Anti-inflammatory action of two novel peptides derived from peanut worms (Sipunculus nudus) in lipopolysaccharide-induced RAW264.7 macrophages
Papassara
Sangtanoo
 a,
Piroonporn
Srimongkol
a,
Piroonporn
Srimongkol
 a,
Tanatorn
Saisavoey
a,
Tanatorn
Saisavoey
 a,
Onrapak
Reamtong
a,
Onrapak
Reamtong
 b and
Aphichart
Karnchanatat
b and
Aphichart
Karnchanatat
 *a
*a
aResearch Unit in Bioconversion/Bioseparation for Value-Added Chemical Production, Institute of Biotechnology and Genetic Engineering, Chulalongkorn University, 254 Phayathai Road, Pathumwan, Bangkok 10330, Thailand. E-mail: Papassara.S@chula.ac.th; Piroonporn.S@chula.ac.th; Tanatorn.S@chula.ac.th; Aphichart.K@chula.ac.th
bDepartment of Molecular Tropical Medicine and Genetics, Faculty of Tropical Medicine, Mahidol University, 420/6 Ratchawithi Road, Ratchathewi, Bangkok 10400, Thailand. E-mail: onrapak.rea@mahidol.edu
First published on 11th December 2019
Abstract
Peanut worm (Sipunculus nudus Linn.) protein was hydrolyzed by three proteases, and NO scavenging activity of the protein hydrolysates was evaluated. The hydrolysate obtained using Alcalase® showed the highest NO scavenging activity. This hydrolysate was fractionated using 10-, 5-, and 3 kDa molecular weight cut-off membranes, and the lowest MW fraction (<3 kDa) exhibited the highest NO scavenging activity. The <3 kDa fraction was further purified by gel filtration and high-performance liquid chromatographies. The peptides in the HPLC fraction with the strongest anti-NO activity were identified by quadrupole-time-of-flight mass spectrometry as LSPLLAAH (821.48 Da) and TVNLAYY (843.42 Da). Both peptides and the corresponding pure synthetic peptides inhibited NO production by RAW 264.7 macrophages without cytotoxicity. Quantitative real-time RT-PCR analysis showed that peptides LSPLLAAH and TVNLAYY reduced expression of proinflammatory cytokine genes iNOS, IL-6, TNF-α, and COX-2 in RAW 264.7 macrophages, suggesting that these peptides are novel anti-inflammatory candidates.
1. Introduction
Inflammation can be explained as a host defense process which responds to injuries to tissue, the threat of exposure to toxic chemicals, and the presence of pathogens. The typical symptoms of inflammation are swelling, reddening of the skin, pain, and heat, which arise due to the influence of cytokines upon the blood vessels, and the condition can be categorized as either chronic or acute.1 When the body is exposed to dangerous stimuli, the immediate response is acute inflammation, and this occurs when the movement from the blood into the areas of injury of plasma and leukocytes, including neutrophils and macrophages, is significantly increased. When inflammation is chronic, and endures over longer periods, the consequences can be problematic as the pathogenesis of a number of conditions can be affected to the detriment of the patient. Such conditions include atherosclerosis, cancer, arthritis, and ischemic heart disease.2,3 When activated by pathogens and pro-inflammatory cytokines, the macrophages subsequently trigger the generation of a number of pro-inflammatory mediators which include cytokines, chemokines, prostaglandins, and matrix metalloproteinases. Also produced are ROS (reactive oxygen species) and NO (nitric oxide).4Nitric oxide is able to permeate membranes, and plays a significant role in the regulation of a wide range of biological and physiological processes.5,6 Nitric oxide synthases (NOSs) support the synthesis of NO from L-arginine, with three different NOS types having been determined. Two of these are the constitutively expressed neural NOS (nNOS) and endothelial NOS (eNOS) which are able to generate NO in low concentrations for the purpose of mediating tissue homeostasis. In particular, blood vessel tone is regulated by eNOS derived NO, while nNOS derived NO serves as a neurotransmitter and neuromodulator. The final form of NO, which is the inducible NOS (iNOS), can offer protection against pathogens which cause infection and can control immunity. However, when NO is produced in excessive quantities, it tends to undergo oxidization to form more reactive nitrogen species. These can in turn cause the death of the cells in the nearby tissues as well acting to destroy tissue homeostasis.6,7 This process typically occurs in pathogenic conditions, and the excessive production of NO in inflammatory cells is usually mediated by iNOS. In order to treat inflammation, it may therefore prove effective to focus upon the regulation of iNOS.8
In cases where the initial inflammatory response is not treated, the condition may develop into both chronic and acute inflammatory disease. To avoid this scenario, the use of anti-inflammatory drugs can be important to support the immune response. Research has shown that NSAIDs (non-steroidal anti-inflammatory drugs) can also provide further benefits in the form of their ability to prevent gastrointestinal system cancers. This enhances the overall potential of this kind of anti-inflammatory treatment.9,10 Recently, however, interest is growing in natural compounds, since these address the shortcomings of NSAIDs, which can lead to cardiovascular problems, or damage to the renal or gastrointestinal systems.11 Many studies have shown that proteins and peptides offer anti-inflammatory properties. Bioactive peptides which are produced through the enzymatic digestion of different proteins obtained from plants and animals can serve as anti-inflammatory agents.12
The peanut worms (Sipunculus nudus Linnaeus) of the phylum Sipuncula, are non-segmented coelom worms which inhabit the marine environment, often burrowing into sand, or taking shelter in empty shells or within coral.13 They are edible and considered a delicacy in the south-east of China. Furthermore, it is claimed that they are able to relieve a number of conditions including hypertension, neurosis, coughing with dyspnea, and excessive urination. Technological advances have allowed research to reveal that peanut worms may be used as a form of seafood which offers further advantages through its bioactivities, which have been shown to include anti-hypoxia, immunomodulatory, antithrombotic, antitumor, antiviral, anti-inflammatory and antinociceptive properties.14 The molecules which govern such mechanisms have not, however, yet been identified.
