Screening of Lactobacillus salivarius strains from the feces of Chinese populations and the evaluation of their effects against intestinal inflammation in mice†
Qixiao
Zhai
 abc,
Xudan
Shen
ab,
Shi
Cen
ab,
Chengcheng
Zhang
ab,
Fengwei
Tian
abc,
Xudan
Shen
ab,
Shi
Cen
ab,
Chengcheng
Zhang
ab,
Fengwei
Tian
 abd,
Jianxin
Zhao
ab,
Hao
Zhang
abd,
Jianxin
Zhao
ab,
Hao
Zhang
 abdef,
Yuzheng
Xue
*g and
Wei
Chen
abdef,
Yuzheng
Xue
*g and
Wei
Chen
 *abeh
*abeh
aState Key Laboratory of Food Science and Technology, Jiangnan University, Wuxi, Jiangsu 214122, China. E-mail: chenwei66@jiangnan.edu.cn; Tel: +(86)510-85912155
bSchool of Food Science and Technology, Jiangnan University, Wuxi, Jiangsu 214122, China
cInternational Joint Research Laboratory for Probiotics at Jiangnan University, Wuxi, Jiangsu 214122, China
dWuxi Translational Medicine Research Center and Jiangsu Translational Medicine Research Institute Wuxi Branch, China
eNational Engineering Research Centre for Functional Food, Wuxi, Jiangsu 214122, China
f(Yangzhou) Institute of Food Biotechnology, Jiangnan University, Yangzhou 225004, China
gDepartment of Gastroenterology, Affiliated Hospital of Jiangnan University, Wuxi, Jiangsu 214122, China. E-mail: xueyz001@163.com; Tel: +(86)0510-81195162
hBeijing Innovation Centre of Food Nutrition and Human Health, Beijing Technology and Business University (BTBU), Beijing 100048, China
First published on 4th January 2020
Abstract
Lactobacillus salivarius is a species of lactic acid bacteria with probiotic potency. Compared to such well-known probiotics as L. rhamnosus and L. casei, the genomic characteristics and health-beneficial effects of L. salivarius are inadequately researched. For this study, a medium with enhanced selectivity for the isolation of L. salivarius was developed by optimizing the carbon source and antibiotics in the medium. Seventy-three L. salivarius strains were isolated from 472 fecal samples from Chinese populations, and their pan-genomic and phylogenetic characterizations were analyzed. Three strains (L. salivarius HN26-4, NT4-8, and FXJCJ7-2) that were clearly categorized in different sub-phylotypes of the phylogenetic tree were randomly selected for further studies. Compared to the other two tested strains, L. salivarius FXJCJ7-2 showed higher tolerance to simulated gastrointestinal tract conditions and more significant anti-inflammatory effects in lipopolysaccharides (LPS)-treated RAW264.7 murine macrophages. This strain was also more effective in reversing LPS-induced alterations in gut barrier function, colonic histopathology, Treg/Th-17 balance, immunomodulatory indicators, nuclear factor kappa B pathway activation, and the intestinal microenvironment of the mice than the other two tested strains. Comparative genomic analysis indicated that these protective effects may be related to the specific genes of L. salivarius FXJCJ7-2 that were involved in the tolerance to the gastrointestinal environment, short-chain fatty acid production, and host–bacterium interaction.
1. Introduction
Lactobacillus salivarius was originally isolated from human saliva and reported in 1953.1 This species was reported to have two subspecies as L. salivarius subsp. salivarius and L. salivarius subsp. salicinius, but a later study based on polyphasic analysis indicated that the assignment of subspecies status in L. salivarius species was without merit.2L. salivarius has been isolated from the oral cavity, intestines, and vagina of humans.3–5 This strain is also widely distributed in food, environments, and the gut of animals.6–8L. salivarius comprises both homofermentative and facultatively heterofermentative strains,2 and can produce metabolites such as short-chain fatty acids (SCFAs) and bacteriocins.9,10 This species is generally regarded as safe and has been listed as a recommended biological agent intentionally added to food by the European Food Safety Authority and Ministry of Public Health of China.11Based on the genome database of the National Center for Biotechnology Information (NCBI), United States, the genomes of 85 L. salivarius strains had been sequenced as of April 2019, far fewer than the genomes of other Lactobacillus species such as L. plantarum (361 strains), L. paracasei (164 strains) and L. rhamnosus (162 strains). The small number of isolated L. salivarius strains have limited research on the genomic and genetic characteristics of L. salivarius, and the available information is not as comprehensive as that about other Lactobacilli strains.12,13L. salivarius is a facultative anaerobe and can utilize a range of carbon sources including glucose, rhamnose, and trehalose.2,14 To the best of our knowledge, no medium with high selectivity for the isolation of L. salivarius has yet been developed.
