Metagenomic analysis of gut microbiota modulatory effects of jujube (Ziziphus jujuba Mill.) polysaccharides in a colorectal cancer mouse model†
Xiaolong
Ji
 *a,
Chunyan
Hou
a,
Yonggang
Gao
b,
Yuqiang
Xue
b,
Yizhe
Yan
*a,
Chunyan
Hou
a,
Yonggang
Gao
b,
Yuqiang
Xue
b,
Yizhe
Yan
 a and
Xudan
Guo
*b
a and
Xudan
Guo
*b
aSchool of Food and Biological Engineering, Zhengzhou University of Light Industry, Zhengzhou 450002, P.R. China. E-mail: xiaolongjiytu@163.com; cyhouaaf@163.com; yanyizhe@mail.ustc.edu.cn
bBasic Medical College, Hebei University of Chinese Medicine, Shijiazhuang 050200, PR China. E-mail: guoxudan123@126.com; gyg3177@163.com; xue@cnstat.org
First published on 9th December 2019
Abstract
Accumulating evidence has reported that the gut microbiota could play important roles in the occurrence and progression of colorectal cancer. The nondigestible plant polysaccharides have always been fermented by the intestinal microbiota. Polysaccharides, the predominant functional composition found in jujube fruit, has been shown to inhibit carcinogenesis in animal models. However, the molecular mechanisms involved in polysaccharides preventing carcinogenesis are still uncharacterized. The aim of this study was to investigate the modulatory effects of jujube polysaccharides (JP) on intestinal microbiota, and the influence of JP on the gut flora structure was then analyzed using an AOM/DSS-induced colitis cancer mouse model, using high-throughput sequencing. Contrasted with control group, the addition of JP could ward off colon cancer by ameliorating colitis cancer-induced gut dysbiosis. In addition, there was a significant decrease in Firmicutes/Bacteroidetes post JP treatment. What's more, KEGG pathways of metabolic pathways, ATP-binding cassette (ABC) transporters and two-component system enriched the most differentially expressed genes after JP intervention for 13 weeks. These results suggested that JP showed prebiotic-like activities by positively modulating intestinal microbiota and affecting certain metabolic pathways contributing to host health. In conclusion, our results demonstrated an appreciable capability of JP to restore the gut microbiota profile altered by AOM/DSS, indicating the potential of jujube polysaccharides as promising prebiotic candidates for the prevention and treatment of colorectal cancer.
1 Introduction
Colorectal cancer (CRC) has become a serious global public health concern, as worldwide, it is the second most common cancer in women, and the third most common in men.1 Epidemiological studies have suggested that the disease could be caused by the consumption of high quantities of processed meat (western diet), providing an imbalance between dietary intake and energy expenditure.2 Therefore, greater attention has been paid to natural plant products as potential sources of functional ingredients, with potential anti-colon cancer activities.3,4 More recently, the relationship between colon cancer and intestinal flora or their metabolites has gained considerable tractions.5,6In the human gastrointestinal tract, the vast majority of microbes form stable microbial ecosystems, playing important roles in the development and life-long maintenance of host health.7 These microbes not only digest food and produce key metabolites, but they also enhance the immune functions. They could regulate disordered intestinal microorganisms associated with CRC and metabolic syndromes.8 The disrupted composition of gut microbiota in high fat, high meat, and low dietary fiber diets could lead to metabolic endotoxemia, inflammation, inflammatory bowel disease and CRC.2 Recent research has indicated significant intestinal flora differences between normal and CRC individuals.9 Especially, there were increases adverse bacteria, including Fusobacteria, Porphyromonadaceae, Coriobacteridae, and beneficial microorganisms, including Methanobacteriales whereas Bifidobacterium, Lactobacillus, Ruminococcus and Faecalibacterium spp. diminished as well as Roseburia and Treponema in CRC.10 Additionally, CRC-reduction may be achieved by balancing the diet and manipulating the microbiota.11,12 Recent studies have indicated that dietary fiber and plant polysaccharide-rich sources could modulate intestinal microbiota in vivo, promoting the proliferation and diversity of beneficial bacteria in the gut.2,13 Thus, the development of an effective dietary regimen could have huge significance for the prevention of CRC.
In China, jujube fruit are generally recognized as valuable sources of biologically active compounds, possessing higher nutritional and pharmacological effects.14 Dried jujube has been commonly used as a food, a food additive, a flavoring and also as a traditional Chinese medicine used for thousands of years. It has been claimed that the jujube has a wide range of health benefits, with anti-obesity, anti-epileptic, anti-insomnia, neuroprotective, antioxidant, anti-inflammatory and anticancer properties, etc.15 Jujube based polysaccharides are the most abundant component of the fruit, and could be responsible for its chemo-preventive properties (e.g. immunomodulation, antioxidant, antitumor, hepatoprotective and hypoglycemic activities and gastrointestinal-protective effects).16,17 In a previous report, orally administered JP effectively shortened gastrointestinal transit time, reduced caecum ammonia levels, elevated total short-chain fatty acids (SCFA) concentrations in the cecum, increased fecal moisture and reduced daily fecal ammonia outputs from feces, in different intestinal and fecal indices in a hamster model.18 Interestingly, we recently observed that wild jujube polysaccharides protected mice against experimentally induced inflammatory bowel disease by enhancing intestinal barrier functions.19 In addition, our previous studies have also suggested that Muzao polysaccharides benefitted the stability of certain gut microbiota, especially under environmentally triggered microbial imbalance, where azoxymethane (AOM) and dextran sodium sulfate (DSS) were used to induce a CRC mice model.20
The biotransformation of plant polysaccharides has been extensively investigated in gut microbiota, suggesting that metabolic and colonic microbiota transformation could enhance dietary oligosaccharide bioactivity.21,22 While, it has been shown that polysaccharides could relieve the occurrence of colon cancer in rats by modulating certain genes, the potential mechanisms in preventing gut dysbiosis and anti-CRC are complex.23 Recently, metagenomic based molecular techniques were widely used to identify rare and unculturable bacterial communities, as well as identifying functional genes enriched by external intervention.24,25 In this study, male C57BL/6 mice were induced to develop colitis-associated colon cancer by AOM, a carcinogen and DSS, a proinflammatory reagent. Once established, this cancer induced mice were orally administrated with JP.20 These mice feces were determined by high-throughput sequencing, and genes enriched in several metabolic pathways were investigated in the gut microbiome.