Peanut worms have been the subject of numerous studies examining their metabolism, reproduction, physiology, and aquaculture. They are important as a protein source in certain regions of Asia.15 This study seeks to achieve the purification of anti-inflammatory peptides which might be obtained from peanut worms via proteases, in order to characterize their amino acid sequences with electrospray liquid chromatography quadrupole-time-of-flight mass spectrometry (Q-TOF-ESI-MS/MS). Once identified, synthesis of the peptides was carried out in order to assess the anti-inflammatory capabilities in lipopolysaccharide (LPS)-stimulated RAW 264.7 mouse macrophages. This research study might serve to encourage the use of hydrolyzed peptides derived from peanut worm proteins to serve as the bioactive components of functional foods.
2. Materials and methods
2.1 Peanut worm samples
Peanut worms of size 6 ± 2 mm in diameter and 10 ± 2 cm in length were obtained from the Sichang Marine Science Research and Training Station on Sichang Island, Chonburi Province, Thailand, under the auspices of Chulalongkorn University. Prior to anesthetizing the peanut works ice for 15 minutes, cleaning was carried out under tap water before homogenization using a mixer. The fat was then removed from the resulting mixture using a technique explained by Prakot et al.16 The dry homogenate was then placed in a mixer for grinding to form a fine powder, before sieving through a 150 inch particle size mesh. The resulting product is known as peanut worm powder (PWP). In this case, the PWP was then stored in a desiccator until required.2.2 Determination of the PWP amino acid composition
The amino acid content of the PWP was assessed by AOAC 994.12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 17 and 988.15
17 and 988.15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 18 methods. In this technique, 25 mg of PWP is dissolved in a test tube containing 5 mL of 6 M HCl and then placed in a heating block for 24 h at 110 °C in order that the amino acids can be released. The internal standard (10 μL of 2.5 mM L-amino-n-butyric acid in 0.1 M HCl) is then added before dilution of the solution to 250 μL using deionized water. This diluted solution is then placed in a heating block for 10 min at 55 °C. Reversed-phase high-performance liquid chromatography (RP-HPLC) is then used for the analysis. A Hypersil GOLD column C18 (4.6 mm × 150 mm, 3 μM) is used for elution with a sodium acetate buffer (pH 4.90) and 60% (v/v) ACN at 0.3 mL min−1. Determination of the amino acid content was performed by ALS Laboratory Group (Thailand) Co., Ltd, Suan Luang, Bangkok, Thailand.
18 methods. In this technique, 25 mg of PWP is dissolved in a test tube containing 5 mL of 6 M HCl and then placed in a heating block for 24 h at 110 °C in order that the amino acids can be released. The internal standard (10 μL of 2.5 mM L-amino-n-butyric acid in 0.1 M HCl) is then added before dilution of the solution to 250 μL using deionized water. This diluted solution is then placed in a heating block for 10 min at 55 °C. Reversed-phase high-performance liquid chromatography (RP-HPLC) is then used for the analysis. A Hypersil GOLD column C18 (4.6 mm × 150 mm, 3 μM) is used for elution with a sodium acetate buffer (pH 4.90) and 60% (v/v) ACN at 0.3 mL min−1. Determination of the amino acid content was performed by ALS Laboratory Group (Thailand) Co., Ltd, Suan Luang, Bangkok, Thailand.
2.3 Preparation of PWP hydrolysates
The PWP (0.5 g) is first of all stirred overnight at 4 °C in 10 mL of 20 mM phosphate buffer (pH 7.2) with 150 mM NaCl (PBS) after the addition of 0 (as the control) and 5 wt% of Alcalase, Flavourzyme or Neutrase in a shaker (150 rpm) for 4 h at 50 °C. The pH value was then fine-tuned to 8 in the case of Alcalase and 7 for the Flavourzyme and Neutrase. The reaction was then stopped by heating to a temperature of 80–90 °C for 10–15 min, before placement in a centrifugation for 15 min at 9880g. The resulting supernatant was then collected and stored at a temperature of 4 °C. The experiment was carried out in triplicate.2.4 NO scavenging activity assay
Using a modified technique proposed by Saisavoey et al.,19 Inkanuwat et al.,20 and Suttisuwan et al.21 an estimation of the assay of NO radical scavenging was obtained. In this case, peptide hydrolysates (5 μL) underwent mixing with 10 mM SNP in PBS before incubation for 150 min at room temperature. The addition of 100 μL of Griess reagent (0.33% sulfanilamide in 5% phosphoric acid) was followed by further incubation of 5 min. Incubation of this mixture following the further addition of 100 μL of 12.5 mM NED was carried out for 30 min at room temperature. The absorbance reading was 540 nm, obtained through the use of a microplate reader (model Multiskan GO, Thermo Fisher Scientific Inc., San Jose, CA). Positive regulation has been achieved using curcumin. The NO inhibition percentage is calculated as follows:in which Abs control denotes the control sample absorbance in the absence of PWP. Abs sample represents the PWP absorbance with NED, and Abs background denotes PWP absorbance in the absence of NEW. Finally, Abs blank shows the deionized water absorbance.