Compared to well-known probiotics such as L. rhamnosus and L. casei, the beneficial effects of L. salivarius on human health are still largely unknown. Existing reports have confirmed that the probiotic properties of L. salivarius include the modulation of gut microbiota,15 the protection of the gut barrier,16 the prevention of colon cancer,17 and the induction of intestinal immune responses.18 Several studies have shown that oral administration of L. salivarius is effective against intestinal inflammation.19,20 The underlying mechanism may be related to the regulation by L. salivarius of the production and activity of regulatory T cells (Tregs), macrophages, natural killer cells, and dendritic cells.20–22
Based on these analyses, the aims of the study were to isolate L. salivarius strains from the fecal samples from Chinese populations using a modified medium with enhanced selectivity for L. salivarius. The genomic and functional characteristics of L. salivarius were further demonstrated by analyzing the genome sequences of the 73 isolated and 29 publicly available strains. Finally, the strains’ anti-inflammatory effects in the gut were evaluated in a mouse model, and the potential protective mechanisms were investigated.
2. Materials and methods
2.1. Bacterial strains
L. salivarius, L. plantarum, L. reuteri, L. paracasei, L. mucosae, L. antri, L. rhamnosus, L. fermentum, L. paralimentarius, and Weissella cibaria strains were isolated from the fecal samples of Chinese populations and preserved in the in-house Culture Collections of Food Microbiology, Jiangnan University (Wuxi, China). The strains were cultured in basic de Man, Rogosa, and Sharpe (MRS) broth. The MRS broth (pH 7.0) contained 20 g L−1 glucose, 10 g L−1 peptone, 5 g L−1 yeast extract, 10 g L−1 beef extract, 2 g L−1 anhydrous sodium acetate, 2 g L−1 hydrogen diamine citrate, 2.6 g L−1 K2HPO4·3H2O, 0.5 g L−1 MgSO4·7H2O, 0.25 g L−1 MnSO4·H2O, 1 g L−1 Tween-80, 20 g L−1 agar, and 15 g L−1 bromcresol purple. The chemicals for the MRS broth were obtained from Sinopharm Chemical Reagent Company, Shanghai, China.2.2. Development of a medium with enhanced selectivity for L. salivarius isolation
To screen out an optimized carbon source for the isolation of L. salivarius, the glucose in basic MRS was replaced by mannitol, raffinose, sorbitol, or rhamnose (Sinopharm Chemical Reagent Company, Shanghai, China). The L. salivarius strains were cultured in basic MRS broth and carbon source–modified MRS media at 37 °C under an anaerobic condition for 12 to 18 h. The growth of strains was determined by the color changes of bromcresol purple in the medium or by colony counting.23To screen for the most suitable antibiotics for the isolation of L. salivarius, gentamicin, streptomycin, chloramphenicol, linezolid, trimethoprim, and clindamycin (Sangon Biological Co., Ltd, Shanghai, China) were added to the basic MRS medium at the different doses shown in Table 1. The antibiotic tolerances of strains were determined by the minimum inhibitory concentration (MIC) approach.24
| Antibiotics | Concentrations (μg ml−1) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| The antibiotics were added to the media after sterilization at a temperature <55 °C and pH of 5.0 ± 0.1. | |||||||||||
| Gentamicin | 0 | 0.50 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 |
| Streptomycin | 0 | 0.50 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 |
| Chloramphenicol | 0 | 0.13 | 0.25 | 0.50 | 1 | 2 | 4 | 8 | 16 | 32 | 64 |
| Linezolid | 0 | 0.03 | 0.06 | 0.13 | 0.25 | 0.50 | 1 | 2 | 4 | 8 | 16 |
| Trimethoprim | 0 | 0.13 | 0.25 | 0.50 | 1 | 2 | 4 | 8 | 16 | 32 | 64 |
| Clindamycin | 0 | 0.03 | 0.06 | 0.13 | 0.25 | 0.50 | 1 | 2 | 4 | 8 | 16 |
LAMVAB medium is an MRS-based medium containing an additional dose of vancomycin at 0.02 g L−1 and with a lower pH at 5.0 ± 0.1.25 This medium has been reported to be highly selective for the isolation of lactobacilli from fecal samples.25 LAMVAB medium was modified in the present study by replacing the carbon sources and adding antibiotics. To determine the selectivity of the modified media for the isolation of L. salivarius, 12 fecal samples from people in Jilin Province, China, were incubated at 37 °C for 18 h under an anaerobic condition in basic and modified LAMVAB broth, respectively. The cultured bacterial suspension was then incubated on a basic MRS plate at 37 °C for 48 h. For the identification of bacteria, colony PCR was performed with primers 27F AGAGTTTGATCCTGGCTCAG and 1492R TACGGCTACCTTGTTACGACTT using 2× Taq Master mix. The selectivity of the modified medium for L. salivarius isolation was expressed as the percentage of isolated L. salivarius colony counts on the plate using the following expression:
