alignment with Bowtie 2, Nat. Methods, 2012, 9, 357–359 CrossRef CAS PubMed.
- X. Xu, Y. Zhang, J. Williams, E. Antoniou, W. R. McCombie, S. Wu, W. Zhu, N. O. Davidson, P. Denoya and E. Li, Parallel comparison of Illumina RNA-Seq and Affymetrix microarray platforms on transcriptomic profiles generated from 5-aza-deoxy-cytidine treated HT-29 colon cancer cells and simulated datasets, BMC Bioinf., 2013, 14(Suppl 9), S1 CrossRef PubMed.
- M. G. Langille, J. Zaneveld, J. G. Caporaso, D. McDonald, D. Knights, J. A. Reyes, J. C. Clemente, D. E. Burkepile, R. L. Vega Thurber, R. Knight, R. G. Beiko and C. Huttenhower, Predivtive functional profiling of microbial communities using 16S rRNA marker gene sequences, Nat. Biotechnol., 2013, 31, 814–821 CrossRef CAS PubMed.
- B. Li and C. N. Dewey, RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome, BMC Bioinf., 2011, 12, 323 CrossRef CAS PubMed.
- M. D. Robinson, D. J. McCarthy and G. K. Smyth, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data, Bioinformatics, 2010, 26, 139–140 CrossRef CAS PubMed.
- D. J. Jenkins, C. Newton, A. R. Leeds and J. H. Cumming, Effect of pectin, guar gum, and wheat fibre on serum-cholesterol, Lancet, 1975, 1, 1116–1117 CrossRef CAS.
- B. K. Popivanova, K. Kitamura, Y. Wu, T. Kondo, T. Kagaya, S. Kaneko, M. Shima, C. Fujii and N. Mukaida, Blocking TNF-alpha in mice reduces colorectal carcinogenesis associated with chronic colitis, J. Clin. Invest., 2008, 118, 560–570 CAS.
- S. B. Bapuji and J. A. Sawatzky, Understanding weight loss in patients with colorectal cancer: a human response to illness, Oncol. Nurs. Forum, 2010, 37, 303–310 CrossRef PubMed.
- J. M. Kocarnik, X. Hua, S. Hardikar, J. Robinson, N. M. Lindor, A. K. Win, J. L. Hopper, J. C. Figueiredo, J. D. Potter, P. T. Campbess, S. Gallinger, M. Cotterchio, S. V. Adams, S. A. Cohen, A. I. Phipps and P. A. Newcomb, Long-term weight loss after colorectal cancer diagnosis is associated with lower survival: the colon cancer family registry, Cancer, 2017, 123, 4701–4708 CrossRef PubMed.
- J. P. Zackular, N. T. Baxter, K. D. Iverson, W. Sadler, J. F. Perosino, G. Y. Chen and P. D. Schloss, The gut microbiome modultates colon tumorigenesis, mBio, 2013, 4, e00692–e00613 CrossRef PubMed.
- T. L. Weir, D. K. Manter, A. M. Sheflin, B. A. Barnett, A. L. Heuberger and E. P. Ryan, Stoll microbiome and metabolome differences between colorectal cancer patients and healthy adylts, PLoS One, 2013, 8, e70803 CrossRef CAS PubMed.
- N. T. Baxter, J. P. Zackular, G. Y. Chen and P. D. Schloss, Structure of the gut microbiome following colonization with human feces determines colonic tumor burden, Microbiome, 2014, 2, 20 CrossRef PubMed.
- S. M. Wolff, M. J. Ellison, Y. Hao, R. R. Cockrum, K. J. Austin, M. Baraboo, K. Burch, H. J. Lee, T. Maurer, R. Patil, A. Ravelo, T. M. Taxis, H. Truong, W. R. Lamberson, K. M. Cammack and G. C. Conant, Diet shifts provoke complex and variables changes in the metabolic networks of the ruminal microbiome, Microbiome, 2017, 5, 60 CrossRef PubMed.