The GraphPad Crystal v. 6.01 program for Windows (GraphPad Program Inc., San Diego, CA) was used to calculate the 50% inhibition (IC50) of the hydrolysate concentration by plotting the graph to show the inhibition proportion against the PWP hydrolysate concentration. Bradford's method22 was used for the assay determining the PWP hydrolysate concentration, making use of bovine albumin serum as standard. In order to generate the calibration curve, the graph of absorbance at 595 nm was plotted against the concentration of protein recorded at the levels of 5–20 μg mL−1.
2.5 NO-inhibitory peptide isolation and enrichment
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0% (v/v) A
0% (v/v) A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B, declining to 90
B, declining to 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (v/v) A
10 (v/v) A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B over a period of 18 min, to the ratio of 65
B over a period of 18 min, to the ratio of 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35 (v/v) A
35 (v/v) A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B over a period of 30 min, before the final ratio of 55
B over a period of 30 min, before the final ratio of 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45 (v/v) A
45 (v/v) A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B over a period of 40 min where the flow rate was 0.7 mL min−1. Mobile phase A comprised 0.1% (v/v) trifluoroacetic acid while B comprised 70% (v/v) acetonitrile with 0.05% (v/v) trifluoroacetic acid. ChromQuest Software (Thermo Fisher Scientific) was used to carry out the chromatographic analyses. The peptide peaks were monitored at 280 nm. Harvesting of the principal antioxidant peptide peaks was followed by analysis using Q-TOF-ESI-MS/MS.
B over a period of 40 min where the flow rate was 0.7 mL min−1. Mobile phase A comprised 0.1% (v/v) trifluoroacetic acid while B comprised 70% (v/v) acetonitrile with 0.05% (v/v) trifluoroacetic acid. ChromQuest Software (Thermo Fisher Scientific) was used to carry out the chromatographic analyses. The peptide peaks were monitored at 280 nm. Harvesting of the principal antioxidant peptide peaks was followed by analysis using Q-TOF-ESI-MS/MS.
2.6 Q-TOF-ESI-MS/MS and de novo peptide sequencing
RP-HPLC produced a fraction which presented significant NO radical scavenging activity in addition to adequate yield (fraction F1-2); this fraction was identified using ESI-Q-TOF-MS/MS when the tool was calibrated to identify those peptide chains falling within the mass range of m/z 50–25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (Model Amazon SL, Bruker, Germany). Evaluation using de novo sequencing and Mascot was employed for all of the available ESI-Q-TOF-MS/MS data, before comparisons were drawn between the sequences obtained and those of the NCBI database using BLASTp.
000 (Model Amazon SL, Bruker, Germany). Evaluation using de novo sequencing and Mascot was employed for all of the available ESI-Q-TOF-MS/MS data, before comparisons were drawn between the sequences obtained and those of the NCBI database using BLASTp.
2.7 Peptide synthesis
Those peptides for which the sequences could be obtained through MS/MS were synthesized by a 433A Synergy solid phase peptide synthesizer (Applied Biosystems, CA). Synthetic peptides such as Leu–Ser–Pro–Leu–Leu–Ala–Ala–His (LSPLLAAH), and Thr–Val–Asn–Leu–Ala–Tyr–Tyr (TVNLAYY) could then undergo verification using analytical mass spectrometry (model Finnigan™ LXQ™, Thermo Fisher Scientific Inc., MA) which was coupled with a Surveyor HPLC. The NO radical scavenging activity of these pure synthesized peptides could then be compared to the activity of the peptides acquired using RP-HPLC fractionation.2.8 Cell culture
Complete medium (CM; DMEM supplemented with 10% [v/v] FBS, 100 U mL−1 penicillin G, 0.4 mg mL−1 streptomycin sulfate, 1% [w/v] sodium pyruvate, and 1% [w/v] HEPES) was used to maintain the RAW 264.7 cell line at a temperature of 37 °C in a humidified 5% (v/v) CO2 atmosphere. Routine maintenance in culture requires that the cells be seeded in non-tissue culture treated dishes with around 10% confluency growing to reach around 80% confluency19–21 in the course of a two-day process. Aspiration of the medium then took place, before rinsing the cells with PBS. The cells were then dislodged using a spatula for gentle scraping, and gathered using a centrifuge (Hettich, Tuttilingen, Germany) at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 min at a temperature of 4 °C.
000g for 5 min at a temperature of 4 °C.