| Isolated L. salivarius (%) = 100% × C1/C0; |
2.3. Isolation and genome sequencing of L. salivarius from fecal samples of Chinese populations
Fecal samples of Chinese populations residing in 15 provinces or municipalities were incubated in the modified medium under an anaerobic condition for the isolation of L. salivarius. Each single colony of L. salivarius was picked and cultured in normal MRS broth, and the genomic DNA was then extracted using a bacterial DNA extraction kit (OMEGA, Biotech, Doraville, GA, USA). Whole-genome sequencing was performed using the Illumina HiSeq Platform to generate 350 bp pair-end reads following the manufacturer's instructions. The software SOAPdenovo v2.0.4 was used to assemble paired-end reads de novo into high-quality sequences, and scaffolds shorter than 200 bp were discarded.27 The inner gaps within each scaffold were filled, and single-base errors were corrected by the software GapCloser (http://sourceforge.net/projects/soapdenovo2/files/GapCloser/). The genome assemblies of 73 L. salivarius strains sequenced in this work have been deposited in the NCBI under the project number PRJNA561366.2.4. Pan-genome analysis, phylogeny reconstruction, and clusters of orthologous groups (COG) annotation of L. salivarius
All 102 L. salivarius strains (29 from NCBI and 73 sequenced in this work) was re-annotated using Prokka,28 and the pan-genome analysis was conducted via Roary to identify the core genes of the L. salivarius species.29 The single-nucleotide polymorphisms (SNPs) of all included genomes were detected as previously reported.30 Each genome was aligned to the reference genome (L. salivarius UCC118) using MUMmer.31 The neighbor-joining tree was constructed based on the sequences of concatenated bi-allelic SNPs using the TreeBeST tool (http://treesoft.sourceforge.net/treebest.shtml). For the identification of differences in the functional genes among strains with different probiotic phenotypes, the COG annotation was conducted against the COG database by BLASTp.322.5. Determination of the tolerance of L. salivarius strains to simulated gastrointestinal tract conditions
Simulated gastric and intestinal juices were prepared as previously reported with minor modifications.33 The cells of the L. salivarius strains (1 × 109 cfu mL−1) were first incubated in simulated gastric juices at 37 °C for 3 h, then transferred to simulated intestinal juices and incubated at 37 °C for 4 h. The tolerance to simulated gastrointestinal tract conditions was expressed as the survival of strains after incubation.342.6. Determination of the anti-inflammatory effects of L. salivarius strains in lipopolysaccharides-treated RAW264.7 murine macrophages
The in vitro cell trial was performed as previously reported with minor modifications.35 Briefly, RAW264.7 cells were cultured until 80 to 90% confluency, and then 1 × 106 cells were seeded in each well of a six-well culture plate for 24 h. Then, 250 ng of lipopolysaccharides (LPS, Sigma-Aldrich, St Louis, MO, USA) and/or 1 × 108 cfu of L. salivarius strains and L. rhamnosus GG (as a positive control strain) were administered to each well. The cells were incubated at 37 °C in a 5% CO2 atmosphere for 24 h and the levels of interleukin-10 (IL-10) in the supernatants were measured via an enzyme-linked immunosorbent assay (ELISA) kit obtained from Fcmacs, Nanjing, China.2.7. Animal experiment
C57BL/6 mice (male, 8 weeks old, weighing 26–28 g, Shanghai Laboratory Animal Center, Shanghai, China) were used in the animal trials. The mice were kept in stainless-steel cages in a room with a temperature at 25 ± 2 °C and humidity of 50 ± 5%. The mice had free access to commercial mouse food and drinking water. The mice were acclimatized for one week before the experiment.All of the protocols for the animal trials in the study were approved by the Ethics Committee of Jiangnan University, China (JN. No20180615b1200901[142]). The trial procedures were carried out in accordance with European Community guidelines (directive 2010/63/EU) for the care and use of experimental animals.
The anti-inflammatory effects of three L. salivarius strains including L. salivarius HN26-4, L. salivarius NT4-8, and L. salivarius FXJCJ7-2 were evaluated in mice treated with LPS (Sigma-Aldrich, St Louis, MO, USA). Mice were randomly assigned to one of the following groups: control, LPS, HN26-4, NT4-8, FXJCJ7-2, LPS + HN26-4, LPS + NT4-8, and LPS + FXJCJ7-2. The animal trial lasted for 5 days. The procedure for the animal experiment is shown in Table 2. The dose of LPS was selected based on a previous study to induce inflammation in the gut of mice.36 At the end of the experiment, mice were weighted, fasted for 6 h, and sacrificed under light ether anesthesia.
| Group (no. of mice) | Protocola |
|---|---|
| a Saline, 0.2 mL saline once daily via intraperitoneal injection. PBS, 0.2 mL phosphate buffered solution once daily via oral gavage. LPS, 0.2 mL saline containing 0.15 mg kg−1 lipopolysaccharides once daily via intraperitoneal injection. L. salivarius NT4-8/HN26-4/FXJCJ7-2, 0.2 mL phosphate buffered solution containing L. salivarius NT4-8/HN26-4/FXJCJ7-2 at 5 × 109 cfu mL−1 once daily via oral gavage. The oral gavage was performed 1 h after the intraperitoneal injection. | |
| Control (n = 10) | Saline + PBS |
| LPS (n = 10) | LPS + PBS |
| HN26-4 (n = 10) | Saline + L. salivarius HN26-4 |
| NT4-8 (n = 10) | Saline + L. salivarius NT4-8 |
| FXJCJ7-2 (n = 10) | Saline + L. salivarius FXJCJ7-2 |
| LPS + HN26-4 (n = 10) | LPS + L. salivarius HN26-4 |
| LPS + NT4-8 (n = 10) | LPS + L. salivarius NT4-8 |
| LPS + FXJCJ7-2 (n = 10) | LPS + L. salivarius FXJCJ7-2 |
2.8. Determination of the intestinal permeability of mice
The intestinal permeability of the mice was evaluated using 4000Da-fluorescent-dextran-FITC (DX-4000-FITC, Sigma-Aldrich, St Louis, MO, USA) as previously described.372.9. Determination of the histopathological changes in the colon of mice
Colonic tissues were collected and stained with hematoxylin and eosin (H&E) as previously described.38 The histopathological changes in the colonic section were examined by light microscopy.2.10. Determination of the levels of CD4+CD25+Foxp3+ regulatory T cells in the spleen of mice
The levels of CD4+CD25+Foxp3+ Tregs in the spleens of the mice were measured as previously described.36 Briefly, fresh spleen was immediately homogenized after isolation and splenocytes were harvested. The cells were stained by CD4, CD25, and Foxp3 using a Mouse Regulatory T Cell staining kit (eBioscience, Shanghai, China) and the levels of CD4+CD25+Foxp3+ Tregs were analyzed by flow cytometry (FACSCalibur, BD Biosciences, USA).2.11. Determination of the levels of immunomodulatory indicators in the serum and colon of mice
The levels of IL-17 and IL-6 in the serum, and the levels of IL-10, IL-1β, tumor necrosis factor alpha (TNF-α), and secretory immunoglobulin A (sIgA) in the colon of the mice were determined by ELISA kits obtained from SBJ-Bio, Nanjing, Jiangsu, China.2.12. Determination of the inflammation-related proteins in the colon of mice
The colonic tissues (40 mg) were homogenized with RIPA lysis buffer (400 μL, Beyotime Biotechnology, Jiangsu, China). The mixture was centrifuged at 4 °C for 20 min and the supernatant was collected for analysis. The expression of iκBα and phosphor-iκBα in the colon of mice was determined by western blot assay as previously described.362.13. Profiling of the gut microbiota of mice
Fecal DNA extraction, sequencing (16S rRNA V3–V4 region) and data processing were performed as previously described.392.14. Determination of SCFAs in colonic contents of mice
The levels of SCFAs in the colonic contents of the mice were analyzed by gas chromatography–mass spectrometry (GCMS-QP2010 Ultra system, Shimadzu Corporation, Japan) as previously described.402.15. Statistical analysis
Data are expressed as the mean ± standard deviation (SD). A minimum of three independent experiments were carried out for each in vitro assay. Differences between means were analyzed using two-tailed Student's t tests or one-way analysis of variance, followed by Tukey's post-hoc test. A P value of <0.05 was considered to be statistically significant. Statistical analyses of the obtained data were performed and visualized using GraphPad (GraphPad Software, San Diego, CA, USA).3. Results
3.1. Selection of carbon sources and antibiotics for the isolation of L. salivarius from fecal samples
Ten species of lactic acid bacteria showed varying utilizations of rhamnose, raffinose, mannitol, and sorbitol (Table 3). No carbon source was exclusively utilized by L. salivarius. Compared to the greater numbers of species utilizing raffinose (n = 9) or mannitol (n = 7), only 4 species of lactic acid bacteria (L. salivarius, L. plantarum, L. paracasei, and L. rhamnosus) can use rhamnose or sorbitol as the only carbon sources. Rhamnose was less favorable for the growth of L. plantarum and L. paracasei than sorbitol (Table 4, P < 0.05). Taking the resistances of these four species of lactic acid bacteria to antibiotics into consideration (Table 5), rhamnose was selected as the only carbon source of the medium for further study.| Carbon sources | LS | LP | LR | LPC | LM | LA | LRH | LF | LPL | WC |
|---|---|---|---|---|---|---|---|---|---|---|
| The utilization of each carbon source was evaluated in at least five strains in each species. + indicates that at least one strain in the species can utilize the carbon source and induce a color change in the medium. − indicates that no strain in the species can utilize the carbon source. LS, L. salivarius; LP, L. plantarum; LR, L. reuteri; LPC, L. paracasei; LM, L. mucosae; LA, L. antri; LRH, L. rhamnosus; LF, L. fermentum; LPL, L. paralimentarius; WC, W. cibaria. | ||||||||||
| Rhamnose | + | + | − | + | − | − | + | − | − | − |
| Raffinose | + | + | + | + | + | + | + | + | + | − |
| Mannitol | + | + | + | + | − | − | + | + | − | + |
| Sorbitol | + | + | − | + | − | − | + | − | − | − |
| Carbon sources | Bacterial concentrations (108 cfu ml−1 broth) | |||
|---|---|---|---|---|
| LS | LP | LPC | LRH | |
| The lactic acid bacteria were inoculated in the broth at 106 cfu mL−1 and incubated at 37 37 °C for 18 h. The assay was performed in at least five strains in each species. The superscript letters a, b, and c indicate statistically significant differences at P values of <0.05 among the different carbon source groups. LS, L. salivarius; LP, L. plantarum; LPC, L. paracasei; LRH, L. rhamnosus. | ||||
| Glucose | 1.01 ± 0.09a | 23.30 ± 1.67a | 9.32 ± 0.86a | 8.70 ± 0.48a |
| Sorbitol | 2.18 ± 0.23b | 25.38 ± 4.15a | 28.42 ± 1.79b | 6.28 ± 0.39b |
| Rhamnose | 1.13 ± 0.14a | 2.82 ± 0.22b | 4.38 ± 0.28c | 17.00 ± 1.76c |
| Antibiotics (μg ml−1) | LS | LP | LPC | LRH |
|---|---|---|---|---|
| The lactic acid bacteria were inoculated in the broth at 106 cfu ml−1 and incubated at 37 37 °C for 18 h. The lowest concentration of each antibiotic that completely inhibited the growth of strains was considered the MIC. MIC was evaluated in at least five strains in each species. LS, L. salivarius; LP, L. plantarum; LPC, L. paracasei; LRH, L. rhamnosus. | ||||
| Gentamicin | >64 | >64 | >128 | >16 |
| Streptomycin | >256 | >256 | >128 | >8 |
| Chloramphenicol | >0.50 | >1 | >1 | >2 |
| Linezolid | >0.13 | >0.25 | >0.50 | >1 |
| Trimethoprim | >2 | >4 | >32 | >64 |
| Clindamycin | >0.03 | >0.13 | >0.06 | >0.13 |
Based on the analysis of the MIC of lactic acid bacteria for antibiotics (Table 5), we did not find a dose of antibiotic that can guarantee the viability of L. salivarius but prohibit the growth of the other three lactic acid bacteria. Streptomycin at 256 μg mL−1 was most suitable for the isolation of L. salivarius, in that it specifically inhibited the growth of L. paracasei and L. rhamnosus.
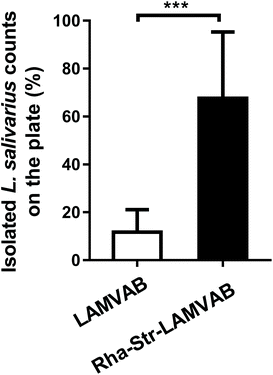
We therefore replaced the glucose in the LAMVAB medium with rhamnose and added 256 μg mL−1 of streptomycin into the medium to formulate a modified medium (Rha-Str-LAMVAB) for L. salivarius isolation. Compared to basic LAMVAB, the rate for L. salivarius isolation in the Rha-Str-LAMVAB medium (67.61 ± 27.67%) was significantly higher than in basic LAMVAB medium (11.75 ± 9.38%, Fig. 1, P < 0.001).
3.2. The genomic and genetic characteristics of L. salivarius strains isolated from fecal samples of Chinese populations
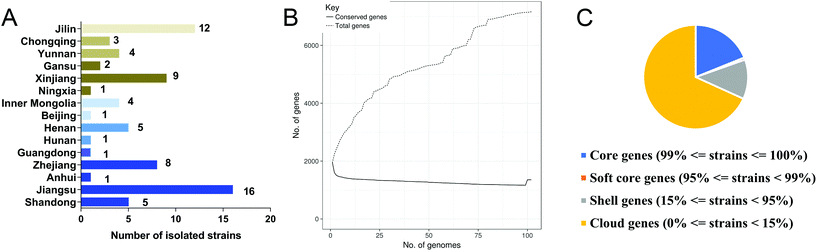
Isolation of 73 L. salivarius strains from fecal samples of Chinese populations from 15 provinces or municipalities was carried out in the modified medium (Fig. 2A). Total genome sizes for these strains ranged from 1.69 to 2.13 Mb, with a mean value of 1.96 ± 0.07 Mb. The GC content varied from 32.57 to 33.05%, with an average value of 32.74 ± 0.09%. The genome sequences of the 73 isolated strains and 29 strains retrieved from NCBI database (Table S1†) were annotated, and the pan-genome of L. salivarius species was analyzed. Of the 7163 genes identified, 1353 of them were core genes shared by more than 99% of the strains, showing a high genomic diversity characterized by a large accessory genome (81% of total gene number, Fig. 2B, C and S1†). These included L. salivarius strains appeared to capture the whole genomic imprints of the species, as indicated by the plateau of total genes with the accruing genomes (Fig. 2B). A phylogenetic tree based on the bi-SNPs in the core genome of these strains was constructed (Fig. S2†), indicating that all isolated L. salivarius strains were classified into three separate sub-phylotypes.3.3. Selection of three L. salivarius strains for the animal trial
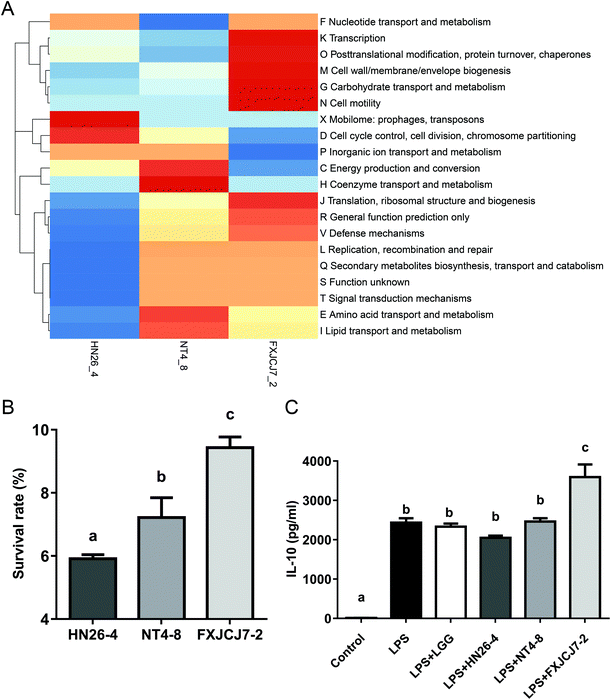
Three L. salivarius strains (L. salivarius HN26-4, NT4-8, and FXJCJ7-2) that were clearly categorized in different sub-phylotypes were randomly selected for further animal trial and genomic analysis. Three strains harbored different COG repertoires (Fig. 3A). As shown in Table 6, a total of 19 genes was uniquely observed in L. salivarius FXJCJ7-2 when compared to the other two analyzed strains.| Geno no.a | Hit nameb | COG ID | Functional description | COG |
|---|---|---|---|---|
| a The corresponding gene tag in L. salivarius FXJCJ7-2. b The hit gene name corresponded with each gene of FXJCJ7-2 in COG database. | ||||
| LS_0669 | TM_0244 | COG4656 | Predicted NADH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ubiquinone oxidoreductase, subunit RnfC ubiquinone oxidoreductase, subunit RnfC |
[C] Energy production and conversion |
| LS_0660 | BMEI0257 | COG3938 | Proline racemase | [E] Amino acid transport and metabolism |
| LS_0670 | Cj1504c | COG0709 | Selenophosphate synthase | [E] Amino acid transport and metabolism |
| LS_0674 | yvbG | COG2095 | Multiple antibiotic transporter | [E] Amino acid transport and metabolism |
| LS_0621 | SPy_1676 | COG0021 | Transketolase | [G] Carbohydrate transport and metabolism |