2 Materials and methods
2.1 Reagents and chemicals
JP were collected from the Innovation Laboratory of Molecular Nutrition & Health Food, Northwest A&F University. Structural characterization confirmed that the JP had average molecular weight of 89.90 kDa, and consisted of arabinose, galactose, glucose, rhamnose, and mannose with a molar percentage of 49.67![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 29.01
29.01![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 11.43
11.43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5.38
5.38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4.51, respectively, suggesting it may have belonged to the type I rhamnogalacturonan family.26 AOM was purchased from Sigma-Aldrich (St Louis, MO, USA), while DSS (molecular weight of 36
4.51, respectively, suggesting it may have belonged to the type I rhamnogalacturonan family.26 AOM was purchased from Sigma-Aldrich (St Louis, MO, USA), while DSS (molecular weight of 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–50
000–50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1) was purchased from MP Biomedicals (Santa Ana, CA, USA). All other chemicals and reagents were analytical grade.
000 g mol−1) was purchased from MP Biomedicals (Santa Ana, CA, USA). All other chemicals and reagents were analytical grade.
2.2 Animals and experimental design
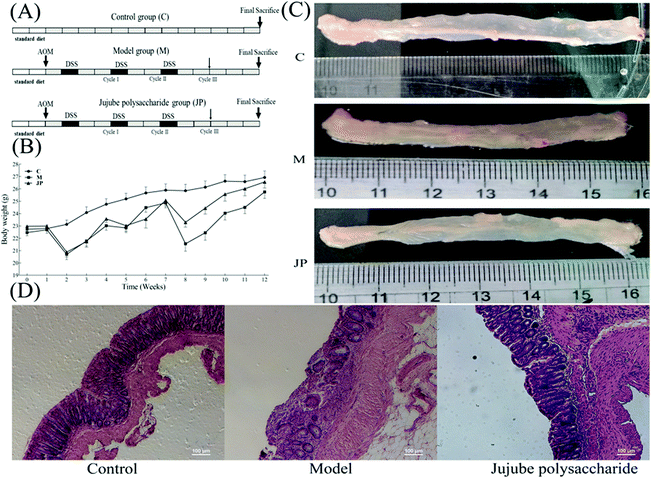
Approximately 6–8 weeks-old male C57BL/6 mice, weighing approximately 20 g, were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. Animal experiments were performed in accordance with the National Institute of Health Guides for the Care and Use of Laboratory Animals (NIH Publication No. 80-23), revised and approved by the Northwest A&F University Animal Care and Use Committee (Yangling, Shaanxi, China). Mice were housed under standard temperatures and humidity conditions, with free access to water and food and were subjected to a 12 h light/dark cycle.20The AOM/DSS induced mouse model is widely used to investigate the occurrence and progression, and chemoprevention interventions in colon cancer (Fig. 1A).20,27 Briefly, a single intraperitoneal injection of AOM was performed on day 0. One week later, mice were exposed to 2.0%–2.5% DSS in drinking water, for seven consecutive days, followed by 14 days of regular drinking water. This cycle was repeated twice.28 For the JP group: AOM/DSS-induced mice were gavaged with 1000 mg kg−1 JP once a day, from day 0 to the study endpoint. Body weights were recorded once a week throughout the experiment (Fig. 1A). Mice in the control group (C) were gavage with sterile saline alone. All mice were sacrificed by cervical dislocation at the end of week 13, and the entire colon, from the ileocecal junction to the anus was excised. The colon length was recorded and afterwards histopathology was performed using samples fixed in 10% phosphate-buffered formalin.20,28
2.3 DNA extraction and intestinal microbiota analysis
Total genomic DNA was extracted from fecal samples using DNA extraction kits in accordance with manufacturer's instructions.29 Once extracted, DNA sample were fragmented by sonication to approximately 350 bp, then fragments were end-polished, A-tailed and ligated with a full-length adaptor for Illumina sequencing, with further PCR amplifications. PCR products were purified (AMPure XP system) and libraries were analyzed for size distribution using the Agilent-2100 Bioanalyzer and quantified using real-time PCR. The clustering of index-coded samples was performed on a c-Bot Cluster Generation System according to manufacturer's instructions. After cluster generation, library preparations were sequenced on an Illumina HiSeq platform and paired-end reads were generated.24Quality trimming is an essential step to generate high confidence of variant calling. Raw reads were processed to generate high quality clean reads according to four stringent filtering standards: (i) removing reads with ≥10% unidentified nucleotides; (ii) removing reads with >50% bases having low quality scores of ≤20; and (iii) removing reads aligned to the barcode adapter. The high-quality reads were clustered into operational taxonomic units (OTUs) using CD-HIT software. The OTUs that reached a 97% nucleotide similarity level were used for further analysis.20,25
2.4 De novo assembly and gene predictions
Open Reading Frame (ORFs) were predicted based on final assembly contigs using MetaGeneMark. Predicted ORFs ≥ 300 bp in length from all samples were pooled and combined based on ≥95% identity and 90% read coverage using CD-HIT to reduce the number of redundant genes for downstream assembly. The reads were re-arrayed to predict genes using BWA to count reads numbers. Finally, gene catalogues were obtained from non-redundant genes with gene reads counts >2.30,312.5 Taxonomical assignment and functional classification
After read filtering, clean reads were used to generate taxonomic profiles using a k-Mer-based taxonomic classifier. This approach employs a novel data structure called l-Othello, which supports efficient querying of taxonomic information from NCBI, using reads k-Mer signatures of 31 bp in length. All unique ORFs were annotated using DIAMOND to the functional databases: Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO).312.6 Statistical analysis