2.9 Pre-treatment of RAW 264.7 macrophages
The RAW 264.7 macrophages underwent seeding in 96-well plates (104 cells per well) in 100 μL CM prior to overnight incubation at 37 °C under a humidified 5% CO2 atmosphere. CM alone was used as a negative control to replace the medium, while in other cases, CM was used which contained varying concentrations of the peptides LSPLLAAH or TVNLAYY, or CM which contained budesonide (2.5 μg mL−1) to serve as the positive control, which was incubated for 1 h. The addition of 100 ng mL−1 LPS before incubation for 12 h served to stimulate NO production.2.10 MTT assay to measure cell viability/proliferation
A slightly modified approach described Saisavoey et al.,19 Inkanuwat et al.,20 and Suttisuwan et al.21 were used to determine the cytotoxicity of peptides LSPLLAAH and TVNLAYY. Under this technique, RAW 264.7 macrophages were placed in 96-well plates (104 cells per well) before incubation for 24 h at a temperature of 37 °C with 5% (v/v) CO2. The cells were then handled using separate test sample (peptide) and LPS levels. Supplementation of each of the wells then took place using 100 μL of a 5 mg mL−1 MTT solution (in PBS). The cells were incubated at a temperature of 37 °C for 4 h in a humidified 5% CO2 atmosphere. Removal of the medium followed, before the addition of DMSO in order to solubilize the formazan crystals which formed at 100 μL per well, before the eventual evaluation of A540 nm.2.11 Analysis of NO reduction using RAW 264.7 macrophages
The Griess reaction serves to determine the extent of NO production through the measurement of NO production in CM. Incubation of the RAW 264.7 macrophages was carried out at varying concentrations with the test sample (control 0 mM). Subsequently, the Griess reagent was added in quantities of 50 μL to 50 μL of supernatant culture placed in a 96-well plate before incubation at room temperature for 10 min. The a further 50 μL of 12.5 mM of NED solution was added, followed by 10 min of incubation. A microplate reader was then employed to take measurements at A540 nm.2.12 Detection of iNOS, tumor necrosis factor α (TNF-α), interleukin 6 (IL-6), and cyclooxygenase-2 (COX-2) mRNA levels through real-time quantitative PCR (qRT-PCR) analysis
The pre-treatment of RAW 264.7 macrophages was carried out at various levels using solvent as the negative control, or synthesized peptide or 2.5 μg mL−1 budesonide to serve as the positive control. This process took one hour. Stimulation of these pre-treated cells then took place through the addition of 100 ng mL−1 LPS and 12 h incubation. The total RNA was then extracted through a MasterPure™ complete DNA and RNA purification kit (Epicentre; Lucigen, under LGC, Biosearch Technologies, Middleton, WI) by adhering to the guidelines of the manufacturer. Measurements of the RNA concentration were taken at 260 nm by NanoDrop 2000 UV-Vis spectrophotometer (Thermo Fisher Scientific, Inc.). The process of reverse transcription of the total RNA (1 μg) involved oligo-dT primers employing a precision nanoScript II reverse transcription kit (PrimerDesign, Camberley, UK) on the basis of the guidelines set by the manufacturer.For the purposes of qRT-PCR analysis, a PCR mixture comprising 1 μL of cDNA, 1 μL of each primer (10 mM), 7 μL of ultrapure water, and 10 μL of 2× qPCRBIO SyGreen Mix (PCR Biosystems Ltd, London, UK) was used. The reactions of qRT-PCR which were carried out separately in the case of each different gene underwent amplification by MyGo Pro® Real time PCR apparatus (IT-IS International Ltd, Stokesley, UK) before thermal treatment for 2 min at a temperature of 95 °C. The next step involved 40 cycles of 10 s at 95 °C, 20 s at 68 °C (or 60 °C in the case of TNF-α and IL-6) for 20 s, and 30 s at 72 °C, before finally melting at 55–95 °C for 1 min. The RT-PCR was carried out with selective primers for mouse β-actin (5′-GATCAAGATCATTGCTCCTCCTG-3′ and 5′-CGCAGCTCAGTAACAGTCCG-3′), iNOS (5′-CGGCAAACATGACTTCAGGC-3′ and 5′-TAGGTCGATGCACAACTGGG-3′), TNF-α (5′-GGGCAGGTCTACTTTGGAGTCA-3′ and 5′-ACAGACTGGGGGCTCTGAGG-3′), IL-6 (5′-CTCTCTGCAAGAGACTTCCATCC-3′ and 5′-ACAGGTCTGTTGGGAGTGGTATC-3′), and COX-2 (5′-CTGACCCCCAAGGCTCAAAT-3′ and 5′-AAGTCCACTCCATGGCCCAG-3′). β-Actin served as the internal reference gene. The Ct (threshold cycle) value was used as a means of determining the relative gene expression level through calculations based on relative gene expression = 2−ΔΔCt, in which ΔΔCt represents the rise in the gene threshold cycle. When the score is 1, this is indicative of no change, but when the score exceeds 1, this represents an increase in gene expression. Scores below 1 indicate the opposite, that gene expression has decreased.