| LS_0679 | lin2981 | COG1070 | Sugar (pentulose and hexulose) kinases | [G] Carbohydrate transport and metabolism |
| LS_0681 | lin2980 | COG4806 | L-Rhamnose isomerase | [G] Carbohydrate transport and metabolism |
| LS_0665 | SMa0015 | COG3276 | Selenocysteine-specific translation elongation factor | [J] Translation, ribosomal structure and biogenesis |
| LS_0667 | aq_1031 | COG1921 | Selenocysteine synthase [seryl-tRNASer selenium transferase] | [J] Translation, ribosomal structure and biogenesis |
| LS_0799 | kdgR | COG1414 | Transcriptional regulator | [K] Transcription |
| LS_0694 | MA_2364 | COG4930 | Predicted ATP-dependent Lon-type protease | [O] Posttranslational modification, protein turnover, chaperones |
| LS_0663 | AF_0165 | COG0425 | Predicted redox protein, regulator of disulfide bond formation | [O] Posttranslational modification, protein turnover, chaperones |
| LS_0653 | Z2789 | COG2897 | Rhodanese-related sulfurtransferase | [P] Inorganic ion transport and metabolism |
| LS_1820 | BH2935 | COG1228 | Imidazolonepropionase and related amidohydrolases | [Q] Secondary metabolites biosynthesis, transport and catabolism |
| LS_0651 | yeeE | COG2391 | Predicted transporter component | [R] General function prediction only |
| LS_0675 | BH2011 | COG1062 | Zn-Dependent alcohol dehydrogenases, class III | [R] General function prediction only |
| LS_0551 | yhai | COG3152 | Predicted membrane protein | [S] Function unknown |
| LS_1650 | CAC0024 | COG4713 | Predicted membrane protein | [S] Function unknown |
| LS_1803 | fraH | COG1716 | FOG: FHA domain | [T] Signal transduction mechanisms |
L. salivarius FXJCJ7-2 showed significantly higher tolerance to the simulated gastrointestinal environment than the other two tested strains (Fig. 3B, P < 0.05). This strain also offered more significant effects on improving the level of IL-10 in LPS-treated RAW264.7 cells than the other tested strains (Fig. 3C, P < 0.05).
3.4. Effects of L. salivarius supplementation on the gut barrier function of LPS-treated mice
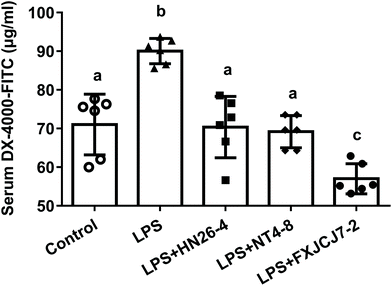
Oral administration of L. salivarius strains themselves did not affect the gut barrier function of the mice, as no significant differences in serum FITC levels were observed among control and L. salivarius-only treated groups (Fig. S3,†P > 0.05). Compared to the control group, LPS treatment markedly increased serum FITC levels in mice. These alterations were recovered by the co-treatment with LPS and L. salivarius (Fig. 4, P < 0.05). Among the three tested strains, L. salivarius FXJCJ7-2 showed the most significant protective effects against LPS-induced increase in intestinal permeability (Fig. 4).3.5. Effects of L. salivarius supplementation on the colonic histopathology of LPS-treated mice