The results were expressed as mean ± SEM where applicable. GraphPad Prism 6.0 software was used for statistical analysis. Statistical significances for group differences were analyzed by ANOVA followed by Duncan's test.3 Results
3.1 Influence of JP on mouse body weight and colon length
To examine the effects of JP in the CRC mouse model, the mutagen AOM was used to initiate colon tumors, followed by a triple dose of DSS to induce chronic inflammation (Fig. 1A). Mice receiving normal saline alone were used as a control group. After AOM/DSS treatment, mice were orally administrated 1000 mg kg−1 JP once a day for 13 weeks. As shown in Fig. 1B, significant body weight loss was observed in AOM/DSS induced mice when compared with the control group. This was significantly alleviated by JP treatment by the end of the study. These observations were similar to a previous study where it was shown that a Rhizopus nigricans polysaccharide increased the weight of AOM/DSS treated mice.32Tumor formation was analyzed at the end of the study. As shown in Fig. 1C, in the absence of JP treatment, AOM/DSS-induced mice exhibited a high tumor burden in their colons, while JP treatment markedly reduced AOM/DSS-induced tumors. These results agreed with a previous study reporting that wild JP inhibited tumor growth in an AOM/DSS-induced CRC model.19 Moreover, decreased colon length was observed in AOM/DSS-induced mice when compared with control mice. Such a significant length decrease was subsequently relieved by JP treatment (Fig. 1C). Hematoxylin and eosin staining of colon tissues was performed to analyze the pathology of AOM/DSS-induced colons. The results showed that JP decreased the number and sizes of tumors and provided protection against the AOM/DSS-induced dysplasia/adeno-carcinoma lesion via surface tumor necrosis. These data indicated that JP exhibited strong suppressive effects on colitis and colorectal tumorigenesis induced by AOM/DSS.20
3.2 Influence of JP on bacterial diversity
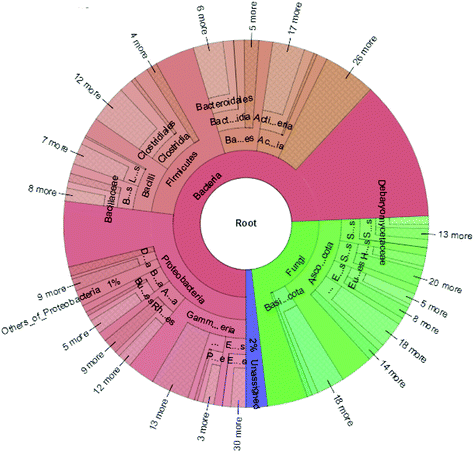
The high-quality clean reads were >98.50% for all six samples, indicating a good sequencing depth for fecal microbiota gene analysis (Table S1†). The flat curves of the base sequence and mass distribution demonstrated higher accuracy (Fig. S1a†). Using MEGAHIT software to assemble effective reads, the default length of KMER should be 21, 41, 61, 81, 99, and the length of contigs could exceed 600 in each group when the sum value reaches 90% of the total length of fragment (Table S2†). Using the ReefSeq database in NCBI, fecal microbial genes were indexed and the species annotated using Meta-Othello. The species annotation and abundance information for each sample were counted, and a species abundance information table obtained in Fig. 2. Among the classifiable sequences in different groups, Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, Ascomycota, and Basidiomycota were represented the dominant lineages in the samples. To better understand the shared richness of each group, a Venn diagram displaying overlaps between groups was produced. This approach showed that a total richness of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 579
579![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 444 OTUs was shared among all samples in Fig. S1c.† These data demonstrated that approximately 43.68% of OTUs were reduced after AOM/DSS-treated mice were fed JP, suggesting that JP decreased microbial community diversity and pathogenic bacteria levels, but increased probiotic diversity.20
444 OTUs was shared among all samples in Fig. S1c.† These data demonstrated that approximately 43.68% of OTUs were reduced after AOM/DSS-treated mice were fed JP, suggesting that JP decreased microbial community diversity and pathogenic bacteria levels, but increased probiotic diversity.20
In keeping with our previous observations,20,24,30 the most abundant phyla included Proteobacteria, Firmicutes, Ascomycota and Bacteroidetes in different groups of mice (Fig. 3A). The most abundant phyla included Proteobacteria, Firmicutes, Ascomycota and Bacteroidetes in different groups of mice. Other phyla were represented in relatively lower abundances; Mucoromycota, Spirochaetes, Verrucomicrobia, Cyanobacteria and a number of unclassified bacteria. After JP treatment, there was a significant increase in the relative abundance of Bacteroidetes, while there was a significant decrease in Firmicutes. In addition, the ratio of Bacteroidetes to Firmicutes (B/F) increased from 0.27 (M) to 0.39 (JP). To investigate fecal microbiota communities in the different mouse groups, a family level analysis was performed (Fig. 3B). At this level, the following families showed an increasing trend during JP treatment: Lactobacillaceae, Bacteroidaceae and Debaryomycetaceae (P < 0.05). Lactobacillaceae was the dominant beneficial bacteria at the family level during JP treatment, their relative abundance increased significantly from 0.010 to 0.017 at the 13th week. The relative abundance of Bacteroidaceae showed a similar trend, which reached 0.022 in the JP group after 13 weeks from 0.015 in the M group. Herpotrichiellaceae, Enterobacteriaceae, Aspergillaceae and Lachnospiraceae families showed less abundance after JP intervention, at 13 weeks. Bacteroidetes and Firmicutes abundance were similar at the phylum level.
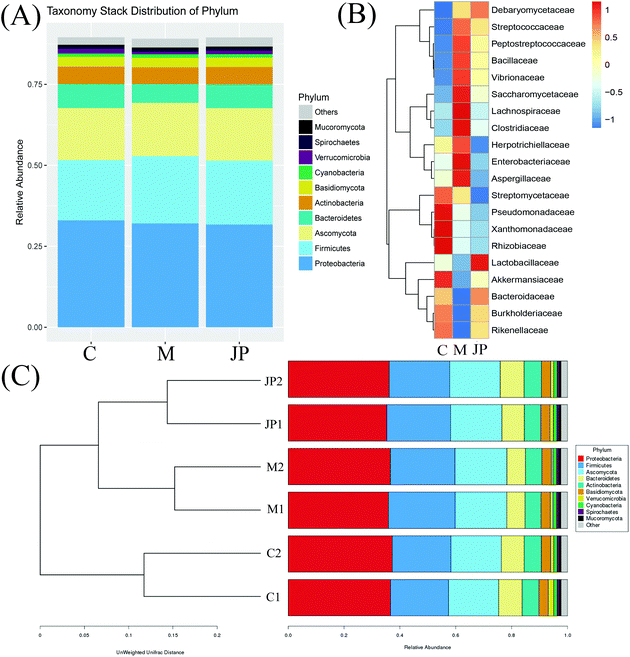 | ||
| Fig. 3 The relative abundance of the top phylum (A), heatmap of the genera (B) with the highest frequency and relative abundance and bar graph of the top families (C) from different groups. | ||
At deeper taxonomic levels, more significant differences were observed (Fig. 3C). Bar graphs showed genera at different levels in samples; Bacteroidetes and Actinobacteria were increased when fed JP for 13 weeks. Significant reductions were observed in the relative abundance of Firmicutes, Cyanobacteria, Deferribacteres, Spirochaetes and Mucoromycota. Bar graphs showed genera at different levels in samples; Bacteroidetes and Actinobacteria were increased when fed JP for 13 weeks. Significant reductions were observed in the relative abundance of Firmicutes, Cyanobacteria, Deferribacteres, Spirochaetes and Mucoromycota. Our data indicated that intestinal dysbiosis by AOM/DSS was extensively alleviated by JP treatment, and that certain genera belonging to two predominant phyla may be used as distinguishing biomarkers. Despite high inter-individual variability after JP intervention for 13 weeks, Firmicutes were significantly less abundant, while Bacteroidetes were significantly more abundant in fecal microbiota, when compared to the M group. To measure the similarity between microbial communities, principal coordinate analysis, principal co-ordinates analysis and non-metric multidimensional scaling analyses were performed (Fig. S1b†). Despite significant inter-individual variation, the fecal microbiota from AOM/DSS-induced mice, fed with JP were separated clearly by these different analytical methods.
3.3 The influence of JP on the gut microbiome
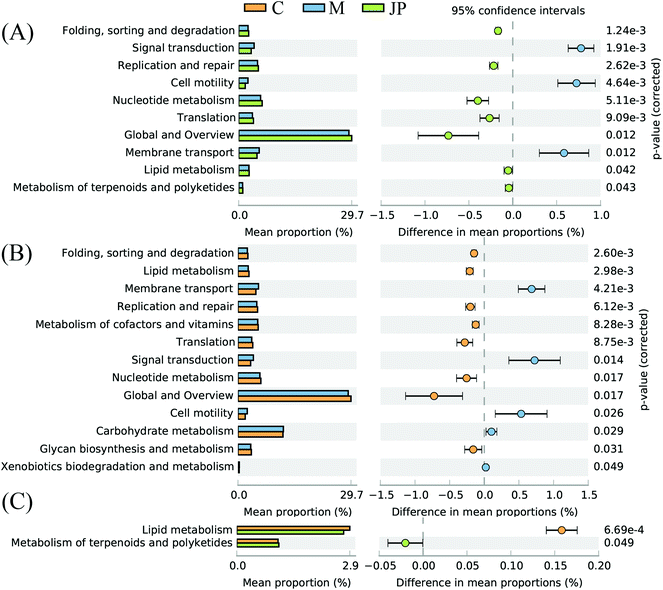
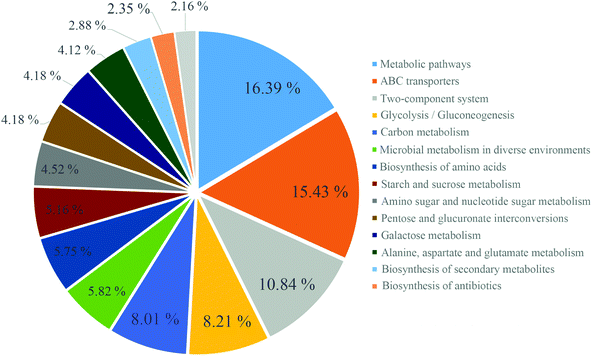
The imputed relative abundances of KEGG pathways in each sample were used to predict changes in metabolic function in microbiomes supplemented with JP (Fig. 4). KEGG pathway analyses showed that membrane transport, carbohydrate metabolism, lipid metabolism and metabolism of terpenoids and polyketides accounted for the top five highest functions. The greatest statistical difference between JP and M groups were translation, nucleotide metabolism and cell motility.After 13 weeks, JP-associated differentially expressed genes were enriched for different KEGG pathways, involved in metabolism, ATP-binding cassette (ABC) transporters, and two-component system (Fig. 5), and ABC transporters, including those predicted to be involved in sugar and amino acid. In addition, KEGG pathway analysis of differentially expressed genes between C and M groups, C and JP groups (data not shown, Fig. S2†) were mainly enriched in two-component system, after JP supplementation for 13 weeks. Although the ranking varied in different groups, the numbers of differentially expressed genes were dramatically changed within the different groups.
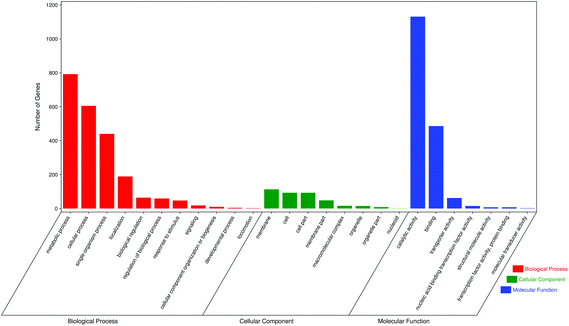
The GO database provides three different types of systematic definition for describing the different functions of gene products. GO functions include biological processes, cellular components and molecular function. Biological processes consist of molecular functions with multiple steps;24 cellular components refer to gene products located in cellular organelles, while molecular functions describe molecular biological activities, such as catalytic and binding activities. For each GO function, we selected seven of the largest GO terms. GO analysis of differentially expressed genes between the M and JP groups showed that most genes were in the biological processes group, including metabolic processes, cellular processes, single organism processes, localization, biological regulation and regulation of biological processes. Cellular components included membranes, cells, cell parts and membrane parts; and molecular functions included catalytic activity, binding, transporter activity and nucleic acid binding transcription factor activity (Fig. 6). GO analysis of differentially expressed genes between the C and M groups and C and JP groups are shown in Fig. S2† and the numbers of differentially expressed genes showed a significant difference in different groups.
4 Discussion
Colorectal cancer and its associated comorbidities are major health issues.2 It has been reported that CRC is associated with several metabolic syndromes, which affect the quality of CRC patients’ lives.33,34 More and more evidence has shown that CRC, accompanied by high mortality rates and increasing incidences among adults, is the third most common cancer globally.1 Therefore, the implementation of a dietary regimen, without side effects, to prevent CRC is a public health goal.Polysaccharides are a major component in jujube and have significant gastrointestinal-protective effects, including promoting tissue repair, regulating intestinal microorganisms, producing anti-inflammatory effects and regulating intestinal microorganisms.17,19 A previous report suggested that JP reduced inflammatory responses by attenuating the activities of tumor necrosis factor α, IL-1b, IL-6, and myeloperoxidase in CRC rats.20 JP also protects against inflammatory bowel disease by enhancing intestinal barrier functions via the activation of AMPK.19 However, due to low bioavailability, JP is not completely absorbed from the gastrointestinal tract. In previous studies, JP can be degraded to oligosaccharides and SCFA by colonic bacteria, which was consistent with previous reports.18,20 These SCFA and oligosaccharides interact with microbiota in the colon, and their resulting metabolites appear to impact on intestinal micro-ecology, in addition to modulating microbial populations.35,36 Several studies have assessed the bioavailability of plant polysaccharides, which is the fraction of ingested oligosaccharides reaching the systemic circulation and specific sites where it exerts biological actions, especially at the colon and rectum.13,37 These metabolic derivatives could play important roles in vivo, and show increased biological activities.38,39
The human gut has been viewed as a bioreactor, where interactions between plant polysaccharides and gut microbiota occur.13 Prebiotics are nondigestible food ingredients, mostly oligosaccharides, which benefit the host by stimulating the growth and/or activity of specific intestinal bacteria.24 It has been reported that certain plant polysaccharides, with prebiotic-like activities, have major influences on gut microbial profiles and helps reestablish healthy intestinal microbial communities.37,40 In our previous study, JP exhibited similar proliferative effects, in combination with fructo-oligosaccharides, on the growth of beneficial bacteria, including Bifidobacterium spp. and Lactobacillus/Enterococcus groups. Furthermore, they displayed inhibitory effects on Enterorhabdus, Odoribacter, Ruminiclostridium, Anaerotruncus, and Blautia groups, and appeared to decrease total bacterial numbers.20 It has been reported Herpotrichiellaceae, Enterobacteriaceae, Aspergillaceae and Lachnospiraceae could promote tumorigenesis, whereas Bifidobacterium spp. and Lactobacillus/Enterococcus could degrade macromolecular plant polysaccharides and have anti-inflammatory and anti-tumorigenic properties.33 By using the AOM/DSS mouse model, we also investigated the modulatory effects of JP on intestinal microbiota. JP showed significant protective effects against AOM/DSS-induced CRC, and a strong activity in regulating dysbiosis and maintaining a balanced microbial ecology.20Bacteroidetes and Firmicutes are the main bacterial groups involved in the colonic metabolism of undigested plant polysaccharides, where they use a complex metabolic energy-harvesting mechanism.41 It is widely believed that an increased ratio of Bacteroidetes/Firmicutes can prevent the development of CRC in both dietary and genetic mouse CRC models in regulating the gut microbiota following the JP treatments.42,43 The increased abundance of Bacteroides has been reported to be associated with immunosuppression and carcinogenesis. The AOM and DSS feeding may not only affect gut morphology, mucin production, gut permeability and SCFA production, but also promote bacterial dysbacteriosis.32,43 In this study, a large decrease in Firmicutes, with an expansion in Bacteroidetes and Proteobacteria was observed in our CRC mouse model, upon JP intervention. These results were consistent with previous reports suggesting that Firmicutes are largely repressed by plant polysaccharides and their metabolites, thus tilting the balance towards Bacteroidetes in the gut.44,45
Dietary polysaccharides could cause variations in specific microorganism populations, whereas microbial conversion of polysaccharides affects other colonic pathways and processes, such as SCFA production.13,24,41 SCFA production is a metabolic consequence of intestinal microbiota. Their ratios and concentrations are dramatically influenced by gut flora and the diet.2,21 With elevated fecal SCFA concentrations, SCFA-producing bacteria could promote energy intake from plant polysaccharides, and prevent host colonic diseases e.g. CRC.46 Increased generation of acids, upon fermentation, is considered desirable, as acidic colonic environments protect against pathogenic bacteria and are helpful towards mineral absorption.20,24 High levels of acetic acid may be correlated with the metabolic activities of Bifidobacterium, Lactobacillus, Bacteroides and Enterococcus.4,46 In this study, at the phylum level, Bacteroides were dramatically increased upon JP intervention, which may be correlated with the production of acetic acid. Faecalibacterium and Roseburia, which are strongly correlated with butyrate production, also counterbalance the dysbiosis of intestinal microbiota.20,47 In our study, their relative abundance was decreased, along with the intake of JP. Recently, a metagenomic study suggested that butyric acid is not predictable at the family level of bacterial taxonomy, but may be predictable at the genus level or deeper.24,48 It is known that butyric acid-producing bacteria belong to the phylum Firmicutes, but other phyla may have potential candidates, such as the Actinobacteria and Proteobacteria.24,37 As is well-known, SCFA is the main end product of carbohydrate metabolism, which can inhibit the proliferation of tumor cells and protect the intestine from colitis and colorectal cancer.27 Indeed, most taxa correlated positively with the fecal concentrations of butyric acid and were from the phylum Firmicutes.1,22 The production of butyric acid occurs at the later stages of fermentation, indicating it may be correlated with the transformation of other bacterial metabolites formed during this fermentation.49 The production of butyric acid occurs at the later stages of fermentation, indicating it may be correlated with the transformation of other bacterial metabolites formed during this fermentation. However, the corresponding pathways and underlying mechanisms must be studied further using integrative genomic technologies.
An expanding body of preclinical evidence suggests the potential of JP on a variety of human diseases involving signal transduction pathways.16,17 Therefore, this nontoxic natural agent could be extremely useful, either alone or in combination with conventional therapeutics for the prevention of human chronic disease.15,17 In terms of anti-colon cancer activities, previous studies have suggested that the positive impact of JP could be from its ability to suppress chronic inflammation and oxidative stress damage.20 However, the effects of JP on CRC mechanisms remain unclear. At present, next-generation sequencing platforms provide opportunities to explore taxonomic, protein-coding gene or protein expression diversities, by applying more comprehensive and less biased measurements to the diet, microbiota and the host.24,50 The approach has provided new information on the diversity and composition of human gut microbiota. The influence of plant polysaccharides on bacterial growth and metabolism depends on polysaccharide structure, the dose assayed and the microorganism strain.13 Recent findings have suggested a variety of potential mechanisms for plant polysaccharides acting on bacteria. For example, plant polysaccharides and/or oligosaccharide could bind to bacterial cell membranes in a dose-dependent manner, thus disturbing membrane function and inhibiting cell growth.2,22,37 Plant polysaccharides could also act on different bacterial species by generating hydrogen peroxide thereby altering the permeability of the microbial membrane.23,36 Plant polysaccharides could also act on different bacterial species by generating hydrogen peroxide thereby altering the permeability of the microbial membrane.23,49 Microbial exposure to polysaccharides/oligosaccharides could upregulate proteins related to defensive mechanisms, protecting cells while simultaneously downregulating metabolic and biosynthetic proteins.2,4
Direct metagenomic sequencing is unravelling the metabolic potential embedded in selected gene pools of gut microbiota, and is identifying gut bacterial genes from intestinal communities.24,51 In our study, GO analysis of differentially expressed genes showed that JP intervention affected cellular component genes, including those in the cytosol, cytoplasm, plasma membrane, membrane and integral components of membrane, to relieve the negative consequences AOM/DSS induction. For the imputed relative abundances of KEGG pathways at different groups, the greatest statistical differences were observed for the excretory system, transcription and substance dependence. However, for KEGG pathway analyses of differentially expressed genes, the rankings varied in different groups; metabolic pathways, ABC transporters and two-component systems occupied the top categories in the M versus JP group. The previous studies summarized plant polysaccharides alleviating CRC by modulation of immune system and intestinal microorganism.52,53 Our data in the present study indicated this function might result in activation of intestinal microorganism as the up-regulated differential express genes involving in JP degradation to produce anti-inflammatory factors pathways (metabolic pathways, ABC transporters and two-component systems) in JP group. These results indicated that JP treatment significantly affected these pathways in this study, which is worthy of future investigation.
JP bioactivity greatly depends on its transformation in the gut. The different health effects of JP have been shown to be a result of individual variability in intestinal microbial ecology.24 We have demonstrated a better understanding of the two-way interactions between JP and intestinal microbiota. This allows us to evaluate and correlate the contribution of microbial metabolites from JP metabolism, to improvements in host health. More metagenomic approaches are required to clarify metabolic pathways altered by JP intervention.
5 Conclusions
In this study, jujube polysaccharides showed significant protective effects against colorectal cancer induced by AOM/DSS. We also demonstrated strong activity in maintaining the balance of microbial ecology. These results suggested that jujube polysaccharides intervention could not only benefit the stability of certain gut microbiota, especially in environments triggered by microbial imbalance, but it also affected key metabolic pathways, contributing to the maintenance of host gut health.Abbreviations
| AOM | Azoxymethane |
| DSS | Dextran sulfate sodium |
| JP | The polysaccharides from jujube (Ziziphus jujuba Mill.) fruit |
| CRC | Colorectal cancer |
| SCFA | Short-chain fatty acids |
| OTUs | Operational taxonomic units |
| ORF | Open reading frame |
| ABC | ATP-binding cassette |
| KEGG | Kyoto encyclopedia of genes and genomes |
| GO | Gene ontology |
Conflicts of interest
The authors have no conflicts of interest to declare.Acknowledgements
This research was financially supported by National Natural Science Foundation of China (21502177), the PhD Research Fund of Hebei University of Chinese Medicine (BSZ2018010), the Scientific Program of Administration of Traditional Chinese Medicine of Hebei (2019089) and the Doctoral Research Foundation of Zhengzhou University of Light Industry (2019).References
- X. Ji, Q. Peng and M. Wang, Anti-colon-cancer effects of polysaccharides: A mini-review of the mechanisms, Int. J. Biol. Macromol., 2018, 114, 1127–1133 CrossRef CAS PubMed.
- S. J. O'Keefe, Diet, microorganisms and their metabolites, and colon cancer, Nat. Rev. Gastroenterol. Hepatol., 2016, 13, 691–706 CrossRef PubMed.
- X. M. Huang, Z. J. Yang, Q. Xie, Z. K. Zhang, H. Zhang and J. Y. Ma, Natural products for treating colorectal cancer: A mechanistic review, Biomed. Pharmacother., 2019, 117, 109142 CrossRef CAS PubMed.
- M. Song and A. T. Chan, Diet, Gut Microbiota, and Colorectal Cancer Prevention: A Review of Potential Mechanisms and Promising Targets for Future Research, Curr. Colorectal Cancer Rep., 2017, 13, 429–439 CrossRef PubMed.
- J. D. Dahmus, D. L. Kotler, D. M. Kastenberg and C. A. Kistler, The gut microbiome and colorectal cancer: A review of bacterial pathogenesis, J. Gastrointest. Oncol., 2018, 9, 769–777 CrossRef PubMed.
- P. Louis, G. L. Hold and H. J. Flint, The gut microbiota, bacterial metabolites and colorectal cancer, Nat. Rev. Microbiol., 2014, 12, 661–672 CrossRef CAS PubMed.
- G. Falony, S. Vieira-Silva and J. Raes, Microbiology Meets Big Data: The Case of Gut Microbiota-Derived Trimethylamine, Annu. Rev. Microbiol., 2015, 69, 305–321 CrossRef CAS PubMed.
- W. S. Garrett, The gut microbiota and colon cancer, Science, 2019, 364, 1133–1135 CrossRef CAS PubMed.
- C. A. Brennan and W. S. Garrett, Gut Microbiota, Inflammation, and Colorectal Cancer, Annu. Rev. Microbiol., 2016, 70, 395–411 CrossRef CAS PubMed.
- B. C. Marta, P. C. José Pedro, D. R. Mário, F. L. M. Adelino and P. N. Pedro, Role of colonic microbiota in colorectal carcinogenesis: A systematic review, Rev. Esp. Enferm. Dig., 2015, 107, 659–671 Search PubMed.
- C. V. D. A. Almeida, M. R. Camargo, E. Russo and A. Amedei, Role of diet and gut microbiota on colorectal cancer, World J. Gastroenterol., 2019, 14, 151–162 Search PubMed.
- S. J. Bultman, Interplay between diet, gut microbiota, epigenetic events, and colorectal cancer, Mol. Nutr. Food Res., 2017, 61, 1500902 CrossRef PubMed.
- T. P. Nathan and E. C. Martens, The Critical Roles of Polysaccharides in Gut Microbial Ecology and Physiology, Annu. Rev. Microbiol., 2017, 71, 349–369 CrossRef PubMed.
- J. W. Li, L. P. Fan, S. D. Ding and X. L. Ding, Nutritional composition of five cultivars of chinese jujube, Food Chem., 2007, 103, 454–460 CrossRef CAS.
- Q. H. Gao, C. S. Wu and M. Wang, The jujube (Ziziphus jujuba Mill.) fruit: a review of current knowledge of fruit composition and health benefits, J. Agric. Food Chem., 2013, 61, 3351–3363 CrossRef CAS PubMed.
- J. H. Xie, W. Tang, M. L. Jin, J. E. Li and M. Y. Xie, Recent advances in bioactive polysaccharides from Lycium barbarum L., Zizyphus jujuba Mill, Plantago spp., and Morus spp.: Structures and functionalities, Food Hydrocolloids, 2016, 60, 148–160 CrossRef CAS.
- X. Ji, Q. Peng, Y. Yuan, J. Shen, X. Xie and M. Wang, Isolation, structures and bioactivities of the polysaccharides from jujube fruit(Ziziphus jujuba, Mill.): A review, Food Chem., 2017, 227, 349–357 CrossRef CAS PubMed.
- Y. L. Huang, G. C. Yen, F. Sheu and C. F. Chau, Effects of water-soluble carbohydrate concentrate from Chinese jujube on different intestinal and fecal indices, J. Agric. Food Chem., 2008, 56, 1734–1739 CrossRef CAS PubMed.
- Y. Yue, S. Wu, Z. Li, J. Li, X. Li, J. Xiang and H. Ding, Wild jujube polysaccharides protect against experimental inflammatory bowel disease by enabling enhanced intestinal barrier function, Food Funct., 2015, 6, 2568–2577 RSC.
- X. Ji, C. Hou, X. Zhang, L. Han, S. Yin, Q. Peng and M. Wang, Microbiome-metabolomic analysis of the impact of Zizyphus jujuba cv. Muzao polysaccharides consumption on colorectal cancer mice fecal microbiota and metabolites, Int. J. Biol. Macromol., 2019, 131, 1067–1076 CrossRef CAS PubMed.
- Q. Shang, H. Jiang, C. Cai, J. Hao, G. Li and G. Yu, Gut microbiota fermentation of marine polysaccharides and its effects on intestinal ecology: An overview, Carbohydr. Polym., 2018, 179, 173–185 CrossRef CAS PubMed.
- X. Xu, P. Xu, C. Ma, J. Tang and X. Zhang, Gut microbiota, host health, and polysaccharides, Biotechnol. Adv., 2013, 31, 318–337 CrossRef CAS PubMed.
- L. Liu, M. Li, M. Yu, M. Shen, Q. Wang, Y. Yu and J. Xie, Natural polysaccharides exhibit anti-tumor activity by targeting gut microbiota, Int. J. Biol. Macromol., 2019, 121, 743–751 CrossRef CAS PubMed.
- X. Zhang, M. Zhang, C.-T. Ho, X. Guo, Z. Wu, P. Weng, M. Yan and J. Cao, Metagenomics analysis of gut microbiota modulatory effect of green tea polyphenols by high fat diet-induced obesity mice model, J. Funct. Foods, 2018, 46, 268–277 CrossRef CAS.
- H. Koo, J. A. Hakim, M. L. Powell, R. Kumar, P. G. Eipers, C. D. Morrow, M. Crowley, E. J. Lefkowitz, S. A. Watts and A. K. Bej, Metagenomics approach to the study of the gut microbiome structure and function in zebrafish Danio rerio fed with gluten formulated diet, J. Microbiol. Methods, 2017, 135, 69–76 CrossRef CAS PubMed.
- X. Ji, Q. Peng, H. Li, F. Liu and M. Wang, Chemical Characterization and Anti-inflammatory Activity of Polysaccharides from Zizyphus jujube cv. Muzao, Int. J. Food Eng., 2017, 13 DOI:10.1515/ijfe-2016-0382.
- N. Hattori, T. Niwa, T. Ishida, K. Kobayashi, T. Imai, A. Mori, K. Kimura, T. Mori, Y. Asami and T. Ushijima, Antibiotics suppress colon tumorigenesis through inhibition of aberrant DNA methylation in an azoxymethane and dextran sulfate sodium colitis model, Cancer Sci., 2019, 110, 147–156 CrossRef CAS PubMed.
- X. Huo, D. Liu, L. Gao, L. Li and L. Cao, Flavonoids extracted from Licorice, prevents colitis-associated carcinogenesis in AOM/DSS mouse model, Int. J. Mol. Sci., 2016, 17, 1343 CrossRef PubMed.
- W. Wei, M. Yang, Y. Liu, H. Huang, C. Ye, J. Zheng, C. Guo, M. Hao, X. He and S. Zhu, Fertilizer N application rate impacts plant-soil feedback in a sanqi production system, Sci. Total Environ., 2018, 633, 796–807 CrossRef CAS PubMed.
- M. Cheng, X. Zhang, J. Zhu, L. Cheng, J. Cao, Z. Wu, P. Weng and X. Zheng, A metagenomics approach to the intestinal microbiome structure and function in high fat diet-induced obesity mice fed with oolong tea polyphenols, Food Funct., 2018, 9, 1079–1087 RSC.
- X. Zhang, Y. Chen, J. Zhu, M. Zhang, C. T. Ho, Q. Huang and J. Cao, Metagenomics Analysis of Gut Microbiota in a High Fat Diet-Induced Obesity Mouse Model Fed with (-)-Epigallocatechin 3-O-(3-O-Methyl) Gallate (EGCG3′′Me), Mol. Nutr. Food Res., 2018, 62, e1800274 CrossRef PubMed.
- Z. Yu, G. Song, J. Liu, J. Wang, P. Zhang and K. Chen, Beneficial effects of extracellular polysaccharide from Rhizopus nigricans on the intestinal immunity of colorectal cancer mice, Int. J. Biol. Macromol., 2018, 115, 718–726 CrossRef CAS PubMed.
- K. Esposito, P. Chiodini, A. Capuano, G. Bellastella, M. I. Maiorino, C. Rafaniello, D. B. Panagiotakos and D. Giugliano, Colorectal cancer association with metabolic syndrome and its components: a systematic review with meta-analysis, Endocrine, 2013, 44, 634–647 CrossRef CAS PubMed.
- J. Saetang and S. Sangkhathat, Diets link metabolic syndrome and colorectal cancer development (Review), Oncol. Rep., 2017, 37, 1312–1320 CrossRef CAS PubMed.
- A. Czajkowska and B. Szponar, Short chain fatty acids (SCFA), the products of gut bacteria metabolism and their role in the host, Postepy Hig. Med. Dosw., 2018, 72, 131–142 CrossRef.
- J. Fernández, S. Redondo-Blanco, I. Gutiérrez-del-Río, E. M. Miguélez, C. J. Villar and F. Lombó, Colon microbiota fermentation of dietary prebiotics towards short-chain fatty acids and their roles as anti-inflammatory and antitumour agents: A review, J. Funct. Foods, 2016, 25, 511–522 CrossRef.
- C. Tang, R. Ding, J. Sun, J. Liu, J. Kan and C. Jin, The impacts of natural polysaccharides on intestinal microbiota and immune responses-A review, Food Funct., 2019, 10, 2290–2312 RSC.
- M. Choct, Y. D. Li, J. McLeish and M. Peisker, Soy oligosaccharides and soluble non-starch polysaccharides: A review of digestion, nutritive and anti-nutritive fffects in pigs and poultry, Asian-Aust, J. Anim. Sci., 2010, 23, 1386–1398 CAS.
- F. A. D. Moura, F. T. Macagnan and L. P. da Silva, Oligosaccharide production by hydrolysis of polysaccharides: A review, Int. J. Food Sci. Technol., 2015, 50, 275–281 CrossRef.
- H. Liu, L. Ma and Q. Wang, Possible metabolic pathway of a novel bioactive polysaccharide extracted from Dendrobium, aphyllum: an in vivo study, J. Food Sci., 2019, 84, 1216–1223 CrossRef CAS PubMed.
- D. W. Cockburn and N. M. Koropatkin, Polysaccharide Degradation by the Intestinal Microbiota and Its Influence on Human Health and Disease, J. Mol. Biol., 2016, 428, 3230–3252 CrossRef CAS PubMed.
- X. Wang, T. Ye, W. J. Chen, Y. Lv, Z. Hao, J. Chen, J. Y. Zhao, H. P. Wang and Y. K. Cai, Structural shift of gut microbiota during chemopreventive effects of epigallocatechin gallate on colorectal carcinogenesis in mice, World J. Gastroenterol., 2017, 23, 8128–8139 CrossRef CAS PubMed.
- C. Z. Wang, W. H. Huang, C. F. Zhang, J. Y. Wan, Y. Wang, C. Yu, S. Williams, T. C. He, W. Du, M. W. Musch, E. B. Chang and C. S. Yuan, Role of intestinal microbiome in American ginseng-mediated colon cancer prevention in high fat diet-fed AOM/DSS mice, Clin. Transl. Oncol., 2018, 20, 302–312 CrossRef CAS PubMed.
- C. Tang, J. Sun, B. Zhou, C. Jin, J. Liu, J. Kan, C. Qian and N. Zhang, Effects of polysaccharides from purple sweet potatoes on immune response and gut microbiota composition in normal and cyclophosphamide treated mice, Food Funct., 2018, 9, 937–950 RSC.
- M. Marzorati, A. Verhelst, G. Luta, R. Sinnott, W. Verstraete, T. Van de Wiele and S. Possemiers, In vitro modulation of the human gastrointestinal microbial community by plant-derived polysaccharide-rich dietary supplements, Int. J. Food Microbiol., 2010, 139, 168–176 CrossRef CAS PubMed.
- M. L. Sanz, N. Polemis, V. Morales, N. Corzo, A. Drakoularakou, G. R. Gibson and R. A. Rastall, In vitro investigation into the potential prebiotic activity of honey oligosaccharides, J. Agric. Food Chem., 2005, 53, 2914–2921 CrossRef CAS PubMed.
- S. H. Duncan, A. Barcenilla, C. S. Stewart, S. E. Pryde and H. J. Flint, Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine, Appl. Environ. Microbiol., 2002, 68, 5186–5190 CrossRef CAS PubMed.
- Y. Zhong, M. Nyman and F. Fak, Modulation of gut microbiota in rats fed high-fat diets by processing whole-grain barley to barley malt, Mol. Nutr. Food Res., 2015, 59, 2066–2076 CrossRef CAS PubMed.
- D. Świątecka, I. Małgorzata, Ś. Aleksander, K. Henryk and K. Elżbieta, The impact of glycated pea proteins on bacterial adhesion, Food Res. Int., 2010, 43, 1566–1576 CrossRef.
- E. G. Zoetendal, M. Rajilic-Stojanovic and W. M. de Vos, High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota, Gut, 2008, 57, 1605–1615 CrossRef CAS PubMed.
- P. Truchado, E. Hernandez-Sanabria, B. N. Salden, P. Van den Abbeele, R. Vilchez-Vargas, R. Jauregui, D. H. Pieper, S. Possemiers and T. Van de Wiele, Long chain arabinoxylans shift the mucosa-associated microbiota in the proximal colon of the simulator of the human intestinal microbial ecosystem (M-SHIME), J. Funct. Foods, 2017, 32, 226–237 CrossRef CAS.
- G. Chihara, J. Hamuro, Y. Y. Maeda, T. Shiio, T. Suga, N. Takasuka and T. Sasaki, Antitumor and metastasis-inhibitory activities of lentinan as an immunomodulator: An overview, Cancer Detect. Prev. Suppl., 1987, 1, 423–443 CAS.
- J. Luo, C. Zhang, R. Liu, L. J. Gao, S. Ou, L. Liu and X. Peng, Ganoderma lucidum polysaccharide alleviating colorectal cancer by alteration of special gut bacteria and regulation of gene expression of colonic epithelial cells, J. Funct. Foods, 2018, 47, 127–135 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9fo02171j |
| This journal is © The Royal Society of Chemistry 2020 |