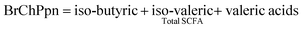Purified plant cell walls with adsorbed polyphenols alter porcine faecal bacterial communities during in vitro fermentation†
Lucas J.
Grant
a,
Deirdre
Mikkelsen
 *a,
Anh Dao T.
Phan
a,
Seungha
Kang
b,
Diane
Ouwerkerk
c,
Athol V.
Klieve
cd,
Michael J.
Gidley
a and
Barbara A.
Williams
*a,
Anh Dao T.
Phan
a,
Seungha
Kang
b,
Diane
Ouwerkerk
c,
Athol V.
Klieve
cd,
Michael J.
Gidley
a and
Barbara A.
Williams
 a
a
aCentre of Nutrition and Food Sciences, Queensland Alliance for Agriculture and Food Innovation, The University of Queensland, St. Lucia, QLD 4072, Australia. E-mail: d.mikkelsen@uq.edu.au
bThe University of Queensland Diamantina Institute, Translational Research Institute, Woolloongabba, QLD 4102, Australia
cRumen Ecology Unit, Agri-Science Queensland, Department of Agriculture and Fisheries, Dutton Park, QLD 4102, Australia
dSchool of Agriculture and Food Sciences, The University of Queensland, Gatton, Queensland, Gatton, QLD 4343, Australia
First published on 19th December 2019
Abstract
A substantial fraction of ingested polyphenols accumulate in the large intestine (LI), attached to undigested plant cell walls (PCW) (dietary fibre). Yet, whether these PCW-bound polyphenols alter the structure and function of the resident microbiota remains unclear. This study characterised bacterial populations during the in vitro fermentation of three standard polyphenols: ferulic acid (FER), (±)-catechin (CAT), and cyanidin-3-glucoside (CYAN), adsorbed individually or in combination to apple cell walls (ACW). During fermentation with porcine faeces, samples were collected at regular time-points (up to 72 hours) for bacterial 16S rRNA gene amplicon sequencing and fermentation end-product analyses (short-chain fatty acids and ammonium). The metabolic end-products differed to only a small extent between substrates, though significantly for propionate (P < 0.0001). Significant differences in microbial populations were noted between substrates tested (P < 0.0001). The presence of cyanidin-3-glucoside resulted in the most significant differences between bacterial communities during fermentation of the ACW substrate. Key microbes identified to be associated with the ACW with adsorbed polyphenols as well as individual polyphenols were: Phascolarctobacterium with ACW + FER and FER, the Lachnospiraceae family with ACW + CYAN, Parabacteroides with ACW + CYAN and CYAN, Collinsella and Coprococcus with ACW + CAT, and the Clostridiales order with ACW + CAT and CAT. This study has demonstrated the use of a simplified model to indicate any microbial effects of polyphenols associated with dietary fibre in whole fruits. This work has shown that individual polyphenols, or those adsorbed to PCW, have potentially very different effects on the gut bacteria. Future work could examine further polyphenols associated with a range of fresh fruits.
Introduction
Polyphenols are an important class of phytonutrients, present in their soluble form primarily within vacuoles of plant cells. They vary in their chemical structure and abundance, and are considered to confer health benefits.1 However, upon consumption of plant-based foods such as fruits and vegetables, immediate bioaccessibility by the host, of those polyphenols not present within the vacuoles, is limited. These polyphenols can either be encapsulated, or continue to be bound to the food matrix,2 therefore requiring degradation of their surrounding structure to enable absorption by the host.3,4 Current estimations are that only 5–10% of polyphenols are absorbed across the small intestinal epithelium.3,5 The remaining 90–95% of polyphenols pass into the large intestine (LI), where they may be subjected to hydrolysis and metabolism by the resident microbiota.3,5 Thereafter, any resulting metabolites can be absorbed across the gut wall, potentially benefiting the host.6 Recently, there has been growing interest in the LI fermentation of polyphenols.4,7–9 It is being realised that it is more likely to be the metabolites resulting from microbial fermentation within the LI, which actually have the beneficial effects ascribed to the consumption of polyphenols.10,11There have been no studies which have investigated the effects of plant cell wall (PCW)-associated polyphenols on the LI bacterial community, particularly in terms of how this community may be altered by PCW-bound polyphenols, both in terms of composition (abundance/diversity) and activity. There are potentially thousands of polyphenols within plants,3 which makes their study, particularly in combination with the LI microbial population, extremely complicated. Therefore, for simplicity, this study investigated three representative polyphenols for fruits/vegetables/cereal products.
Ferulic acid is often used to exemplify the phenolic acid class, found throughout the plant kingdom.12 It is known to be present as a substituent within some PCW, as well as within the cell vacuoles. Catechin is the archetypal member of the catechins, one of the largest groups of polyphenols present in plant-based foods.5 Anthocyanins are abundant in many fruits.13 Cyanidin-3-glucoside is part of the anthocyanin group, (glycosylated anthocyanidins), and is one of six common types of anthocyanidins. Anthocyanins exert antioxidant and anti-inflammatory properties in the host,13 and have been speculated to modify LI bacterial communities.14
This study examined shifts in bacterial populations during in vitro fermentation of apple cell walls (ACW) to which one polyphenol (ferulic acid; (±)-catechin; or cyanidin-3-glucoside) or a mixture of all three, had been adsorbed, compared with the triple polyphenol mixture without ACW, and the ACW alone as controls. A faecal inoculum from pigs fed a controlled low polyphenol and low fibre diet was used, as pigs are recognised as a useful human model for the digestive tract, in terms of anatomy, physiology, and their omnivorous diet.15 In addition, they have recently been shown to share a comparable LI microbial community.16,17
It was hypothesised that the bacterial community structure grown on ACW as the main carbon source, would respond differently to the presence of three diverse polyphenols that had been adsorbed to the ACW. Furthermore, it was anticipated that the use of polyphenols adsorbed to ACW would provide insights into whether polyphenol adsorption to PCW, might have a different outcome, in terms of changes in the microbial community compared with the use of polyphenols alone. This would have potential implications for the consumption of polyphenols in whole foods (particularly fruits, vegetables, and whole grains), versus the addition of polyphenolic extracts to processed food products, in terms of the LI microbial community.
Materials and methods
Substrates for bacterial fermentation
Ferulic acid (FER), (±) catechin (CAT) (HPLC grade, Sigma Aldrich, AUS), and cyanidin-3-glucoside (CYAN; HPLC grade, Biopurify Phytochemicals Ltd, China), were dissolved in citrate/phosphate buffer (0.1 M; pH 4.2). Individual polyphenolic solutions (10 mM) and an equimolar polyphenol mixture (10 mM of FER, CAT and CYAN), were prepared freshly prior to adsorption experiments. “Ready-to-eat” Pink Lady® apples, purchased from a local supermarket, were used to produce apple cell walls (ACW), following a previously published method with modifications.18,19Briefly, 5 g of ACW was soaked in 250 mL of citrate/phosphate buffer (0.1 M; pH 4.2), containing either 10 mM of FER, CAT, CYAN, or the equimolar polyphenol mixture, for 2 h with constant mixing (100 rpm). Importantly, not all polyphenols adsorbed to the same extent, so the final concentrations are shown in Table 1. Post-adsorption, samples were centrifuged (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 10 min at 25 °C; Beckman Coulter – Avanti J-E, USA) to obtain the pelleted fraction, lyophilized and cryoground as previously described.20 Briefly, the 6850 SPEX freezer/mill (SPEX, Metuchen, NJ, USA) cryo-grinding conditions involved precooling for 5 min, followed by two grinding cycles per run at an impactor speed level of 10 s−1 for 5 min each, with an intermediate cooling period of 2 min between grinding cycles. A sample of only ACW, with no adsorbed polyphenols, but which otherwise had been treated in the same way, was used as a positive control, to determine the fermentability of the ACW alone, as well as any shifts in the microbial community. Post-cryogrinding, all substrates were stored in a desiccator until fermentation analysis.
000g, 10 min at 25 °C; Beckman Coulter – Avanti J-E, USA) to obtain the pelleted fraction, lyophilized and cryoground as previously described.20 Briefly, the 6850 SPEX freezer/mill (SPEX, Metuchen, NJ, USA) cryo-grinding conditions involved precooling for 5 min, followed by two grinding cycles per run at an impactor speed level of 10 s−1 for 5 min each, with an intermediate cooling period of 2 min between grinding cycles. A sample of only ACW, with no adsorbed polyphenols, but which otherwise had been treated in the same way, was used as a positive control, to determine the fermentability of the ACW alone, as well as any shifts in the microbial community. Post-cryogrinding, all substrates were stored in a desiccator until fermentation analysis.
| Substrate | Abbreviation | Adsorbed polyphenol |
|---|---|---|
| Apple cell wall | ACW | 0 |
| Apple cell wall & ferulic acid (−) | ACW + FER | 20.0 |
| Apple cell wall & (±)-catechin | ACW + CAT | 61.7 |
| Apple cell wall & cyanidin-3-glucoside (+) | ACW + CYAN | 143.7 |
| Apple cell wall & triple mix | ACW + TM | 15.4 (FER) |
| 42.3 (CAT) | ||
| 79.5 (CYAN) | ||
| Blank & triple mix | Blank + TM | 0.77 mM (FER) |
| 0.77 mM (CAT) | ||
| 0.77 mM (CYAN) |
To determine the fermentability of the test polyphenols, in their unbound form and in the absence of ACW as an energy source, a triple mixture (TM) containing each of the polyphenol solutions (3 mL, 10 mM, pH 4.2) was prepared. This was added (3 mL) to serum bottles containing medium only (Blank + TM), giving a final concentration in each serum bottle of 0.77 mM per polyphenol.
In vitro batch culture fermentation
An in vitro batch culture method21 was used with a porcine faecal inoculum. Briefly, ∼0.12 g of dried and ground substrate was weighed accurately into 60 mL serum bottles, to which 38 mL of anaerobically-prepared medium22 was added. Thereafter, the bottles were closed with butyl rubber stoppers and aluminium crimp seals. In addition, one set of bottles was prepared as a blank, whereby medium was inoculated without substrates. All procedures were conducted under a constant stream of CO2, unless otherwise stated.Pig faecal collection procedures were approved by The University of Queensland Animal Ethics Committee (SAFS/111/13/ARC). Pigs had access to water ad libitum and were fed twice daily. The faecal inoculum was prepared as follows: Faeces were collected from five large white male pigs (∼35 kg), fed a semi-purified diet containing highly-digestible maize starch (primary carbohydrate source) and fish meal, for ∼10 days prior to faeces collection. This diet was designed to be as low as possible in potentially fermentable carbohydrates, and as free as possible of polyphenols, to avoid adaptation of microbiota to any polyphenols in the diet. Faeces was collected per rectum early in the morning (∼7.00 am) and placed immediately in a wide-mouthed vacuum flask, which was pre-warmed and flushed with CO2. The flask was sealed with a butyl rubber stopper containing a fermentation air-lock valve to allow escape of gas, but no entry. It took approximately one hour between collection at the piggery, and arrival at the lab, where the faeces was further processed under a constant stream of CO2. Collected faeces from all pigs were combined and diluted with pre-warmed (39 °C), sterile saline solution (0.9% NaCl), at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 (w/v) (faeces
5 (w/v) (faeces![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) saline). After homogenisation using an electric hand blender, the faecal slurry was filtered through four layers of muslin cloth. The resulting mixture (2.5 mL) was then injected into serum bottles. Post-inoculation, serum bottles were incubated at 39 °C for up to 72 hours. Bottles were removed at individual time-points in duplicate; at 0, 2, 4, 6, 9, 12, 24, 48 and 72 hours.
saline). After homogenisation using an electric hand blender, the faecal slurry was filtered through four layers of muslin cloth. The resulting mixture (2.5 mL) was then injected into serum bottles. Post-inoculation, serum bottles were incubated at 39 °C for up to 72 hours. Bottles were removed at individual time-points in duplicate; at 0, 2, 4, 6, 9, 12, 24, 48 and 72 hours.
Post-fermentation analyses
SCFA and NH4+ analyses
Methods for SCFA and NH4+ analyses have been described previously.23 Gas chromatography with a flame ionisation detector (GC-FID-Hewlett-Packard 6890 Series GC) was used for the analysis of SCFA, with a modified method,24 using a DB-FFAP capillary column (30 × 0.5 mm). Helium was the carrier gas, at a flow rate of 5.3 mL min−1, with an injector volume of 0.5 μL. The internal standard was iso-caproic acid. The standard for SCFA consisted of acetic acid (52.51 mM), propionic acid (13.4 mM), iso-butyric acid (1.07 mM), n-butyric acid (5.45 mM), iso-valeric acid (0.91 mM), n-valeric acid (0.92 mM), n-caproic acid (0.16 mM), and heptanoic acid (0.15 mM) (Sigma-Aldrich, AUS). All results are reported as mmol per litre.Branched-Chain Proportion (BrChPpn) was calculated as:
For NH4+ analysis, a modified method was used,25 and has been reported previously.20 Briefly, the chemical reaction between NH4+ ions with sodium salicylate and nitroprusside in a weakly alkaline buffer was determined colorimetrically at 650 nm, using a UV/visible spectrophotometer (Olympus AU400, Tokyo, Japan), and results are reported as mmol per litre.
Percent dry matter (%DM) of substrates was determined by accurately weighing ∼0.12 g of substrate into pre-dried and -weighed porcelain crucibles (in duplicate). The crucibles were placed in a pre-warmed oven (constant 103 °C ± 2 °C) for 24 hours. Thereafter, the combined crucibles and dried contents were weighed and %DM of each substrate calculated (ISO 6496![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1999). This value, multiplied by the weight of the substrate in each bottle, gave the weight of DM of the substrate weighed into each bottle.
1999). This value, multiplied by the weight of the substrate in each bottle, gave the weight of DM of the substrate weighed into each bottle.
DNA extraction
Samples for gDNA extraction were removed from the −80 °C freezer and thawed on ice. The 2 mL tubes were then centrifuged at 5000g for 10 min at 4 °C (Beckman Coulter – Avanti J-E, USA), after which 50 mg of biomass was weighed out into fresh 2 mL Eppendorf tubes, containing ∼1 g pre-weighed 0.7 mm garnet beads (catalogue number MB-13123-05, Gene Works, Thebarton, AUS). A volume of 400 μL of tissue-lysis buffer (TLB; catalogue number A8261, Promega Corporation, Alexandria, NSW, AUS) was then added, and the tubes were placed in a bead beater for 60 s at 30 Hz (TissueLyser II, Qiagen Pty Ltd, Chadstone, AUS). Samples were placed on ice for 2 min, after which an additional 100 μL of TLB was added and sample tubes vortexed briefly. A volume of 400 μL of each sample was transferred to an individual starting well of a Maxwell® 16 SEV cartridge (catalogue number AS1200, Promega Corporation, Alexandria, NSW, AUS). gDNA was extracted using the Maxwell® 16 automated nucleic acid purification system (Promega Corporation, Alexandria, NSW, AUS), running in standard elution volume (SEV) mode. The gDNA was eluted into 300 μl of elution buffer (Promega Corporation, Alexandria, NSW, AUS), which was then transferred into new 2 mL Eppendorf tubes for storage at −20 °C, pending downstream analysis.16S rRNA gene amplicon sequencing and phylogenetic analysis
Prior to PCR, extracted gDNA quality and quantity was determined by QuantiT PicoGreen dsDNA assay kit (ThermoFisher Scientific Pty Ltd, Scoresby, VIC, AUS). The protocol for PCR and generation of 16S rRNA gene amplicons has been previously described.26 Briefly, 20 ng of gDNA was used as template for producing partial 16S rRNA gene amplicons (V3 to V4 region). Barcoded amplicons were generated by PCR, using Phusion high fidelity DNA polymerase (Thermo Fisher, Waltham, MA, USA), and barcoded forward primer 341F (5′-Fusion A-Barcode-CCTACGGGAGGCAGCAG-3′) and reverse primer 787R (5′-Fusion B-CTACCAGGGTATCTAAT -3′).27 Barcoded primers were unique to each extracted sample. The PCR (Biorad S1000 Thermal Cycler, Gladesville, NSW, AUS) was conducted under the following conditions: lid heated continually at 105 °C, initial denaturation at 98 °C for 30 s, followed by 30 cycles of: denaturation at 98 °C (10 s); primer annealing at 65 °C (20 s); extension at 72 °C (15 s), and a final extension at 72 °C for 10 min, with samples being held at 12 °C until removed from the machine. PCR amplicons were purified by gel electrophoresis on a 2% agarose gel at 100 volts for 50 min in 1× TE buffer. PCR products were visualised under UV, 3× gel red (Jomar Life Research, Welland, SA, AUS) was added to the agarose gel, and bands corresponding to the correct product size (∼450 bp) were excised for purification. Post-PCR, products were purified using the QIAquick® gel purification kit (QIAGEN®, Chatswood, AUS), following manufacturer's instructions. Purified amplicons were standardised to 40 ng for each sample, using the Quant-iT PicoGreen double stranded DNA assay kit (ThermoFisher Scientific Pty Ltd, Scoresby, VIC, AUS), pooled, concentrated and sent to Macrogen Inc. (Seoul, Republic of Korea) for 454 amplicon pyrosequencing. Sequences and sample information was submitted to the European Nucleotide Archive of the European Bioinformatics Institute (EBI), under the study accession number ‘PRJEB33502’.http://www.ebi.ac.uk/ena/data/view/PRJEB33502.
Analysis of pyrosequencing reads
As raw data was returned in standard flowgram file format (.sff), the files were converted to .fasta and .qual files using the QIIME software.28 Sequences were de-noised using Acacia.29 Sequences with quality values of less than 25 across a sliding window of 50 bp, and outside the lengths of 370–600 bp, were discarded. The data was rarefied to 7263 sequences per sample. The QIIME software was then used to assign taxonomy by clustering sequences into Operational Taxonomic Units (OTU) at the 97% identity level (corresponds approximately to species level).32 The default first cluster seed OTU sequence was chosen as the representative sequence, and these sequences were aligned against the 16S rRNA gene core set in Greengenes (http://greengenes.lbl.gov/)31,34. Each representative OTU sequence was taxonomically classified, from phylum to genus level, using Greengenes’ taxonomy in the Ribosomal Database Project.33 Chimera Slayer,30 and the QIIME script filter_otus_from_otu_table.py, were respectively used to remove chimeras and singletons from the OTU table. Alpha diversity indices (Shannon-Weiner, Phylogenetic Distance, equitability and number of observed species) were calculated at a sequence depth of 2800 sequences. The same sampling depth was used to calculate principal co-ordinates analysis (PCoA), using weighted UniFrac distances as a measure of beta diversity.Data analysis
Statistical analysis to detect differences between substrates and time intervals, were conducted for SCFA and NH4+ data using Tukey's Studentized range test of multiple comparisons within Proc GLM of SAS (Statistical Analysis Systems Institute 9.1, 2002/3).Sequencing data was analysed with R (v3.2.2) (http://www.r-project.org) and R Studio (v0.99.486) (http://www.rstudio.org). The PCoA plots were produced using Between Class Analysis (BCA) and the R packages: “ade4”, “gdata”, and “made4”, based on weighted 16S rRNA gene amplicon sequences. Furthermore, Monte-Carlo permutation tests were undertaken on groups within the PCoA and significant P values reported as <0.005. Where sample data is combined, it is reported as the mean and standard deviation. The software program STAMP (v2) was used to perform a multiple group statistical test (analysis of variance), to calculate significantly different OTU between substrate groups.35
Results
Fermentation end-product differences
The fermentation end-products (including acetic, propionic, butyric, and the branched chain acids, both as total values, and as proportion of the total, as well as NH4+) and pH were measured. Fig. 1 shows graphs of acetic acid, propionic acid, butyric acid, Total SCFA, Branched chain acids and NH4+ values for each substrate in time. The complete dataset for all time points per substrate are shown in Table S1.† Additionally, Table 2 presents the statistical analysis of the end-products data (72 h) for each substrate, showing significant differences between substrates.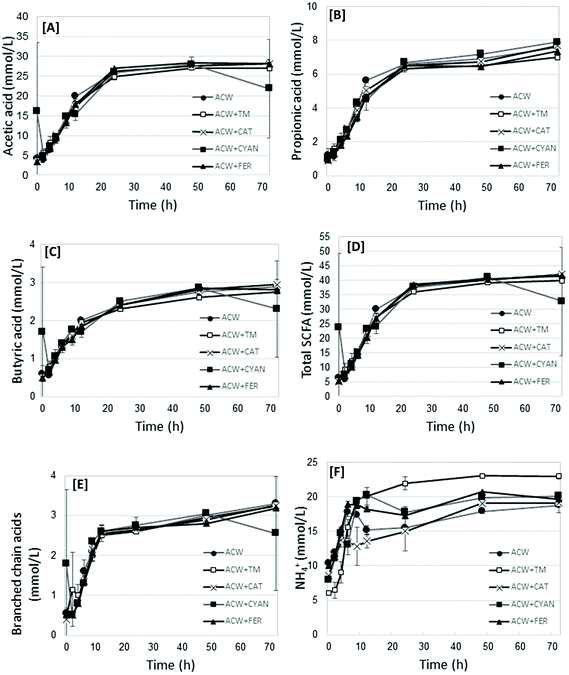 | ||
| Fig. 1 Mean acetic acid [A], propionic acid [B], butyric acid [C], Total SCFA [D], branched chain acids [E] and NH4+ [F] values (mmoles L−1), per substrate at each measured time removal point. | ||
| Acetic acid | Propionic acid | Butyric acid | Total SCFA | TotBrCh | AcTot | PrTot | BuTot | BrChPpn | NH4+ | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Substrate | mmol L−1 | % | mmol L−1 | pH | |||||||
| a, b, c Superscripts in the same column under the same factor indicate significant differences. | |||||||||||
| ACW | 16.7a | 4.53a | 1.80ab | 25.0a | 2.02a | 65.4a | 18.4a | 7.9a | 0.085a | 16.0b | 6.80a |
| ACW + TM | 16.5a | 4.24b | 1.78ab | 24.6a | 2.04a | 66.4b | 17.3b | 7.7a | 0.086a | 17.1a | 6.80a |
| ACW + CAT | 16.7a | 4.47a | 1.81ab | 25.0a | 1.98a | 65.9a | 18a | 7.8a | 0.079a | 14.6c | 6.79a |
| ACW + CYAN | 16.6a | 4.55a | 1.84a | 25.0a | 2.05a | 65.6a | 18.2a | 7.9a | 0.083a | 16.7ab | 6.78a |
| ACW + FER | 16.7a | 4.22b | 1.76b | 24.6a | 1.96a | 66.7b | 17.3b | 7.8a | 0.083a | 17.0a | 6.78a |
| Prob substrate | 0.95 | <0.0001 | 0.03 | 0.34 | 0.33 | <0.0001 | <0.0001 | 0.21 | 0.41 | <0.0001 | 0.32 |
| MSD | 0.63 | 0.171 | 0.075 | 0.88 | 0.142 | 0.74 | 0.45 | 0.24 | 0.0101 | 0.85 | 0.037 |
All end-product values increased in time, before generally plateauing between 24 and 72 hours. Acetic, propionic and butyric acid concentrations increased in time up to 48 hours (P < 0.0001) (Table 2). Acetic acid was not significantly different between substrates. Propionic acid concentrations were lower (P < 0.0001) for ACW + FER and ACW + TM, compared with the other substrates, but not from each other, though this difference was not marked. Butyric acid differed (P = 0.03) between ACW + FER and ACW + CYAN, but not from other substrates. Of all the substrates, ACW + CAT had the lowest overall concentration of NH4+, being significantly different from the other groups (P < 0.0001). ACW + TM, ACW + FER and ACW + CYAN had the highest overall concentrations of NH4+, with no significant differences between them. The values for Blank + TM, while not included in the analysis of variance, had significantly lower mean values for SCFA but comparable values for branched chain fatty acids, NH4+ and pH. The mean values for all timed removals of Blank + TM are reported in Table 2.
Total SCFA showed a general trend of increasing SCFA between 0 and 24 hours, after which values generally plateaued. In contrast, NH4+ concentrations fluctuated between timed removals for all substrates (except ACW + TM), particularly up to 12 hours. For propionic acid, the main differences occurred at 12, 48 and 72 hours, though these differences while significant (P < 0.0001), were not large.
Bacterial richness and diversity based on 16S rRNA gene sequencing analysis
Raw 16S rRNA gene amplicon sequences totalled 760![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 923. After quality control (removal of chimeric and single sequences), 668
923. After quality control (removal of chimeric and single sequences), 668![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 198 sequences remained, with a mean of 7263 sequences per sample. Fig. S1† shows the rarefaction plot indicating the depth of coverage obtained for each sample.
198 sequences remained, with a mean of 7263 sequences per sample. Fig. S1† shows the rarefaction plot indicating the depth of coverage obtained for each sample.
Table 3 indicates the number of OTU per substrate (mean OTU), as well as the mean species richness and alpha diversity indices of sequenced samples including the Shannon (equitability/evenness) and Simpson indices. Ultimately, these sequences were assigned to 2506 unique OTU, classified to 14 phyla, 68 families and 118 genera. In terms of the mean OTU and Chao 1 richness estimator, there were few differences between the different substrates, though the number of OTU for the ACW substrate were generally higher than for those ACW with adsorbed polyphenols. Furthermore, when these overall values at the different removal times were compared with the inoculum (0 hour), the latter had a higher mean OTU and Chao 1 value, suggesting that the original inoculum had higher diversity than the batch cultures. This was to be expected, given that the inoculum came from the diverse environment of the pig GIT. However, once inoculated, the microbial populations were exposed to a single energy source forcing species selection, promoting those bacteria able to utilise the substrates provided, in this case ACW with and without adsorbed polyphenols.
| Substrate | Time | Mean OTU | Chao1 | Shannon's index | Simpson's index |
|---|---|---|---|---|---|
| Inoculum | 0 | 620 | 1072 | 5.32 | 0.87 |
| ACW | 2 | 495 | 718 | 5.38 | 0.85 |
| 4 | 550 | 919 | 5.71 | 0.90 | |
| 6 | 513 | 851 | 6.06 | 0.93 | |
| 9 | 578 | 1002 | 6.03 | 0.94 | |
| 12 | 565 | 942 | 6.15 | 0.95 | |
| 24 | 573 | 1003 | 5.88 | 0.92 | |
| 48 | 521 | 848 | 5.64 | 0.90 | |
| 72 | 530 | 857 | 5.79 | 0.91 | |
| ACW + TM | 2 | 446 | 596 | 6.04 | 0.93 |
| 4 | 478 | 726 | 6.34 | 0.96 | |
| 6 | 447 | 726 | 6.57 | 0.97 | |
| 9 | 477 | 804 | 6.46 | 0.97 | |
| 12 | 396 | 611 | 6.16 | 0.96 | |
| 24 | 477 | 824 | 6.22 | 0.95 | |
| 48 | 491 | 802 | 6.64 | 0.97 | |
| 72 | 482 | 860 | 6.43 | 0.96 | |
| ACW + CAT | 4 | 342 | 513 | 5.74 | 0.91 |
| 6 | 417 | 542 | 5.78 | 0.91 | |
| 9 | 421 | 624 | 6.30 | 0.95 | |
| 12 | 367 | 467 | 6.19 | 0.95 | |
| 24 | 355 | 527 | 5.91 | 0.94 | |
| 48 | 415 | 580 | 5.76 | 0.92 | |
| 72 | 353 | 575 | 6.02 | 0.95 | |
| ACW + CYAN | 4 | 393 | 565 | 5.85 | 0.92 |
| 6 | 335 | 430 | 6.30 | 0.97 | |
| 9 | 408 | 575 | 6.31 | 0.96 | |
| 12 | 372 | 523 | 6.31 | 0.96 | |
| 24 | 285 | 426 | 6.26 | 0.97 | |
| 48 | 310 | 482 | 6.37 | 0.97 | |
| 72 | 432 | 654 | 6.48 | 0.96 | |
| ACW + FER | 4 | 348 | 475 | 5.57 | 0.89 |
| 6 | 288 | 410 | 5.60 | 0.90 | |
| 9 | 381 | 520 | 5.96 | 0.94 | |
| 12 | 422 | 604 | 6.19 | 0.95 | |
| 24 | 433 | 663 | 5.90 | 0.93 | |
| 48 | 382 | 537 | 5.62 | 0.90 | |
| 72 | 371 | 570 | 5.46 | 0.90 | |
| BLANK + TM | 2 | 444 | 270 | 5.52 | 0.90 |
| 4 | 475 | 367 | 6.45 | 0.98 | |
| 6 | 473 | 332 | 6.47 | 0.98 | |
| 9 | 536 | 267 | 5.84 | 0.95 | |
| 12 | 583 | 306 | 6.27 | 0.97 | |
| 24 | 435 | 295 | 6.18 | 0.97 | |
| 48 | 454 | 294 | 5.80 | 0.95 | |
| 72 | 449 | 299 | 6.12 | 0.97 |
Bacterial profile differences within individual polyphenol substrate groups
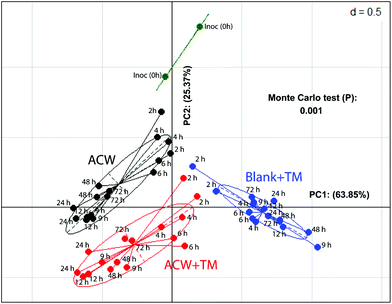
In a comparison of bacterial composition between the ACW, ACW + TM and Blank + TM substrates, sequence abundance classified to phylum level (Fig. S2†), showed that Firmicutes dominated in all samples. However, for ACW, the ratio between the largest two phyla, Firmicutes and Bacteroidetes, changed between 4 and 48 hours. Bacteroidetes increased during this period whilst Firmicutes decreased, although by 48 hours, the abundance of Firmicutes was comparable to that of the original inoculum. Additionally, such data for the individual polyphenols combined with ACW have been reported separately (Fig. S3†).To illustrate the overall community structure, a PCoA plot (Fig. 2) summarises the clustering of those samples with and without added TM across all times. The ACW and the Blank + TM groups clustered furthest apart, while ACW + TM clustered between these two groups, but was still well separated from both. The majority of variation (64%) was explained by the difference between the Blank + TM and the other two substrates containing ACW. This is most likely related to the fact that the Blank + TM, while containing the same medium and inoculum, had no carbohydrate present as an energy source, but only the TM mix containing the three polyphenols. When the removal times are considered, an interesting picture emerges (Fig. 2). At the earlier time removals (2, 4, and 6 hours), Blank + TM and ACW + TM are relatively close together, but already distant from Inoc. ACW, on the other hand, is closer to Inoc at these times (2, 4, and 6 hours). Thereafter, values for each substrate move both further away from Inoc and from each other (according to substrate). The proximity of values for ACW + TM and Blank + TM (2 and 4 hours) suggest that there is potentially an early bactericidal or bacteriostatic effect associated with the presence of polyphenols.
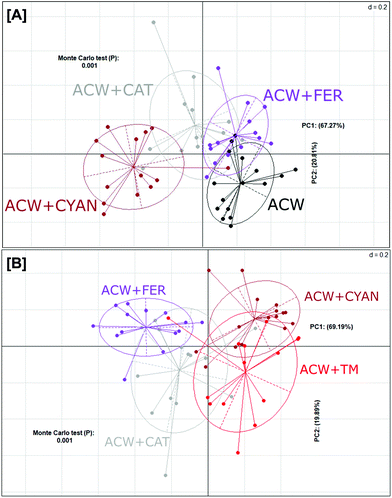
In contrast, a PCoA plot of the individual polyphenols adsorbed to ACW (Fig. 3A) showed that overall, these groups failed to fully differentiate from one another. Both ACW + FER and ACW + CAT still had some overlap with ACW alone, unlike ACW + CYAN, which was completely separated from both ACW and ACW + FER. However, ACW + CYAN had more comparative overlap with ACW + TM (Fig. 3B) than the other ACW and polyphenol substrate combinations, indicating that cyanidin-3-glucoside had a greater effect on shifting bacterial communities within these inoculated bottles away from that of ACW alone, which potentially might be from a sugar moiety in its initial molecular structure that is cleaved off and utilised.36
Bacterial profiles through time and associated fermentation end-products
Samples containing ACW with or without polyphenols were analysed by PCoA based on time (ESI Fig. S4†), confirming that the bacterial community structure was strongly influenced by removal time. This analysis included all samples that had the addition of polyphenols adsorbed to ACW or ACW alone, but did not include Blank + TM.Fermentation end-products were also influenced by time (Fig. S3†). Between 4 and 12 hours, there was an increased association with acetic, propionic, and butyric acids. While NH4+ and the BrChPpn (both associated with protein fermentation),37 were more associated with the later time-points of 24, 48 and 72 hours
Significantly different OTU between substrates and time removals
Table 4 shows those OTU (of all detected) which were either unique to different substrate groups, or differed significantly between the groups, and their nearest known classifications. The ACW + CYAN and ACW + FER substrates had a greater number of different OTU compared with ACW + CAT. The family Lachnospiraceae and the genus Parabacteroides were the main differential OTU classifications for ACW + CYAN. For the ACW + FER, differentiated OTU predominantly belonged to the genus Phascolarctobacterium, with the 72 hours time point being the most significantly different, while ACW + CAT had differentiated OTU belonging to the genera Collinsella, Coprococcus and the order Clostridiales.| Closest taxonomic classification | Denovo ID | Substrate | P value | Abundance | Time (h) |
|---|---|---|---|---|---|
| Collinsella genus | 9268 | ACW + CAT | <0.0001 | 0.4% | 9 |
| Coprococcus genus | 6815 | <0.0001 | 0.2% | 9 | |
| Clostridiales order | 7472 | <0.0001 | 1.3% | 9 | |
| Ruminococcaceae family | 6390 | ACW + FER | <0.0001 | 3.1% | 6 |
| Clostridiales order | 7472 | <0.0001 | 1.3% | 48 | |
| Phascolarctobacterium genus | 1544 | <0.0001 | 0.53% | 72 | |
| 4662 | <0.0001 | 35.2% | 72 | ||
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 826 826 |
<0.0001 | 0.8% | 72 | ||
| 2701 | <0.0001 | 0.4% | 72 | ||
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 644 644 |
<0.0001 | 0.8% | 72 | ||
| Coriobacteriaceae family | 9492 | <0.0001 | 1.1% | 72 | |
| Lachnospiraceae family | 5968 | ACW + CYAN | <0.0001 | 1.4% | 6 |
| Prevotella copri | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150 150 |
<0.0001 | 0.8% | 6 | |
| Parabacteroides genus | 7488 | <0.0001 | 0.7% | 6 | |
| Parabacteroides genus | 8470 | <0.0001 | 0.7% | 6 | |
| Prevotella copri | 8001 | <0.0001 | 6.1% | 24 | |
| Prevotella copri | 1018 | <0.0001 | 4.6% | 48 | |
| Parabacteroides genus | 4312 | <0.0001 | 1.4% | 48 | |
| 7098 | <0.0001 | 0.9% | 48 | ||
| Clostridiales order | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 211 211 |
<0.0001 | 0.4% | 48 |
Polyphenol disappearance in the medium
In order to identify the timescale over which the disappearance of polyphenols occurred during fermentation, residual levels of each of the three polyphenols were determined as a function of fermentation time. The results (Fig. 4) show that in all cases, the original polyphenols added initially, had disappeared by 9 hours, including the Blank + TM. The rate of disappearance was similar for all substrates regardless of adsorption to ACW.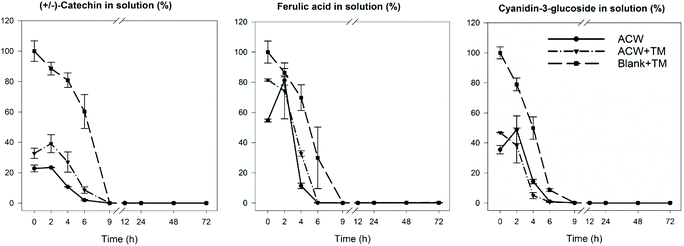 | ||
| Fig. 4 Polyphenol disappearance during the 72 hours fermentation. Reported as % of the original polyphenolic compound remaining. The values were calculated based on the initial adsorbed amounts of polyphenols described in Table 1. Reproduced from Phan, 2016.19 | ||
Discussion
During in vitro fermentation, the bacterial community composition was examined across nine time-points during 72 hours. ACW alone was compared with ACW with adsorbed polyphenols, including: ferulic acid (ACW + FER), catechin (ACW + CAT), and cyanidin-3-glucoside (ACW + CYAN). In addition, a combination of the three polyphenols adsorbed simultaneously to ACW (ACW + TM), and the three together with no ACW carbohydrate source (Blank + TM) were included. The last two combinations were included to detect the possibility of the three chosen polyphenols interacting either in the presence or absence of ACW.The ACW + FER group showed a decrease in propionic acid concentration compared to that of the ACW alone (P < 0.0001). In a previous in vitro study of wheat arabinoxylans, there was an overall decreased SCFA, which was correlated with increased freely available ferulic acid.38 The lower propionic acid may be a result of slightly reduced fermentation of ACW in the presence of ferulic acid, possibly due to its potential as an antimicrobial agent.39 Interestingly, despite this small reduction in propionic acid, the most dominant genus detected in these samples at 72 hours was Phascolarctobacterium, which is known to be a propionic acid producer.40
The differences between end-products of substrate groups, even when significant, were not large, suggesting that the overall conversion of ACW into SCFA was not greatly affected by the presence of polyphenols. Interestingly, when comparing the data shown in Table 2 with the graphs shown in Fig. 4, polyphenol disappearance occurred within the first 9 hours, while the bacterial fermentation end-product production occurred within the first 24 hours. This contrasts with an in vitro study examining the biotransformation of dissolved rutin, quercetin, chlorogenic and caffeic acids, where all four compounds had disappeared from the medium within 30 min.41 However, these polyphenols were freely available in solution, whereas in our study the adsorbed polyphenols had been gradually released over time. Slower release was due to bacteria fermenting and altering the ACW matrix which released strongly bound polyphenols (Fig. 4). Furthermore, this latter study used a human faecal inoculum from a donor who ate a fruit- and vegetable-rich diet, and would therefore be expected to have a microbiota which was well adapted to rapidly ferment polyphenols. In contrast, in this study the inoculum was from pigs who had been fed a low polyphenol diet, consistent with the microbial population requiring longer to adapt to the polyphenol substrate.
Despite limited differences in end-products of fermentation, there were clear differences in microbial population structure with substrate (Fig. 3), suggesting that a similar end-product profile may be obtained from different microbial consortia as a result of the inclusion of polyphenols. This aligns with research that suggests polyphenol metabolism pathways can be completed by a varied group of bacterial species.42,43 Comparing populations with a mix of three polyphenols either separately or adsorbed to ACW (Fig. 2), showed early time-points (hours 2, 4, 6) for the polyphenol-containing substrates, had a similar change of position from the initial inoculum. The ACW bacterial community was thus markedly affected by the presence of polyphenols, even though polyphenols are present at only about 15% of the level of ACW (Table 1).
The three groups of separately adsorbed polyphenols to ACW also showed shifts in bacterial populations (Fig. 3A). Separation of these four groups differed depending on the amount and types of polyphenol adsorbed (Table 1). Cyanidin-3-glucoside led to the greatest separation and was adsorbed to the ACW in the largest quantities, while ferulic acid was the least adsorbed and showed less separation from the ACW group. In addition, the analysis in Fig. 3B shows that cyanidin-3-glucoside created similar bacterial community profile differences for ACW + TM. Cyanidin-3-glucoside appears to have had the largest effect on both these groups, leading to the principal variance explained across the PC1 axis (69% Fig. 3B), with ACW + CYAN and ACW + TM split from other groups. This correlation with adsorbed polyphenol levels suggests that dose–response studies of different amounts of individual or mixed polyphenols in combination with plant cell walls would provide a more nuanced explanation for polyphenol-specific effects on microbial populations.
Fig. 2A & B indicated that the ACW-containing samples were more closely clustered, than the groups (±ACW), shown in Fig. 2. Given that ACW was present in large quantities (polyphenols generally present as ∼2% of the fermentation solutions), it is noteworthy that the small additions of polyphenols adsorbed to ACW (Table 1) in their respective quantities, have significantly shifted the bacterial community overall.
The time of fermentation was also a major factor in bacterial community differences, as can be seen for all substrates in Fig. S3.† Combined with the SCFA and NH4+ data in this figure, there seems to be a distinct shift at ∼12 hours from a carbohydrate-dominant to a protein-dominant pattern of fermentation. The association of bacterial community profiles with end-products and time, suggest that readily available carbohydrates started to become limiting around 9 to 12 hours, after which protein fermentation became more prevalent. Given the known presence of protein in the medium, it is well established that as fermentable carbohydrates are depleted, protein fermentation may increase, further altering the environment of the batch fermenter, by the substantial change in metabolites.44 This changeover is also likely to be related to the metabolically co-operative environment of the bacterial community, where the metabolites of polyphenols can be better utilised by other members of the community compared with those first involved in their degradation.45,46
Previously, cyanidin has been shown to have antimicrobial properties against the Gram-negative bacteria E.coli, at concentrations of 50 mg L−1 (0.005%) or greater,47 which is lower than the concentration of 0.24% of cyanidin used in this study. It has also been shown that Helicobacter pylori can be inhibited by cyanidin-3-O-glucoside at 100 μM concentration.48 At least partially, this previously published data may explain the reduced Gram-negative Bacteroidetes seen for the control groups of Blank + TM and ACW + TM that both contained cyanidin-3-glucoside, compared to that of the ACW alone. The OTU from the genus Parabacteroidetes, was the predominant classification for significantly different OTU within the ACW + CYAN substrate group, accounting for 1.4% of the overall community at 6 hours, and 2.3% at 48 hours. Interestingly, Parabacteroides has been shown to have bacteriocin pathways in some species,49 and has also been identified as being involved in difficult polymer degradation such as cellulose and type IV resistant starch.50 Both of these characteristics would help to explain its dominance in the ACW + CYAN group.
Within the ACW + FER group, the genus Phascolarctobacterium was a predominant classification for significantly different OTU increasing to 37.73% of the community at 72 hours. Although Phascolarctobacterium is a producer of propionic acid,40 propionic acid concentrations were lower in comparison to other substrates overall (Table 2). Phascolarctobacterium is common in human and porcine GI tracts, most commonly utilising succinic acid.40,51
The ACW + CAT group had significantly different OTU, which were associated with the order Clostridiales and the genera Collinsella and Coprococcus, with these bacterial identifications accounting for 1.3, 0.4 and 0.2% respectively, of the total proportion of the community at 9 hours. A previous study also reported increased Collinsella when incubated with catechin in vitro at a concentration of 0.15 g L−1.52 Whilst, Coprococcus increased in rats fed a catechin supplement to a high fat diet.53Coprococcus can produce a wide range of fatty acids including acetic, propionic and butyric acids, and/or lactic acid.54
Overall, it is clear that the bacterial community structure responded differently to the three different polyphenols that had been adsorbed to ACW. However, when added as a “triple mix” with no accompanying carbohydrates/cell walls in the form of ACW, the effect seems to have been quite different, particularly in terms of bacterial population structure. There are also indications of a bactericidal or bacteriostatic effect. Further investigation is required to examine the potential for these effects more thoroughly.
Conclusions
The adsorption of three different polyphenols to ACW led to bacterial community changes during in vitro fermentation. At concentrations of 2–15% of the initial ACW substrate for fermentation, the polyphenols used in this study had significant effects on the abundance of community members. Interestingly, the polyphenols adsorbed to PCW, rather than in isolation, impacted the GIT bacterial population differently, compared with their isolated counterparts. Bactericidal or bacteriostatic properties may have contributed to polyphenol-mediated changes in microbial populations, but did not markedly affect overall ACW fermentation end-products in the short term. This approach of using polyphenols adsorbed to isolated PCW is useful as a simplified and representative model for testing the large intestinal microbial effects of polyphenols derived from fruit- or vegetable-based foods, than polyphenols alone. However, future work is needed to resolve the effects of individual polyphenols from that of the adsorbed dose. A dose–response curve using different initial concentrations of adsorbed polyphenols could be one approach for this.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors thank Peter Isherwood for SCFA analysis; Brian Burren for NH4+ analysis; Barbara Gorham and Gabi Netzel for technical assistance with the in vitro experiment. This study was supported by the ARC Centre for Excellence in Plant Cell Walls (CE110001007).References
- B. A. Acosta-Estrada, J. A. Gutierrez-Uribe and S. O. Serna-Saldivar, Bound phenolics in foods, a review, Food Chem., 2014, 152, 46–55 CrossRef CAS PubMed.
- J. Pérez-Jiménez and F. Saura-Calixto, Macromolecular antioxidants or non-extractable polyphenols in fruit and vegetables: Intake in four European countries, Food Res. Int., 2015, 74, 315–323 CrossRef PubMed.
- C. Manach, A. Scalbert, C. Morand, C. Rémésy and L. Jiménez, Polyphenols: food sources and bioavailability, Am. J. Clin. Nutr., 2004, 79, 727–747 CrossRef CAS PubMed.
- J. Van Duynhoven, E. E. Vaughan, D. M. Jacobs, R. A. Kemperman, E. J. van Velzen, G. Gross, L. C. Roger, S. Possemiers, A. K. Smilde, J. Dore, J. A. Westerhuis and T. Van de Wiele, Metabolic fate of polyphenols in the human superorganism, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 4531–4538 CrossRef CAS PubMed.
- F. Cardona, C. Andres-Lacueva, S. Tulipani, F. J. Tinahones and M. I. Queipo-Ortuno, Benefits of polyphenols on gut microbiota and implications in human health, J. Nutr. Biochem., 2013, 24, 1415–1422 CrossRef CAS PubMed.
- P. Mena, L. Calani, R. Bruni and D. Del Rio, Bioactivation of high-molecular-weight polyphenols by the gut microbiome Diet-microbe interactions, in Diet-microbe interactions in the gut, ed. K. Tuohy and D. Del Rio, Academic Press, San Diego USA, 2015, pp. 73–101 Search PubMed.
- G. Williamson and M. N. Clifford, Colonic metabolites of berry polyphenols: the missing link to biological activity?, Br. J. Nutr., 2010, 104, S48–S66 CrossRef CAS PubMed.
- J. Fleschhut, F. Kratzer, G. Rechkemmer and S. E. Kulling, Stability and biotransformation of various dietary anthocyanins in vitro, Eur. J. Nutr., 2006, 45, 7–18 CrossRef CAS PubMed.
- A. M. Aura, P. Martin-Lopez, K. A. O'Leary, G. Williamson, K. M. Oksman-Caldentey, K. Poutanen and C. Santos-Buelga, In vitro metabolism of anthocyanins by human gut microflora, Eur. J. Nutr., 2005, 44, 133–142 CrossRef CAS PubMed.
- D. Wang, L. Ho, J. Faith, K. Ono, E. M. Janle, P. J. Lachcik, B. R. Cooper, A. H. Jannasch, B. R. D'Arcy, B. A. Williams, M. G. Ferruzzi, S. Levine, W. Zhao, L. Dubner and G. M. Pasinetti, Role of intestinal microbiota in the generation of polyphenol-derived phenolic acid mediated attenuation of Alzheimer's disease beta-amyloid oligomerization, Mol. Nutr. Food Res., 2015, 59, 1025–1040 CrossRef CAS PubMed.
- C. Tsang, C. Auger, W. Mullen, A. Bornet, J.-M. Rouanet, A. Crozier and P.-L. Teissedre, The absorption, metabolism and excretion of flavan-3-ols and procyanidins following the ingestion of a grape seed extract by rats, Br. J. Nutr., 2007, 94, 170–181 CrossRef PubMed.
- M. J. Hopkins, H. N. Englyst, S. Macfarlane, E. Furrie, G. T. Macfarlane and A. J. McBain, Degradation of cross-linked and non-cross-linked arabinoxylans by the intestinal microbiota in children, Appl. Environ. Microbiol., 2003, 69, 6354–6360 CrossRef CAS PubMed.
- J. He and M. M. Giusti, Anthocyanins: natural colorants with health-promoting properties, Annu. Rev. Food Sci. Technol., 2010, 1, 163–187 CrossRef CAS PubMed.
- M. Hidalgo, M. J. Oruna-Concha, S. Kolida, G. E. Walton, S. Kallithraka, J. P. Spencer and S. de Pascual-Teresa, Metabolism of anthocyanins by human gut microflora and their influence on gut bacterial growth, J. Agric. Food Chem., 2012, 60, 3882–3890 CrossRef CAS PubMed.
- E. R. Miller and D. E. Ullrey, The pig as a model for human nutrition, Annu. Rev. Nutr., 1987, 7, 361–382 CrossRef CAS PubMed.
- S. N. Heinritz, R. Mosenthin and E. Weiss, Use of pigs as a potential model for research into dietary modulation of the human gut microbiota, Nutr. Res. Rev., 2013, 26, 191–209 CrossRef PubMed.
- P. Guilloteau, R. Zabielski, H. M. Hammon and C. C. Metges, Nutritional programming of gastrointestinal tract development. Is the pig a good model for man?, Nutr. Res. Rev., 2010, 23, 4–22 CrossRef PubMed.
- K. A. Bindon, P. A. Smith and J. A. Kennedy, Interaction between grape-derived proanthocyanidins and cell wall material. 1. Effect on proanthocyanidin composition and molecular mass, J. Agric. Food Chem., 2010, 58, 2520–2528 CrossRef CAS PubMed.
- A. D. Phan, Interactions between dietary polyphenols and plant cell wall models, P.h.D Thesis, The University of Queensland, 2016 Search PubMed.
- D. Mikkelsen, M. J. Gidley and B. A. Williams, In vitro fermentation of bacterial cellulose composites as model dietary fibers, J. Agric. Food Chem., 2011, 59, 4025–4032 CrossRef CAS PubMed.
- B. A. Williams, M. W. Bosch, H. Boer, M. W. A. Verstegen and S. Tamminga, An in vitro batch culture method to assess potential fermentability of feed ingredients for monogastric diets, Anim. Feed Sci. Technol., 2005, 123–124, 445–462 CrossRef CAS.
- S. E. Lowe, M. K. Theodorou, A. P. J. Trinci and R. B. Hespell, Growth of anaerobic ruman fungi on defined and semi-defined media lacking rumen fluid, J. Gen. Microbiol., 1985, 131, 2225–2229 Search PubMed.
- B. A. Williams, D. Zhang, A. T. Lisle, D. Mikkelsen, C. S. McSweeney, S. Kang, W. L. Bryden and M. J. Gidley, Soluble arabinoxylan enhances large intestinal microbial health biomarkers in pigs fed a red meat-containing diet, Nutrition, 2015, 32, 491–497 CrossRef PubMed.
- H. J. Vreman, J. A. Dowling, R. A. Raubach and M. W. Weiner, Determination of acetate in biological material by vacuum microdistillation and gas chromatography, Anal. Chem., 1978, 50, 1138–1141 CrossRef CAS PubMed.
- W. E. Baethgen and M. M. Alley, A manual colorimetric procedure for measuring ammonium nitrogen in the soil and plant kjeldahl digests, Commun. Soil Sci. Plant Anal., 1989, 20, 961–969 CrossRef CAS.
- L. M. Gulino, D. Ouwerkerk, A. Y. Kang, A. J. Maguire, M. Kienzle and A. V. Klieve, Shedding light on the microbial community of the macropod foregut using 454-amplicon pyrosequencing, PLoS One, 2013, 8, e61463 CrossRef CAS PubMed.
- G. C. Baker, J. J. Smith and D. A. Cowan, Review and re-analysis of domain-specific 16S primers, J. Microbiol. Methods, 2003, 55, 541–555 CrossRef CAS PubMed.
- J. G. Caporaso, J. Kuczynski, J. Stombaugh, K. Bittinger, F. D. Bushman, E. K. Costello, N. Fierer, A. Gonzalez Pena, J. K. Goodrich, J. I. Gordon, G. A. Huttley, S. T. Kelley, D. Knights, J. E. Koenig, R. E. Ley, C. A. Lozupone, D. McDonald, B. D. Muegge, M. Pirrung, J. Reeder, J. R. Sevinsky, P. J. Turnbaugh, W. A. Walters, J. Widmann, T. Yatsunenko, J. Zaneveld and R. Knight, QIIME allows analysis of high-throughput community sequencing data, Nat. Methods, 2010, 7, 335–336 CrossRef CAS PubMed.
- L. Bragg, G. Stone, M. Imelfort, P. Hugenholtz and G. W. Tyson, Fast, accurate error-correction of amplicon pyrosequences using Acacia, Nat. Methods, 2012, 9, 425–426 CrossRef CAS PubMed.
- B. J. Haas, D. Gevers, A. M. Earl, M. Feldgarden, D. V. Ward, G. Giannoukos, D. Ciulla, D. Tabbaa, S. K. Highlander, E. Sodergren, B. Methe, T. Z. DeSantis, Human Microbiome Consortium, J. F. Petrosino, R. Knight and B. W. Birren, Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons, Genome Res., 2011, 21, 494–504 CrossRef CAS PubMed.
- T. Z. DeSantis, P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu and G. L. Andersen, Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB, Appl. Environ. Microbiol., 2006, 72, 5069–5072 CrossRef CAS PubMed.
- P. D. Schloss and J. Handelsman, Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness, Appl. Environ. Microbiol., 2005, 71, 1501–1506 CrossRef CAS PubMed.
- J. R. Cole, Q. Wang, E. Cardenas, J. Fish, B. Chai, R. J. Farris, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, T. Marsh, G. M. Garrity and J. M. Tiedje, The Ribosomal Database Project: improved alignments and new tools for rRNA analysis, Nucleic Acids Res., 2009, 37, D141–D145 CrossRef CAS PubMed.
- D. McDonald, M. N. Price, J. Goodrich, E. P. Nawrocki, T. Z. DeSantis, A. Probst, G. L. Andersen, R. Knight and P. Hugenholtz, An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea, ISME J., 2012, 6, 610–618 CrossRef CAS PubMed.
- D. H. Parks, G. W. Tyson, P. Hugenholtz and R. G. Beiko, STAMP: Statistical analysis of taxonomic and functional profiles, Bioinformatics, 2014, 30, 3123–3124 CrossRef CAS PubMed.
- A. Braune and M. Blaut, Bacterial species involved in the conversion of dietary flavonoids in the human gut, Gut microbes, 2016, 7, 216–234 CrossRef CAS PubMed.
- G. T. Macfarlane, J. H. Cummings and C. Allison, Protein degradation by human intestinal bacteria, J. Gen. Microbiol., 1986, 132, 1647–1656 CAS.
- S. A. Hughes, P. R. Shewry, L. Li, G. R. Gibson, M. L. Sanz and R. A. Rastall, In vitro, fermentation by human fecal microflora of wheat arabinoxylans, J. Agric. Food Chem., 2007, 55, 4589–4595 CrossRef CAS PubMed.
- A. Borges, C. Ferreira, M. J. Saavedra and M. Simoes, Antibacterial activity and mode of action of ferulic and gallic acids against pathogenic bacteria, Microb. Drug Resist., 2013, 19, 256–265 CrossRef CAS PubMed.
- Y. Watanabe, F. Nagai and M. Morotomi, Characterization of Phascolarctobacterium succinatutens sp. nov., an asaccharolytic, succinate-utilizing bacterium isolated from human feces, Appl. Environ. Microbiol., 2012, 78, 511–518 CrossRef PubMed.
- S. G. Parkar, T. M. Trower and D. E. Stevenson, Fecal microbial metabolism of polyphenols and its effects on human gut microbiota, Anaerobe, 2013, 23, 12–19 CrossRef CAS PubMed.
- I. Rowland, G. Gibson, A. Heinken, K. Scott, J. Swann, I. Thiele and K. Tuohy, Gut microbiota functions: metabolism of nutrients and other food components, Eur. J. Nutr., 2018, 57, 1–24 CrossRef CAS PubMed.
- F. A. Tomas-Barberan, R. Garcia-Villalba, A. Gonzalez-Sarrias, M. V. Selma and J. C. Espin, Ellagic acid metabolism by human gut microbiota: consistent observation of three urolithin phenotypes in intervention trials, independent of food source, age, and health status, J. Agric. Food Chem., 2014, 62, 6535–6538 CrossRef CAS PubMed.
- S. W. Gratz, A. J. Richardson, S. H. Duncan, W. R. Russell, C. Fyfe, A. M. Johnstone, H. J. Flint and G. Holtrop, Influence of dietary carbohydrate and protein on colonic fermentation and endogenous formation of N-nitroso compounds, Proc. Nutr. Soc., 2015, 74, E44 CrossRef.
- W. R. Russell, L. Hoyles, H. J. Flint and M. E. Dumas, Colonic bacterial metabolites and human health, Curr. Opin. Microbiol., 2013, 16, 246–254 CrossRef CAS PubMed.
- A. Duda-Chodak, T. Tarko, P. Satora and P. Sroka, Interaction of dietary compounds, especially polyphenols, with the intestinal microbiota: a review, Eur. J. Nutr., 2015, 54, 325–341 CrossRef CAS PubMed.
- R. Puupponen-Pimia, L. Nohynek and C. Meier, Antimicrobial properties of phenolic compounds from berries, J. Appl. Microbiol., 2001, 90, 494–507 CrossRef CAS PubMed.
- S. H. Kim, M. Park, H. Woo, N. Tharmalingam, G. Lee, K. J. Rhee, Y. B. Eom, S. I. Han, W. D. Seo and J. B. Kim, Inhibitory effects of anthocyanins on secretion of Helicobacter pylori CagA and VacA toxins, Int. J. Med. Sci., 2012, 9, 838–842 CrossRef CAS PubMed.
- V. Nakano, A. Ignacio, M. R. Fernandez, M. H. Fukugaiti and M. J. Avila-Campos, Intestinal Bacteroides, and Parabacteroides species producing antagonistic substances, Microbiology, 2006, 1, 61–64 Search PubMed.
- I. Martinez, J. Kim, P. R. Duffy, V. L. Schlegel and J. Walter, Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects, PLoS One, 2010, 5, e15046 CrossRef CAS PubMed.
- Y. Sun, L. Zhou, L. Fang, Y. Su and W. Zhu, Responses in colonic microbial community and gene expression of pigs to a long-term high resistant starch diet, Front. Microbiol., 2015, 6, 877 Search PubMed.
- B. Xue, J. Xie, J. Huang, L. Chen, L. Gao, S. Ou, Y. Wang and X. Peng, Plant polyphenols alter a pathway of energy metabolism by inhibiting fecal Bacteroidetes and Firmicutes in vitro, Food Funct., 2016, 7, 1501–1507 RSC.
- J. Huang, X. Lin, B. Xue, J. Luo, L. Gao, Y. Wang, S. Ou and X. Peng, Impact of polyphenols combined with high-fat diet on rats’ gut microbiota, J. Funct. Foods, 2016, 26, 763–771 CrossRef CAS.
- L. V. Holdeman and W. E. C. Moore, New genus, Coprococcus, twelve new species, and emended descriptions of four previously described species of bacteria from human feces, Int. J. Syst. Evol. Microbiol., 1974, 24, 260–277 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9fo02428j |
| This journal is © The Royal Society of Chemistry 2020 |