2.13 Statistical analysis
In this study, the numerical data are presented in the form of mean ± standard deviation, with each test performed in triplicate. Analysis of variance was carried out along with Duncan's multiple range post hoc tests. The significance level was set to be P < 0.05, and all analyses were completed with SPSS statistical software.3. Results and discussion
3.1 Amino acid content of peanut worms
The amino acid components and their arrangement within the peptide are believed to be intimately linked to the bioactivity of a peptide. In terms of physicochemical properties, operative properties, and bioactivities, the hydrophobicity of amino acids, peptides, and proteins have a significant part to play. The levels of eighteen amino acids in peanut worms are exhibited in the data of this research, with alanine, arginine, aspartic acid, glutamic acid, glycine, leucine, phenylalanine, proline, tryptophan and tyrosine comprising the most abundant amino acids (Table 1). Ordinarily, glycine occurs equivalently in the α-chain of collagen molecules in recurring motif Gly–X–Y (where X is primarily proline and Y is primarily hydroxyproline). Similarly, collagen molecules typically comprise alanine, proline and hydroxyproline, while cystine and tyrosine are rare.23,24 Comparatively high content of hydrophobic amino acids and positively charged amino acids comprise PWP. A significant impact on the anti-inflammatory activities of peptides was reported by hydrophobic amino acids (such as leucine, phenylalanine, tryptophan, and tyrosine) and positively charged amino acids.25 Accordingly, the authors hypothesized that protease hydrolysis could be employed to obtain anti-inflammatory peptides from PWP.| Amino acid profile | % (w/w) |
|---|---|
| The results are presented as the mean ± SEM (n = 3). | |
| Alanine (Ala) | 1.40 ± 0.056 |
| Arginine (Arg) | 1.41 ± 0.018 |
| Aspartic acid (Asp) | 1.32 ± 0.015 |
| Cysteine (Cys) | 0.10 ± 0.051 |
| Glutamic acid (Gln) | 2.52 ± 0.009 |
| Glycine (Gly) | 1.22 ± 0.025 |
| Histidine (His) | 0.25 ± 0.010 |
| Isoleucine (Ile) | 0.56 ± 0.003 |
| Leucine (Leu) | 1.17 ± 0.007 |
| Lysine (Lys) | 1.04 ± 0.019 |
| Methionine (Met) | 0.40 ± 0.020 |
| Phenylalanine (Phe) | 1.46 ± 0.125 |
| Proline (Pro) | 1.55 ± 0.035 |
| Serine (Ser) | 0.53 ± 0.019 |
| Threonine (Thr) | 0.59 ± 0.014 |
| Tryptophan (Trp) | 1.09 ± 0.024 |
| Tyrosine (Tyr) | 1.47 ± 0.084 |
| Valine (Val) | 0.58± 0.009 |
| Total | 14.66 ± 0.285 |
3.2 NO radical scavenging activity of PWP hydrolysate
To separately hydrolyze PWP, three enzymes with distinct specificities were used. PWP hydrolysate obtained with 5% Alcalase comprised antioxidant peptides with IC50 value for NO radical generation of 1.21 ± 0.02 μg mL−1 (Table 2). As well to search for antioxidant peptides, Alcalase has been used in many bioactive peptide isolation processes. It has likewise produced results to meet the anticipations of different researchers. Alcalase® 2.4L hydrolysates displayed immense potential in 2,2-diphenyl-1-picrylhydrazyl (DPPH) radical scavenging aptitude and the constraint of lipid peroxidation, as revealed in work of Bougatef et al.26 on sardine derivatives. The Alcalase hydrolysates showed the highest reducing power compared to those generated using proteases from Bacillus licheniformis NH1 (1.98) and Aspergillus clavatus, as presented by the results. Lee et al.27 tested the hydrolysate from short-neck clam (Ruditapes philippinarum) for NO inhibitory activity by Alcalase-derived hydrolysate, which showed the highest reduction.| Enzymes | NO scavenging activity (IC50) μg mL−1 |
|---|---|
| Values followed by the different letter in the same column are significantly different (p < 0.05). | |
| Neutrase | 19.93 ± 0.12c |
| Flavourzyme | 2.07 ± 0.04b |
| Alcalase | 1.21 ± 0.02a |
3.3 Purification of NO-inhibitory peptides from PWP
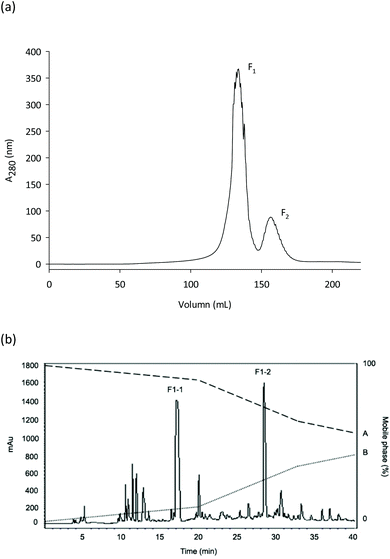
The 5% Alcalase hydrolysate was filtered through 10-, 5-, and 3 kDa membrane filters in order to cleanse NO-inhibitory peptides from PWP. 18.40 ± 0.18, 3.54 ± 0.22, 3.27 ± 0.06, and 2.02 ± 0.04 μg mL−1 comprised the IC50 values for the acquired >10, 5–10, 3–5, and <3 kDa fractions, respectively. Therefore, Sephadex G-10 gel filtration chromatography was used on the <3 kDa fraction and eluted with purified water. The Sephadex G-10 column (fractions F1 and F2) was used to individualize the peptides in the <3 kDa fraction. The fraction F1 had NO inhibitory activity with IC50 value 54.95 ± 1.20 μg mL−1, but the yield of fraction F2 was too low to conclude an IC50 value, as shown by calculation of NO inhibitory activity (Fig. 1a). Low-molecular-weight fractions filtered from protein hydrolysates have high antioxidant, antihypertensive and immuno-modulatory activities, as demonstrated by comparable studies. Because they are less prone to gastrointestinal hydrolysis, these peptides are better for acting in a physiological capacity compared to longer peptides. Further, antioxidant peptides resulting from foods are deemed benign and nourishing compounds that are inexpensive, display robust activity, and are easily absorbed.28Employing a C18 column, the lyophilized fraction F1 was further separated by RP-HPLC, resulting in two fractions acquired (F1-1 and F1-2). Partly due to its enhanced NO-inhibition activity (IC50 = 22.00 ± 0.41 μg mL−1), the lyophilized fraction F1-2 was assessed by Q-TOF-ESI-MS/MS (Fig. 1b). Various peptides were removed on RP-HPLC. Particularly when assessing the structural and functional properties of peptides <5000 Da, this approach can be utilized to fractionate peptides according to their hydrophobic properties.29,30 Hydrolysates with a high amount of peptides comprising hydrophobic amino acids are thought to exhibit elevated antioxidant activity, as offered by Mendis et al.31 Peptides with high hydrophobicity cleansed from hoki frame protein hydrolysate showed exceptional antioxidant activity, as described by Kim et al.32 Antioxidant peptides comprised of 44% hydrophobic amino acids had excessive attraction for linoleic acid by interrelating with lipid molecules and contributing protons to lipid-derived radicals, as noted by the authors.
3.4 Classification of peptides by Q-TOF MS and assessment of the NO radical scavenging activity between synthetic and enzymatically-derived peptides
Employing de novo sequencing, the amino acid structures of the peptides in fraction F1-2 were examined. Accurate elementary composition was utilized to identify peptides with sequence Leu–Ser–Pro–Leu–Leu–Ala–Ala–His (LSPLLAAH; 821.48 Da) and Thr–Val–Asn–Leu–Ala–Tyr–Tyr (TVNLAYY; 843.42 Da), while Q-TOF-MS was used to establish b and y series fragment ions (Fig. 2a and b). To the part of S. nudus NADH dehydrogenase subunit 2 (mitochondrion), LSPLLAAH had 100% amino acid categorization correspondence. To the part of NADH dehydrogenase subunit 2 (mitochondrion) from S. nudus, TVNLAYY displayed 66.7% correspondence, which implies that the peptide could likewise be originated from this protein. Peptides with these categorizations were created and assessed for their NO radical scavenging activity because fraction F1-2 revealed NO radical scavenging action. IC50 values of 367.72 ± 0.96 (LSPLLAAH) and 232.76 ± 1.18 μM (TVNLAYY) were exhibited by the peptides.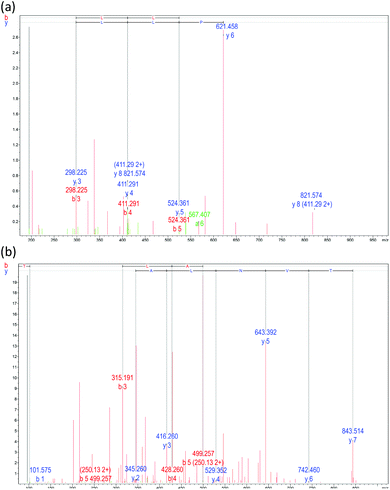 | ||
| Fig. 2 Mass fragmentation spectrum of subfraction F1-2 obtained by reversed-phase high-performance liquid chromatography (see Fig. 1b). (a) Peptides LSPLLAAH and (b) TVNLAYY. | ||
Proteins and peptides from various origins possess NO-scavenging activity, as shown in previous studies. Nonetheless, the active compounds of plants comprise the attention of most studies concerning NO-inhibition activity, with peptides from animal protein only studied by a few. For example, peptides from clam,27 fish,33 and milk34 have been reported. Direct proton-donors such as Tyr exist in the arrangements of our purified peptides, which is significant for the radical searching activity of peptides due to their capacity to satisfy single electrons by contributing protons. Including examples such as Val–Pro–Tyr from soybean,35 and Pro–Ala–Tyr from salmon pectoral fin by-product,33 more than a few low-molecular-weight peptides with Tyr residues have exhibited NO radical scavenging actions.
3.5 Cytotoxicity and its influences on NO production of LSPLLAAH and TVNLAYY
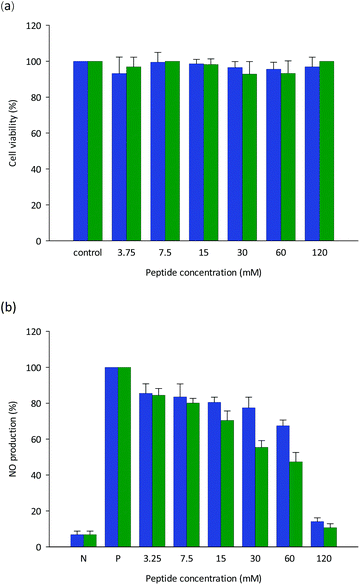
The initial defenders against attacking pathogens are macrophages. A murine macrophage line utilized to exhibit macrophage-mediated inflammation in vitro is RAW 264.7. For assessing anti-inflammatory agents, this cell system is an exceptional model.36 As demonstrated in Fig. 3a, the produced peptides LSPLLAAH and TVNLAYY exhibited no cytotoxicity to RAW264.7 cells in the concentration range employed in this research (3.75–120 mM). A statistically noteworthy surge in NO production compared to the un-stimulated group (P < 0.05) resulted from the inspiration of RAW264.7 cells with E. coli LPS (100 ng mL−1). In a dose-dependent manner with IC50 values of 82.15 ± 0.19 for LSPLLAAH and 67.87 ± 0.54 mM for TVNLAYY, NO production was inhibited (Fig. 3b). For low-molecular-weight peptides obtained from flaxseed protein, which are able to constrain LPS-induced NO production in RAW 264.7 macrophages without obvious cytotoxicity, a comparable observation was previously reported.19–21,36 The findings of the Griess assay propose that LSPLLAAH and TVNLAYY may be potentially successful anti-inflammatory agents, though the uptake systems of LSPLLAAH and TVNLAYY, as well as their influences on cell metabolism, necessitate additional examination.3.6 Influence of LSPLLAAH and TVNLAYY on LPS-excited expression of inflammation-associated genes in RAW 264.7 macrophages established by (qrt)-RT-PCR
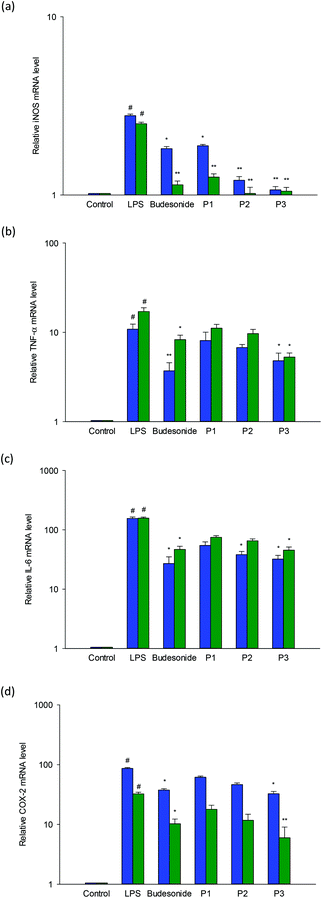
RAW264.7 macrophages were utilized to assess the impact of LSPLLAAH and TVNLAYY on mRNA expression of iNOS, TNF-α, IL-6, and COX-2. The design of specific primers was made possible by the nucleotide sequences of Mus musculus β-actin, iNOS, TNF-α, IL-6 and COX-2. To establish gene expression, (qrt)-RT-PCR was used, as shown in the results in Fig. 4(a–d). LPS stimulation was employed to augment the expression levels of pro-inflammatory cytokines in RAW264.7 macrophages. Both peptides diminished the stimulatory influences of LPS on the expression of iNOS, TNF-α, IL-6 and COX-2 in RAW 264.7 macrophages in a dose dependent manner, as revealed by the treatment of cells with LSPLLAAH or TVNLAYY. The deduction that the synthetic peptides had anti-inflammatory effects in vitro is reinforced by the results.Possessing the capacity to facilitate endogenous pyrogens, control inflammatory reactions, and promote acute phase reactants, TNF-α and IL-6 are pro-inflammatory cytokines. Typically resulting in the creation of NO and prostaglandin, pro-inflammatory cytokines influence macrophages. iNOS and COX-2 expression may also be affected by cytokines.5 The precise amino acid configuration is the primary determinant for the function of any peptide. Further, robust action against intracellular NO and ROS creation has been exhibited by those containing His, Ala, and Lys.37 A 5–10 kDa fraction that constrained LPS-induced prostaglandin and NO production in RAW 264.7 macrophages was reported in vitro study on germinated soybean peptides by González-Montoya et al.38 One of the key factors accountable for the anti-inflammatory influences of peptides is hydrophobicity.39 The contact between the peptide and cell membranes is augmented by the existence of hydrophobic amino acids, which may impact the variation of downstream signaling pathways with anti-inflammatory effects. Nonetheless, clarification of the significance of hydrophobicity and its function in anti-inflammatory effects requires additional examination.
4. Conclusions
In this study, we produced antioxidant peptides from peanut worm protein Alcalase hydrolysates, and identified two novel peptides: LSPLLAAH and TVNLAYY. Our data suggest that both peptides may contribute to preventing inflammation through inhibition of the expression of the inflammatory mediators iNOS, TNF-α, IL-6 and COX-2 in LPS-stimulated RAW 264.7 macrophages without having any effect on cell viability. These findings may indicate the potential utility of PWP peptides as an ingredient in functional foods. However, further research into the in vivo anti-inflammatory effects of the peptides is needed.Funding
We acknowledge financial support from the Ratchadaphiseksomphot Endowment Fund, Chulalongkorn University (R_016_2556), and the Center of Excellence on Medical Biotechnology (CEMB), S&T Postgraduate Education and Research Development Office (PERDO), Office of Higher Education Commission (OHEC), Thailand (SN-60-003-909) for providing financial support for this research.Conflicts of interest
The authors have no conflicts of interest to declare.Acknowledgements
The authors thank the Institute of Biotechnology and Genetic Engineering, Chulalongkorn University for their support and providing access to their facilities.References
- P. Libby, Nutr. Rev., 2007, 65, S140–S146 CrossRef.
- K. J. O'Byrne and A. G. Dalgleish, Br. J. Cancer, 2001, 85, 473–483 CrossRef PubMed.
- A. G. Dalgleish and K. J. O'Byrne, Adv. Cancer Res., 2002, 84, 231–276 CrossRef CAS.
- T. A. Butterfield, T. M. Best and M. A. Merrick, J. Athl. Train., 2006, 41, 457–465 Search PubMed.
- G. Arango Duque and A. Descoteaux, Front. Immunol., 2014, 5, 491 CrossRef PubMed.
- W. K. Alderton, C. E. Cooper and R. G. Knowles, Biochem. J., 2001, 357, 593–615 CrossRef CAS.
- U. Förstermann and W. C. Sessa, Eur. Heart J., 2012, 33, 829–837 CrossRef.
- Q. Xue, Y. Yan, R. Zhang and H. Xiong, Int. J. Mol. Sci., 2018, 19, 3805 CrossRef PubMed.
- S. Chakrabarti, F. Jahandideh and J. Wu, BioMed Res. Int., 2014, 608979 Search PubMed.
- C. Gunaydin and S. S. Bilge, Eurasian J. Med., 2018, 50, 116–121 CAS.
- R. S. Y. Wong, Adv. Pharmacol. Sci., 2019, 3418975 CAS.
- S. Chakrabarti, S. Guha and K. Majumder, Nutrients, 2018, 12, E1738 CrossRef PubMed.
- M. J. Boyle and M. E. Rice, Int. J. Dev. Biol., 2014, 58, 485–499 CrossRef PubMed.
- Y. H. Ge, Y. Y. Chen, G. S. Zhou, X. Liu, Y. P. Tang, R. Liu, P. Liu, N. Li, J. Yang, J. Wang, S. J. Yue, H. Zhou and J. A. Duan, Int. J. Mol. Sci., 2018, 19, 3023 CrossRef PubMed.
- P. W. Hsueh and K. S. Tan, Raffles B. Zool., 2016, 34, 235–240 Search PubMed.
- P. Prakot, N. Chaitanawisuti, P. Sangtanoo, T. Saisavoey and A. Karnchanatat, J. Aquat. Food Prod. Technol., 2018, 27, 811–829 CrossRef CAS.
- Official Methods AOAC. 986.25., Gaithersburg, MD, USA. 2012.
- Official Method AOAC. 988.15, Gaithersburg, MD, USA. 2005.
- T. Saisavoey, P. Sangtanoo, O. Reamtong and A. Karnchanatat, J. Sci. Food Agric., 2019, 99, 5112–5121 CrossRef CAS PubMed.
- A. Inkanuwat, R. Sukaboon, O. Reamtong, P. Asawanonda, A. Pattaratanakun, T. Saisavoey, P. Sangtanoo and A. Karnchanatat, Food Sci. Biotechnol., 2019, 57, 200–211 Search PubMed.
- R. Suttisuwan, S. Phunpruch, T. Saisavoey, P. Sangtanoo, N. Thongchul and A. Karnchanatat, Food Biotechnol., 2019, 33, 303–324 CrossRef CAS.
- M.M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- Y. Li, B. Brodsky and J. Baum, J. Biol. Chem., 2007, 282, 22699–22706 CrossRef CAS PubMed.
- J. H. Muyonga, C. G. B. Cole and K. G. Duodu, Food Chem., 2004, 85, 81–89 CrossRef CAS.
- M. Nikoo, S. Benjakul, M. Bashari, M. Alekhorshied, A. I. Cissouma, N. Yang and X. Xu, Food Biosci., 2014, 5, 19–26 CrossRef CAS.
- A. Bougatef, N. Nedjar-Arroume, L. Manni, R. Ravallec, A. Barkia, D. Guillochon and M. Nasri, Food Chem., 2010, 118, 559–565 CrossRef CAS.
- S. J. Lee, E. K. Kim, Y. S. Kim, J. W. Hwang, K. H. Lee and D. K. Choi, et al. , Food Chem. Toxicol., 2012, 50, 1660–1666 CrossRef CAS PubMed.
- X. Q. He, W. H. Cao, G. K. Pan, L. Yang and C. H. Zhang, J. Sci. Food Agric., 2015, 95, 1544–1553 CrossRef CAS PubMed.
- R. Song, K. Q. Zhang and R. B. Wei, Process Biochem., 2016, 51, 1674–1682 CrossRef CAS.
- B. H. Sarmadi and A. Ismail, Peptides, 2010, 31, 1949–1956 CrossRef CAS PubMed.
- E. Mendis, N. Rajapakse and S. K. Kim, J. Agric. Food Chem., 2005, 53, 581–587 CrossRef CAS PubMed.
- S. Y. Kim, J. Y. Je and S. K. Kim, J. Nutr. Biochem., 2007, 18, 31–38 CrossRef CAS PubMed.
- C. B. Ahn, Y. S. Cho and J. Y. Je, Food Chem., 2015, 168, 151–156 CrossRef CAS PubMed.
- X. Y. Mao, X. Cheng, X. Wang and S. J. Wu, Food Chem., 2011, 126, 484–490 CrossRef CAS.
- J. Kovacs-Nolan, H. Zhang, M. Ibuki, T. Nakamori, K. Yoshiura and P. V. Turner, et al. , Biochim. Biophys. Acta, Gen. Subj., 2012, 1820, 1753–1763 CrossRef CAS PubMed.
- F. O. Martinez, L. Helming and S. Gordon, Annu. Rev. Immunol., 2009, 27, 451–483 CrossRef CAS PubMed.
- B. Wang, L. Li, C. F. Chi, J. H. Ma, H. Y. Luo and Y. F. Xu, Food Chem., 2013, 138, 1713–1719 CrossRef CAS PubMed.
- M. González-Montoya, B. Hernández-Ledesma, J. M. Silván, R. Mora-Escobedo and C. Martínez-Villaluenga, Food Chem., 2017, 242, 75–82 CrossRef PubMed.
- L. Zhao, X. Wang, X. L. Zhang and Q. F. Xie, J. Food Drug Anal., 2016, 24, 376–384 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2020 |