- Y. S. Jung, S. Ryu, Y. Chang, K. E. Yun, J. H. Park, H. J. Kim, Y. K. Cho, C. I. Sohn, W. K. Jeon, B. I. Kim, K. Choi and D. I. Park, Association between parameters of glucose and lipid metabolism and risk of colorectal neoplasm, Dig. Dis. Sci., 2015, 60, 2996–3004 CrossRef CAS PubMed.
- S. Han, Y. Pan, X. Yang, M. Da, Q. Wei, Y. Gao, Q. Qi and L. Ru, Intestinal microorganisms involved in colorectal cancer complicated with dyslipidosis, Cancer Biol. Ther., 2019, 20, 81–89 CrossRef PubMed.
- S. Singh, J. Arcaroli, D. C. Thompson, W. Messersmith and V. Basiliou, Acetaldehyde and retinaldehyde-metabolizing enzemes in colon and pancreatic cancers, Adv. Exp. Med. Biol., 2015, 815, 281–294 CrossRef CAS PubMed.
- N. D. Mathewson, R. Jeng, A. V. Mathew, M. Koenigsknecht, S. Hnash, T. Toubai, K. Oravecz-Wilson, S. R. Wu, Y. Sun, C. Rossi, H. Fujuwara, J. Byun, Y. Shono, C. Lindemans, M. Calafiore, T. M. Schimidt, K. Honda, V. B. Young, S. Pennathur, M. van den Brink and P. Reddy, Gut microbiome0derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease, Nat. Immunol., 2016, 17, 505–513 CrossRef CAS PubMed.
- H. Mollazadeh, A. F. G. Cicero, C. N. Blesso, M. Pirro, M. Majeed and A. Sahebkar, Immune modulation by curcumin: the role of interleukin-10, Crit. Rev. Food Sci. Nutr., 2019, 59, 89–101 CrossRef CAS PubMed.
- J. C. Arthur, E. Perez-Chanona, M. Mühlbauer, S. Tomkovich, J. M. Uronis, T. J. Fan, B. J. Campbell, T. Abujamel, B. Dogan, A. B. Rogers, J. M. Rhodes, A. Stintzi, K. W. Simpson, J. J. Hansen, T. O. Keku, A. A. Fodor and C. Jobin, Intestinal inflammation targets cancer-inducing activity of the microbiota, Science, 2012, 338, 120–123 CrossRef CAS PubMed.
- X. Wang, T. Li, T. Cheng, P. Wang, W. Yuan, Q. Liu, D. Ma, S. Ding, J. Wang and W. Han, CYTL1 inhibits tumor metastasis with decreasing STAT3 phosphorylation, OncoImmunology, 2019, 8, e1577126 CrossRef PubMed.
- S. Sarkar, M. R. O'Connell, Y. Okugawa, B. S. Lee, Y. Toiyama, M. Kusunoki, R. D. Daboval, A. Goel and P. Singh, FOXD3 regulates CSC marker, DCLK1-S, and invasive potential: prognostic implications in colon cancer, Mol. Cancer Res., 2017, 15, 1678–1691 CrossRef CAS PubMed.
- F. Qiu, W. Gao and B. Wang, Correlation of IGFBP-6 expression with apoptosis and migration of colorectal carcinoma cells, Cancer Biomarkers, 2018, 21, 893–898 CAS.
- L. Xu, H. G. Yi, Z. Wu, W. Han, K. Chen, M. Zang, D. Wang, X. Zhao, H. Wang and C. Qu, Activation of mucosal mast cells promotes inflammation-related colon cancer development through recruiting and modulating inflammatory CD11b(+)Gr1(+) cells, Cancer Lett., 2015, 364, 173–180 CrossRef CAS PubMed.
- Y. Ma, J. Peng, W. Liu, P. Zhang, L. Huang, B. Gao, T. Shen, Y. Zhou, H. Chen, Z. Chu, M. Zhang and H. Qin, Proteomics identification of desmin as potential oncofetal diagnostic and prognostic biomarker in colorectal cancer, Mol. Cell. Proteomics, 2009, 8, 1878–1890 CrossRef CAS PubMed.
- J. Yang, Y. Chen, Z. Yu, H. Ding and Z. Ma, Changes in gene expression in lungs of mice exposed to traffic0related air pollution, Mol. Cell. Probes, 2018, 39, 33–40 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |