Exploring the flavor formation mechanism under osmotic conditions during soy sauce fermentation in Aspergillus oryzae by proteomic analysis†
Guozhong
Zhao
 *ab,
Chong
Liu
ab,
Shu
Li
ab,
Xiaowen
Wang
c and
Yunping
Yao
*ab,
Chong
Liu
ab,
Shu
Li
ab,
Xiaowen
Wang
c and
Yunping
Yao
 *ab
*ab
aState Key Laboratory of Food Nutrition and Safety, Key Laboratory of Food Nutrition and Safety, Ministry of Education, College of Food Science and Engineering, Tianjin University of Science & Technology, Tianjin 300457, China. E-mail: zgzjiangnan@163.com; yaoyunping1314@163.com; Fax: +86-22-60912587; Tel: +86-22-60912585
bDemonstration center of food quality and safety testing technology, Tianjin University of Science and Technology, 300457 Tianjin, China
cCollege of Food Science and Engineering, Shanxi Agricultural University, Shanxi 030801, China
First published on 16th December 2019
Abstract
Aspergillus oryzae is a common starter in the soy sauce industry and struggles to grow under complex fermentation conditions. However, little is known about the flavor formation mechanism under osmotic conditions (low-temperature and high-salt) in A. oryzae. This work investigated the flavors and the relative protein expression patterns by gas chromatography-mass spectrometry (GC-MS) and proteomic analysis. Low-temperature and a high-salt content are unfavorable to the secretion of hydrolases and the formation of fragrant aldehydes. The aldehyde contents under osmotic conditions were reduced to 1.4–3.7 times lower than that of the control. Besides, copper amine oxidases which decreased under low-temperature stress and salt stress were shown to be important in catalyzing the oxidative deamination of several amine substrates to fragrant aldehydes. Furthermore, alcohol dehydrogenase and polyketide synthase are beneficial to the formation of alcohols and aromatic flavors under low-temperature stress and salt stress. Particularly, the ethanol content under 16 °C stress was 3.5 times higher than that under 28 °C.
1. Introduction
Soy sauce, a popular condiment in Asia, is widely consumed all over the world for its umami taste and characteristic flavor. Over 400 flavor compounds have been identified in soy sauce. Alcohols, esters, aldehydes, pyrazines, and furans are the most abundant volatile flavors which are mixed at appropriate proportions in soy sauce. The unique flavor of soy sauce is closely related to the particular fermentation environment which regulates the secretion of various enzymes in microorganisms.Aspergillus oryzae is essential for koji and brine fermentation as main processes of soy sauce production as it potentially secretes lots of proteases and glycoside hydrolases for hydrolyzing soybean polysaccharides and proteins.1 Brine fermentation is dominated by bacteria, followed by yeast and A. oryzae.2 The complex formation of flavors might correspond to microorganism interaction. Alcohols, produced by yeast, make a great contribution to the flavor of soy sauce.3 Alcohols and carboxylic acids can be formed in the reaction to esters. Also, alcohols and aldehydes can be transformed into each other under the actions of alcohol/aldehyde dehydrogenase. Aldehydes are responsible for the malty and nutty aroma of soy sauce. Besides, pyrazines come from the Strecker degradation of α-amino acids and reductones.4 Furans probably come from the degradation of glucose, a thermal degradation product of cellulose.
To date, the manipulation of the environment of fermentation is favorable for flavor formation. For example, a high content of salt is necessary for brine fermentation to prevent microorganism contamination and affects the saline taste simultaneously.5 Also, a low-temperature resists the activity of enzymes.6 The fermentation environment has a relationship with the microbe compositions which contribute to the aroma formation during soy sauce fermentation.7 However, other techniques such as low-intensity ultrasound treatment are also capable of enhancing the taste of soy sauce.8
The proteomic technique can identify expressed proteins revealing important metabolic information of microorganisms.9 The isobaric tags for relative and absolute quantitation (iTRAQ) labeling technique can identify and quantify proteins from differently treated samples in complex developmental processes.10 Therefore, the iTRAQ technique is more suitable to gain insights into the changes of proteomic profiles in response to osmotic stresses.
In the present work, we aim to identify A. oryzae stress adaptation proteins under low-temperature stress and salt stress by the iTRAQ technique. Moreover, we intend to compare the flavor compounds to establish the correlation between stresses and flavors. The comparative proteomic analysis is a very effective method to show the mechanisms of A. oryzae against environmental stresses, which may illustrate the global protein expression under stress to gain a deeper understanding of stress regulation and signaling.
2. Materials and methods
2.1. Materials
Tris-HCl, sodium dodecyl sulfate (SDS), phenylmethylsulfonyl fluoride (PMSF) and ethylene diamine tetraacetic acid (EDTA) were purchased from Shanghai Sangon; sequencing grade modified trypsin, DNase I and RNase were supplied by Promega; tetraethylammonium bromide (TEAB), dithiothreitol (DTT) 2-octanol and dichloromethane were purchased from Sigma; 2,5-dihydro-2,4,5-trimethylthiazoline (TMT) kit was purchased from Thermo; formic acid (FA) was supplied by FLUKA; acetonitrile (ACN) was purchased from Fisher Chemical. n-Alkanes standard (C7–C30) (Sigma–Aldrich Chemical Co, St Louis, MO) was used to determine the retention index (RI) of volatile compounds.2.2. Strain and sample preparation
A. oryzae 3.042 was isolated from soil samples by the Shanghai Brewing Science Research Institute and extensively used as a starter for soy sauce fermentation in China. A. oryzae 3.042 was cultured in the rice-juice medium under different osmotic conditions as described previously (Table 1). The rice-juice medium has been described in detail previously.11A. oryzae spores (0.3%) (w![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v) were initially incubated into the medium. Briefly, 1.5 g of the spores (7.5 × 109) was grown in 500 mL of submerged culture medium. Then 15% agar was supplemented in the medium (10 g) for solid culture, and then poured in a plate (90 mm diameter). A. oryzae spores (1.5 g) were mixed with a drop of Tween 80 in 10 mL of ddH2O. Then, 200 μL of spore suspension was added and coated on the surface of the solid plate medium with a cellophane membrane. The temperature was controlled by using a biochemical incubator. Samples were collected when the mold of A. oryzae changed to yellow-green, and this mimicking fermentation lasted for about 68 h. A part of the fermentation medium was selected for the extraction of aroma compounds. Mycelia for the proteomic analysis were collected by centrifugation and ground in a mortar in the presence of liquid nitrogen following a PBS wash.
v) were initially incubated into the medium. Briefly, 1.5 g of the spores (7.5 × 109) was grown in 500 mL of submerged culture medium. Then 15% agar was supplemented in the medium (10 g) for solid culture, and then poured in a plate (90 mm diameter). A. oryzae spores (1.5 g) were mixed with a drop of Tween 80 in 10 mL of ddH2O. Then, 200 μL of spore suspension was added and coated on the surface of the solid plate medium with a cellophane membrane. The temperature was controlled by using a biochemical incubator. Samples were collected when the mold of A. oryzae changed to yellow-green, and this mimicking fermentation lasted for about 68 h. A part of the fermentation medium was selected for the extraction of aroma compounds. Mycelia for the proteomic analysis were collected by centrifugation and ground in a mortar in the presence of liquid nitrogen following a PBS wash.
| Name | Temperature | Saline water | Culture |
|---|---|---|---|
| 0–28C-S | 28 °C | 0% | Solid-state |
| 0–28C-L | Submerged | ||
| 12–28C-S | 28 °C | 12% | Solid-state |
| 12–28C-L | Submerged | ||
| 0–16C-S | 16 °C | 0% | Solid-state |
| 0–16C-L | Submerged |
2.3. Extraction of aroma compounds
The extraction of aroma compounds from the samples was performed by the solid-phase micro-extraction (SPME) technique. The SPME trisplus automated sampler equipped with a 50/30 μm DVB/CAR on PDMS extraction fiber was employed for the extraction. The fiber was firstly aged at 250 °C for 30 min to remove impurities and contaminants and then desorbed for 10 min at 250 °C between injections to prevent cross-contamination. The sample (5 mL) and 0.9 g of NaCl were transferred into a 15 mL gas-tight glass container, and 30 μL of 2-octanol (0.1257 mg L−1 in methanol solution) was added as an internal standard, then shaken and mixed before analysis. The samples were continuously extracted with DVB/CAR on PDMS fibers for 30 min after equilibrating at 45 °C for 20 min. Each sample was extracted in triplicate.Meanwhile, samples were extracted by the liquid–liquid extraction method. The sample (100 mL) was mixed with 100 mL of dichloromethane in a flask, then 2-octanol (30 μL, 200 mg L−1) as the internal standard was added. The mixture was shaken and extracted for 30 min, and centrifuged (8000 rpm, 10 min) for emulsion removal. Then the liquid was separated by using a separating funnel.
2.4. Flavor detection using GC-MS
A gas chromatography-mass spectrometer (GC-MS) (Varian, Walnut Creek, CA, USA) was equipped with a 2010 Ultra GC, a Trisplus automated sampler and a quadrupole MS. Separation was performed with a VF-5 ms capillary column (30 m × 0.25 mm, 0.25 μm). Helium was used as a carrier gas with a constant flow rate of 1.0 mL min−1. The split ratio was 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The analytical conditions were as follows: the temperature of the column was maintained at 40 °C for 3 min, ramped to 150 °C at 4 °C min−1, held for 1 min and then increased to 250 °C at a rate of 8 °C min−1 and held for 6 min. The injection temperature was 250 °C, and the ion source temperature was set at 220 °C. The mass spectrometer was operated in the electron-impact (EI) mode. The ionization energy, detector voltage, scan range and scan rate applied for the analysis were 70 eV, 350 V, 35–500 m/z and 3.00 scans per s, respectively.
1. The analytical conditions were as follows: the temperature of the column was maintained at 40 °C for 3 min, ramped to 150 °C at 4 °C min−1, held for 1 min and then increased to 250 °C at a rate of 8 °C min−1 and held for 6 min. The injection temperature was 250 °C, and the ion source temperature was set at 220 °C. The mass spectrometer was operated in the electron-impact (EI) mode. The ionization energy, detector voltage, scan range and scan rate applied for the analysis were 70 eV, 350 V, 35–500 m/z and 3.00 scans per s, respectively.
Identification analysis was based on Kovats retention indices (RI) and tentatively identified by comparing the mass spectral data of reference standards with the standard NIST 17 library. All aroma components were confirmed based on the similarity index (SI) >80 (the highest value is 100) from the mass spectra library. The RI was sourced from the NIST Standard Reference Database. The RI was calculated using the C7–C30 n-alkane series simultaneously.
2.5. Protein preparation and trypsin digestion
Six samples were selected for the proteomic study. The sample was ground in a mortar in the presence of liquid nitrogen and was suspended in 500 μL of buffer [50 mM Tris-HCl (pH 7.8), 0.3% SDS, 200 mM DTT, 1 mM PMSF, and 1 mM EDTA]. The suspension was then sonicated for 2 min and centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 min at 4 °C. The supernatant was separated and treated with DNase I and RNase (50 μg ml−1). Then, the protein (100 μg for each sample) was digested with trypsin and desalted by using the Strata X C18 SPE column (Phenomenex).
000 rpm for 30 min at 4 °C. The supernatant was separated and treated with DNase I and RNase (50 μg ml−1). Then, the protein (100 μg for each sample) was digested with trypsin and desalted by using the Strata X C18 SPE column (Phenomenex).
2.6. TMT labeling and HPLC fractionation
The mixture was vacuum-dried and then reconstituted in 0.5 M TEAB and processed using a 6-plex TMT kit. The mixture was fractionated over 80 min into 80 fractions by high pH reverse-phase HPLC using an Agilent 300 Extend C18 column (5 μm × 4.6 mm × 250 mm). Then, the peptides were combined into 10 fractions and vacuum-dried.2.7. LC-MS/MS analysis
The samples were dissolved in FA, and separated with a reversed-phase analytical column (Acclaim PepMap RSLC, Thermo Scientific). The gradient was increased from 7% to 22% (0.1% FA in 98% ACN) over 26 min, 22% to 35% in 8 min, and increased to 80% in 3 min, then held at 80% for another 3 min, all at a constant flow rate of 300 nl min−1 on an EASY-nLC 1000 UPLC system. Then the treated peptides were analyzed by using a Q Exactive™ plus hybrid quadrupole-orbitrap mass spectrometer (ThermoFisher Scientific).The peptides were subjected to a NSI source followed by tandem mass spectrometry (MS/MS) in Q Exactive™ plus (Thermo) coupled online to the UPLC. Intact peptides were detected in the orbitrap at a resolution of 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. Peptides were selected for MS/MS using a normalized collision energy (NCE) setting of 30; ion fragments were detected in the orbitrap at a resolution of 17
000. Peptides were selected for MS/MS using a normalized collision energy (NCE) setting of 30; ion fragments were detected in the orbitrap at a resolution of 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500. A data-dependent procedure that alternated between one MS scan followed by 20 MS/MS scans was applied for the top 20 precursor ions above a threshold ion count of 10
500. A data-dependent procedure that alternated between one MS scan followed by 20 MS/MS scans was applied for the top 20 precursor ions above a threshold ion count of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 in the MS survey scan with 30.0 s dynamic exclusion. The electrospray voltage applied was 2.0 kV. Automatic gain control (AGC) was used to prevent overfilling of the orbitrap; 5E4 ions were accumulated for the generation of the MS/MS spectra. For MS scans, the m/z scan range was 350 to 1800. The fixed first mass was set as 100 m/z. The resulting MS/MS data were processed using the Mascot search engine (v.2.3.0). Tandem mass spectra were searched against the Uniprot A. oryzae database.1
000 in the MS survey scan with 30.0 s dynamic exclusion. The electrospray voltage applied was 2.0 kV. Automatic gain control (AGC) was used to prevent overfilling of the orbitrap; 5E4 ions were accumulated for the generation of the MS/MS spectra. For MS scans, the m/z scan range was 350 to 1800. The fixed first mass was set as 100 m/z. The resulting MS/MS data were processed using the Mascot search engine (v.2.3.0). Tandem mass spectra were searched against the Uniprot A. oryzae database.1
To assess the flavor formation mechanism of A. oryzae during soy sauce fermentation, we compared these proteomes in this research. A quantitative ratio over 1.5 was considered up-regulation while a quantitative ratio less than 0.67 was considered as down-regulation.
2.8. Statistical analysis
All experiments were analyzed by one-way ANOVA and performed with n = 3 biological replicates. Proteins were classified by Gene Ontology (GO) annotation into three categories: biological process, cellular compartment and molecular function. GO annotation proteome was derived from the UniProt-GOA database (http://www.ebi.ac.uk/GOA/). The Encyclopedia of Genes and Genomes (KEGG) database was used to identify enriched pathways. For each category of proteins, the InterPro database was researched and two-tailed Fisher's exact test was employed to test the enrichment of the differentially expressed proteins against all identified proteins. Cluster membership was visualized by a heat map using the “heatmap.2” function from the “gplots” R-package. Data were statistically analyzed using SPSS 17.0 (SPSS Statistics, Chicago, IL, USA) by ANOVA and Duncan's multiple comparison tests. P values less than 0.05 were considered significant.3. Results and discussion
3.1. Identification of the changing flavors under osmotic conditions
| No. | Flavors | 0–28C-L/0–16C-L | 0–28C-L/12–28C-L |
|---|---|---|---|
| 1 | Benzaldehyde | 2.1 | 2.9 |
| 2 | Phenylacetaldehyde | 3.2 | 3.0 |
| 3 | Isovaleraldehyde | 1.6 | 2.3 |
| 4 | Furfural | 2.8 | 1.9 |
| 5 | Hexanal | 2.2 | 3.7 |
| 6 | Nonanal | 1.4 | 2.4 |
| 7 | Decanal | 2.2 | — |
| 8 | Dodecanal | 1.7 | — |
| No. | Flavors | 0–16C-L/0–28C-L | 12–28C-L/0–28C-L |
|---|---|---|---|
| 1 | Ethanol | 3.5 | 1.3 |
| 2 | 2-Methyl-1-butanol | 1.5 | 1.4 |
| 3 | 2-Ethyl hexanol | 1.2 | 1.2 |
| 4 | Isoamyl alcohol | 1.5 | 1.3 |
| 5 | Isobutanol | 2.6 | 1.4 |
| 6 | Butyl acetate | 1.4 | 1.6 |
| 7 | Methyl butyrate | 1.2 | 1.3 |
| 8 | Ethyl oleate | 1.3 | 1.4 |
| 9 | Ethyl palmitate | 1.8 | 1.3 |
| 10 | Ethyl stearate | 1.9 | 1.1 |
| 11 | Ethyl linoleate | 1.6 | 1.2 |
| 12 | Isopropyl palmitate | 1.4 | 1.3 |
The contents of butyl acetate and methyl butyrate, short-chain fatty acid esters, were higher under 16 °C stress or 12% salt stress than that of the control. Short-chain fatty acid esters are important to soy sauce because of their typical fruity smell. Palmitic acid, stearic acid, oleic acid, linoleic acid, and ethanol are involved in the synthesis of long-chain fatty acid esters under low-temperature or high-salt stress. Some kinds of pyrazines and furans were unique under 16 °C stress or 12% salt stress, including 2-methyl pyrazine and 2,5-dimethyl-tetrahydrofuran which are the product of the substitution reaction between pentose sugar and amino acid with the baked fragrance of nuts (Table 2).18
Based on the results of this study, the contents of alcohols, esters, pyrazines, and furans were increased about 1.2–3.5 times, and aldehydes were decreased about 1.2–3.7 times under low-temperature stress and salt stress. It can be inferred that low-temperature stress and salt stress are beneficial to the formation of aroma flavor in soy sauce fermentation. Thus, it is beneficial to produce more pleasant soy sauce using the combination of different fermentation conditions.
3.2. Functional groups by proteomic analysis
In the present study, the proteomic technique was utilized to explore the flavor formation mechanism under osmotic conditions. Proteomic is an efficient platform technology that provides global profiles of protein expression patterns and additionally helps us gain further insight into the responses that occur during fungal adaptation to variable conditions. For the comparison of A. oryzae proteomes, we selected the conditions of low-temperature and high-salt during soy sauce fermentation and performed deep proteomic analyses. A total of 4381 proteins were quantified in all samples (90% of all identified proteins) and the average of 25% protein identifications was shared between at least two proteomes (Fig. 1, Tables S2 and S3†).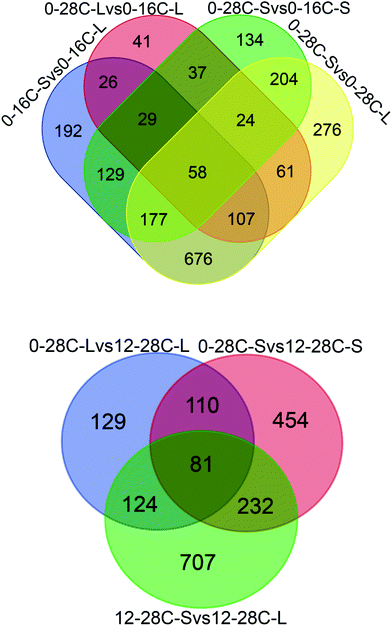 | ||
| Fig. 1 The number of proteins identified by proteomics. The overlapping region shows those proteins present in common. | ||
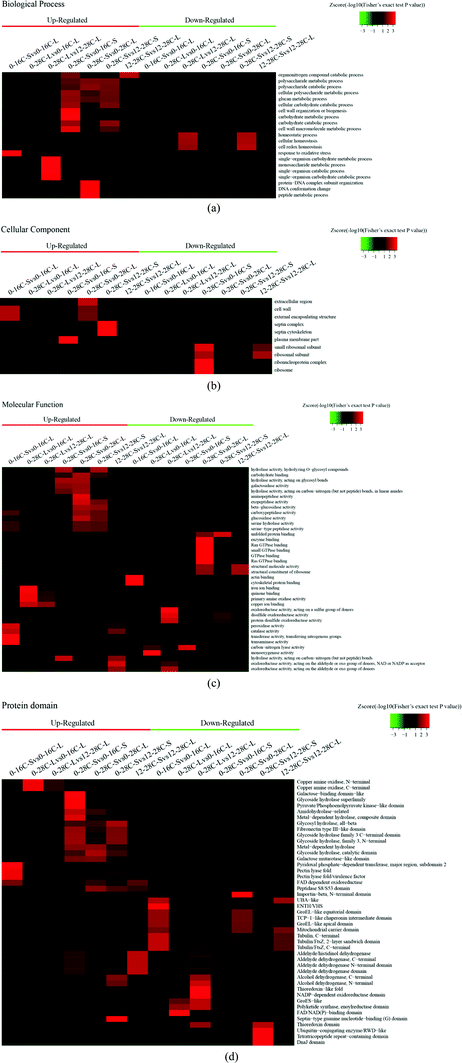
According to the Gene Ontology (GO) annotation information of these identified proteins, functional classifications of differentially quantified proteins were mainly divided into three parts: “biological process”, “cellular component”, and “molecular function”. As shown in Fig. 2, the expression levels of all the proteins detected in this research were compared as a color gradient from low (green) to high (red). The results showed that most of the differently expressed proteins in the group of “biological process” were related to metabolic processes, single-organism processes, and cellular processes that convert materials to others or regulate reactions. In the molecular function group of the proteomes, the identified proteins that created the “catalytic activity” occupied the majority of the category, suggesting that the relevant functions were important in fermentation. Beyond that, some proteins in this category related to “binding” were in the second ratio of the identified proteins, and they may help to locate proteins for binding and performing.19 Few differently expressed proteins were found in the cellular component group, indicating that proteins are stable under low-temperature and high-salt culture conditions. Differentially expressed proteins were mainly located in the cytosol, mitochondria, nucleus, and extracellular matrix according to the subcellular location of identified proteins (Fig. S1–S4†).
3.3. Flavor formation mechanism in low-temperature and high-salt
As we know, a low-temperature keeps lowering the metablolic rate for plants, animals or microorganisms, which affects the expression of enzymes. As shown in Fig. 2d, a low-temperature is unfavorable to the secretion of hydrolases, such as the glycoside hydrolase superfamily, metal-dependent hydrolase, amidohydrolase, and glycosyl hydrolase, and even the galactose binding domain and galactose mutarotase like domain. But a low-temperature of solid fermentation is beneficial to the secretion of the peptidase S8/S53 domain. The peptidase S8/S53 domain is an excellent hydrolytic enzyme which utilizes the catalytic serine residue for cleaving peptide bonds in soybean protein during soy sauce fermentation.20 The higher expression of the major facilitator superfamily (MFS) domain protein than the normal indicates that the movement of a wide range of substrates across biomembranes of A. oryzae is essential under osmotic stresses. MFS transporters target a wide spectrum of substrates including ions, carbohydrates, lipids, amino acids and peptides, nucleosides and other molecules.21
The expression trend of hydrolases under salt stress was the same as the temperature stress, but the salt stress may inhibit the expression of peptidases. We observed a significant up-regulation of the glycoside hydrolase domain by protein domain enrichment analysis (Fig. 2d). The glycoside hydrolase domain exists in a widespread group of enzymes that are classified into many glycoside hydrolase families based on amino acid sequence similarity.22 The expression levels of kinds of glycoside hydrolases at 28 °C were increased obviously than that at 16 °C or 12% salt content, which shows that A. oryzae needs more carbon sources to grow at 28 °C than at 16 °C or 12% salt content.
A. oryzae grow at 12% salt content and also activate the alcohol dehydrogenase, aldehyde dehydrogenase and PKS activities responsible for the oxidative metabolism of alcohol and aromatization. The possible limitation of the salt content is related to the contribution of aromatic flavors found in the soy sauce fermentation process.
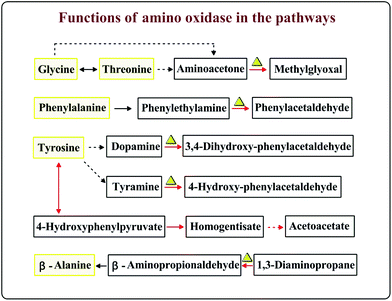
Enzymes indicated with red circles were up-regulated at 16 °C or under 12% salt condition in the amino acid metabolism (Fig. 4). The gene encoding branched-chain amino acid aminotransferase which is shown in Fig. 4 (points 1, 5, 9 and 27) is important for the production of isoamyl alcohol and isobutanol.29 Small branched alcohols are key intermediates in the branched-chain amino acid catabolism.30 2-Ethyl-1-hexanol and 2-methyl-1-butanol can be synthesized through the isoleucine pathway (Table 2).31 However, the metabolic conversion of tyrosine to acetoacetate was unique under the 12% salt condition. The accumulation of acetoacetate may contribute to the formation of acetoacetyl–CoA into the TCA cycle.32 The formation of aromatic compounds proved that A. oryzae needs more of these amino acid transformations at 16 °C or under 12% salt condition. We speculated that the metabolism of amino acids, such as leucine, valine, isoleucine, and glutamate is close related to the formation of aroma compounds under 16 °C or 12% salt condition.
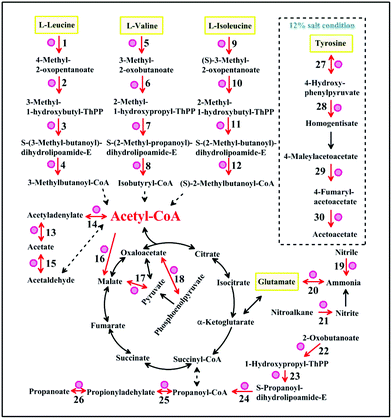 | ||
| Fig. 4 The common metabolic networks under low-temperature and salt stress. The pathway in the dashed box is unique under salt stress. Enzymes (increased) are indicated with red circles. Reaction numbers correspond to the reactions listed in Table S4.† | ||
In addition to the above, the fungi and bacterial community diversity with different enzyme activities can be significantly affected by osmotic stresses during the natural fermentation of soy sauce.33Zygosaccharomyces and Saccharomycetales would be increased and A. oryzae would be decreased. Then the total volatile flavors of soy sauce would be changed dramatically.34
4. Conclusion
It is widely accepted that A. oryzae plays an important role in flavor formation during soy sauce fermentation. Moreover, this study demonstrated that the fermentation conditions of A. oryzae cannot be neglected. Low-temperature is beneficial for the secretion of the peptidase S8/S53 domain. Low-temperature and high-salt conditions will stimulate A. oryzae to produce more unique aromatic flavors, such as esters, pyrazines, and furans, which demonstrated the importance of technological progress in the soy sauce industry. At the same time, we realized that more technological innovations are needed to be explored. Particularly the microbial diversity and the interactions between bacteria and A. oryzae during soy sauce fermentation are unclear and need to be studied soon.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31972194), the Open Project Program of State Key Laboratory of Food Nutrition and Safety, Tianjin University of Science & Technology (No. SKLFNS-KF-201816), the Foundation (No. gxb201905) of Demonstration Center of Food Quality and Safety Testing Technology, Tianjin University of Science and Technology, P. R. China and Shanxi Province Key R&D Plan (No. 201703D211001-06-02).References
- G. Zhao, L. L. Ding, Z. H. Pan, D. H. Kong, H. Hadiatullah and Z. C. Fan, Proteinase and glycoside hydrolase production is enhanced in solid-state fermentation by manipulating the carbon and nitrogen fluxes in Aspergillus oryzae, Food Chem., 2019, 271, 606–613 CrossRef CAS PubMed.
- P. V. P. Devanthi and K. Gkatzionis, Soy sauce fermentation: Microorganisms, aroma formation, and process modification, Food Res. Int., 2019, 120, 364–374 CrossRef CAS PubMed.
- Y. Feng, G. Su, H. Zhao, Y. Cai, C. Cui, D. Sun-Waterhouse and M. Zhao, Characterisation of aroma profiles of commercial soy sauce by odour activity value and omission test, Food Chem., 2015, 167, 220–228 CrossRef CAS PubMed.
- D. R. Xiao, R. S. Liu, L. He, H. M. Li, Y. L. Tang, X. H. Liang, T. Chen and Y. J. Tang, Aroma improvement by repeated freeze-thaw treatment during Tuber melanosporum fermentation, Sci. Rep., 2015, 5, 17120 CrossRef CAS PubMed.
- L. Zhang, R. Zhou, R. Cui, J. Huang and C. Wu, Characterizing Soy Sauce Moromi Manufactured by high-salt dilute-state and low-salt solid-state fermentation using Multiphase Analyzing Methods, J. Food Sci., 2016, 81, C2639–C2646 CrossRef CAS PubMed.
- N. X. Hoang, S. Ferng, C. H. Ting, W. H. Huang, Y. Y. Chiou and C. K. Hsu, Optimizing the initial moromi fermentation conditions to improve the quality of soy sauce, LWT–Food Sci. Technol., 2016, 74, 242–250 CrossRef.
- R. Harada, M. Yuzuki, K. Ito, K. Shiga, T. Bamba and E. Fukusaki, Microbe participation in aroma production during soy sauce fermentation, J. Biosci. Bioeng., 2018, 125, 688–694 CrossRef CAS PubMed.
- X. Gao, J. Zhang, E. Liu, M. Yang, S. Chen, F. Hu, H. Ma, Z. Liu and X. Yu, Enhancing the taste of raw soy sauce using low intensity ultrasound treatment during moromi fermentation, Food Chem., 2019, 298, 124928 CrossRef CAS PubMed.
- Y. Jing, J. Wan, I. Angelidaki, S. Zhang and G. Luo, iTRAQ quantitative proteomic analysis reveals the pathways for methanation of propionate facilitated by magnetite, Water Res., 2017, 108, 212–221 CrossRef CAS PubMed.
- J. Y. Liu, J. L. Men, M. C. Chang, C. P. Feng and L. G. Yuan, iTRAQ-based quantitative proteome revealed metabolic changes of Flammulina velutipes mycelia in response to cold stress, J. Proteomics, 2017, 156, 75–84 CrossRef CAS PubMed.
- G. Zhao, L. Hou, M. Lu, Y. Wei, B. Zeng, C. Wang and X. Cao, Construction of the mutant strain in Aspergillus oryzae 3.042 for abundant proteinase production by the N+ ion implantation mutagenesis, Int. J. Food Sci. Technol., 2012, 47, 504–510 CrossRef CAS.
- X. Meng, Q. Wu, L. Wang, D. Q. Wang, L. Q. Chen and Y. Xu, Improving flavor metabolism of Saccharomyces cerevisiae by mixed culture with Bacillus licheniformis, for Chinese Maotai-flavor liquor making, J. Ind. Microbiol. Biotechnol., 2015, 42, 1601–1608 CrossRef CAS PubMed.
- H. Y. Tian, J. Zhang, B. G. Sun, M. Q. Huang, J. R. Li and X. X. Han, Preparation of natural isovaleraldehyde by the Maillard reaction, Chin. Chem. Lett., 2007, 18, 1049–1052 CrossRef CAS.
- Y. Zhang, H. Song, P. Li, J. Yao and J. Xiong, Determination of potential off-flavour in yeast extract, LWT–Food Sci. Technol., 2017, 82, 184–191 CrossRef CAS.
- D. W. Chen, D. P. Balagiannis and J. K. Parker, Use of egg yolk phospholipids to generate chicken meat odorants, Food Chem., 2019, 286, 71–77 CrossRef CAS PubMed.
- Y. Zhang, S. Lane, J. M. Chen, S. K. Hammer, J. Luttinger, L. Yang, Y.-S. Jin and J. L. Avalos, Xylose utilization stimulates mitochondrial production of isobutanol and 2-methyl-1-butanol in Saccharomyces cerevisiae, Biotechnol. Biofuels, 2019, 12, 223 CrossRef PubMed.
- M. J. Kim, H. S. Kwak, H. Y. Jung and S. S. Kim, Microbial communities related to sensory attributes in Korean fermented soy bean paste (doenjang), Food Res. Int., 2016, 89, 724–732 CrossRef CAS PubMed.
- S. J. Lee and B. Ahn, Comparison of volatile components in fermented soybean pastes using simultaneous distillation and extraction (SDE) with sensory characterisation, Food Chem., 2009, 114, 600–609 CrossRef CAS.
- C. Zhang, W. Zheng, P. L. Freddolino and Y. Zhang, MetaGO: Predicting Gene Ontology of non-homologous proteins through low-resolution protein structure prediction and protein–protein network mapping, J. Mol. Biol., 2018, 430, 2256–2265 CrossRef CAS PubMed.
- P. P. Singh, D. Srivastava, A. Jaiswar and A. Adholeya, Effector proteins of Rhizophagus proliferus: conserved protein domains may play a role in host-specific interaction with different plant species, Braz. J. Microbiol., 2019, 50, 593–601 CrossRef PubMed.
- N. Yan, Structural advances for the major facilitator superfamily (MFS) transporters, Trends Biochem. Sci., 2013, 38, 151–159 CrossRef CAS PubMed.
- M. Hidaka, S. Fushinobu, N. Ohtsu, H. Motoshima, H. Matsuzawa, H. Shoun and T. Wakagi, Trimeric crystal structure of the glycoside hydrolase family 42 beta-galactosidase from Thermus thermophilus A4 and the structure of its complex with galactose, J. Mol. Biol., 2002, 322, 79–91 CrossRef CAS PubMed.
- C. Peter, J. Laliberté, J. Beaudoin and S. Labbé, Copper distributed by Atx1 is available to copper amine oxidase 1 in Schizosaccharomyces pombe, Eukaryotic Cell, 2008, 7, 1781–1794 CrossRef CAS PubMed.
- V. J. Klema, C. J. Solheid, J. P. Klinman and C. M. Wilmot, Structural analysis of aliphatic versus aromatic substrate specificity in a copper amine oxidase from Hansenula polymorpha, Biochemistry, 2013, 52, 2291–2301 CrossRef CAS PubMed.
- B. J. Brazeau, B. J. Johnson and C. M. Wilmot, Copper-containing amine oxidases. Biogenesis and catalysis; a structural perspective, Arch. Biochem. Biophys., 2004, 428, 22–31 CrossRef CAS PubMed.
- Y. Seshime, P. R. Juvvadi, K. Kitamoto, Y. Ebizuka and I. Fujii, Identification of csypyrone B1 as the novel product of Aspergillus oryzae type III polyketide synthase CsyB, Bioorg. Med. Chem., 2010, 18, 4542–4546 CrossRef CAS PubMed.
- J. M. Clomburg, S. C. Contreras, A. Chou, J. B. Siegel and R. Gonzalez, Combination of type II fatty acid biosynthesis enzymes and thiolases supports a functional β-oxidation reversal, Metab. Eng., 2017, 45, 11–19 CrossRef PubMed.
- A. L. Carroll, S. H. Desai and S. Atsumi, Microbial production of scent and flavor compounds, Curr. Opin. Biotechnol., 2016, 37, 8–15 CrossRef CAS PubMed.
- W. Li, S. J. Chen, J. H. Wang, C. Y. Zhang, Y. Shi, X. W. Guo, Y. F. Chen and D. G. Xiao, Genetic engineering to alter carbon flux for various higher alcohol productions by Saccharomyces cerevisiae for Chinese Baijiu fermentation, Appl. Microbiol. Biotechnol., 2018, 102, 1783–1795 CrossRef CAS PubMed.
- E. Danilo, R. Federica, N. Antonella, F. Pasquale and V. Francesco, Mesophilic and psychrotrophic bacteria from meat and their spoilage potential in vitro and in beef, Appl. Environ. Microbiol., 2009, 75, 1990–2001 CrossRef PubMed.
- T. Ji, M. Kang and B. K. Baik, Volatile Organic Compounds of Whole Grain Soft Winter Wheat, Cereal Chem., 2017, 94, 594–601 CrossRef CAS.
- D. Abdurrachim, C. C. Woo, X. Q. Teo, W. X. Chan, G. K. Radda and P. T. H. Lee, A new hyperpolarized (13)C ketone body probe reveals an increase in acetoacetate utilization in the diabetic rat heart, Sci. Rep., 2019, 9, 5532 CrossRef PubMed.
- E. Y. Son, S. M. Lee, M. Kim, J. A. Seo and Y. S. Kim, Comparison of volatile and non-volatile metabolites in rice wine fermented by Koji inoculated with Saccharomycopsis fibuligera and Aspergillus oryzae, Food Res. Int., 2018, 109, 596–605 CrossRef CAS PubMed.
- R. Liang, J. Huang, X. Wu, Y. Xu, J. Fan, C. Wu, Y. Jin and R. Zhou, Characterizing the metabolites and the microbial communities of the soy sauce mash affected by temperature and hydrostatic pressure, Food Res. Int., 2019, 123, 801–808 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: The volatile flavors, proteome, the biochemical reactions related to the carbon and nitrogen metabolism and the subcellular location of proteins. See DOI: 10.1039/c9fo02314c |
| This journal is © The Royal Society of Chemistry 2020 |