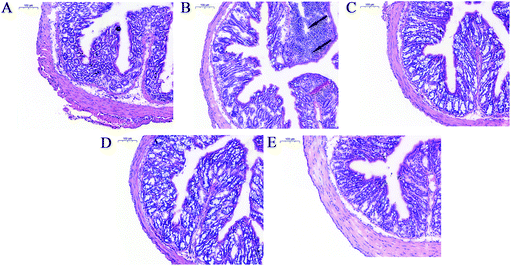
Compared to the animals in the control group, mice receiving LPS treatment showed pathological lesions including inflammatory cell infiltration and crypt architecture disruption in the colon (Fig. 5A and B). Co-treatment with LPS and L. salivarius provided significant protection against these lesions and restored the intestinal mucosa structure (Fig. 5C–E). Oral administration of L. salivarius strains themselves did not induce any pathological alterations in the colon of the mice (Fig. S4†).3.6. Effects of L. salivarius supplementation on the Treg/Th-17 balance of LPS-treated mice
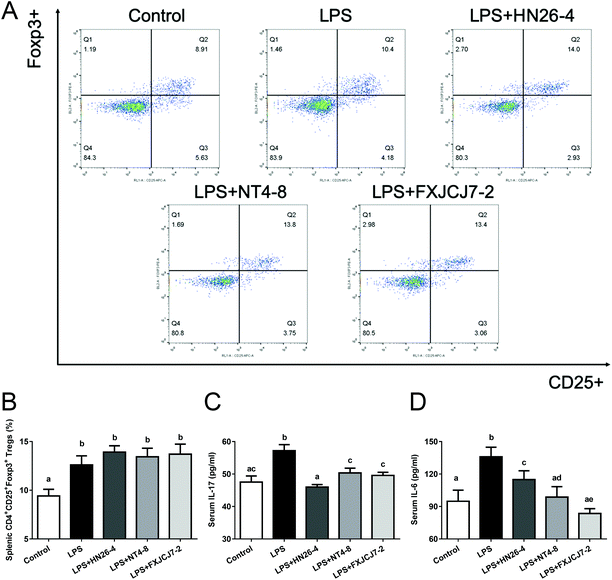
Compared to the LPS-only treatment, co-treatment with LPS and L. salivarius did not significantly altere the percentages of splenic Tregs in mice (Fig. 6A and B). However, the co-treatment markedly recovered LPS-induced changes in serum IL-17 and IL-6 levels (Fig. 6C and D, P < 0.05). Oral administration of the three L. salivarius strains themselves increased the percentages of splenic Tregs when compared to the control group (Fig. S5A and B,†P < 0.05) but showed no significant effects on the levels of serum IL-17 and IL-6 in mice (Fig. S5C and D†).3.7. Effects of L. salivarius supplementation on the levels of colonic immunomodulatory indicators of LPS-treated mice
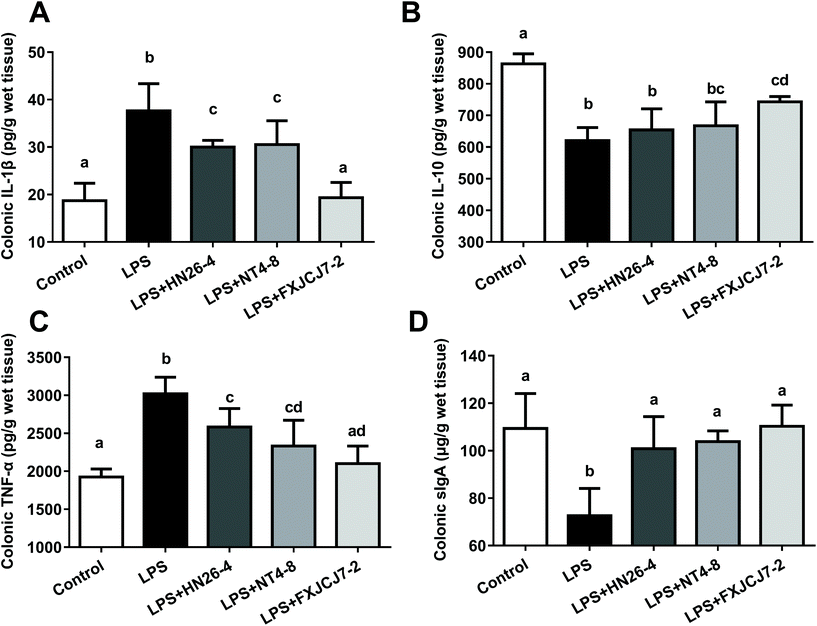
LPS treatment significantly increased the levels of colonic IL-1β and TNF-α but reduced levels of colonic IL-10 and sIgA (Fig. 7, P < 0.05). Co-treatments with LPS and L. salivarius were protective against LPS-induced alterations in these colonic immunomodulatory indicators, and L. salivarius FXJCJ7-2 exhibited more significant recovery effects than the other two strains. Oral administration of L. salivarius strains themselves did not induce alterations in these indicators (Fig. S6†).3.8. Effects of L. salivarius supplementation on the nuclear factor kappa B (NF-κB) pathway of LPS-treated mice
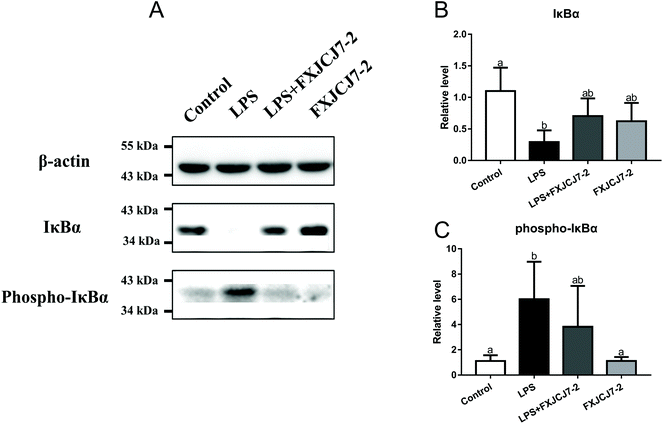
A significant increase in the expression of phosphor-iκBα and a reduction in that of total iκBα were observed in the colon of the mice after treatment with LPS alone (Fig. 8). Co-treatments with LPS and L. salivarius FXJCJ7-2 prohibited the enhancement of the phosphor-iκBα and recovered the original level of total iκBα. Treatment with L. salivarius FXJCJ7-2 alone did not activate the NF-κB pathway, as little iκBα protein was phosphorylated.3.9. Effects of L. salivarius supplementation on the intestinal microenvironment of LPS-treated mice
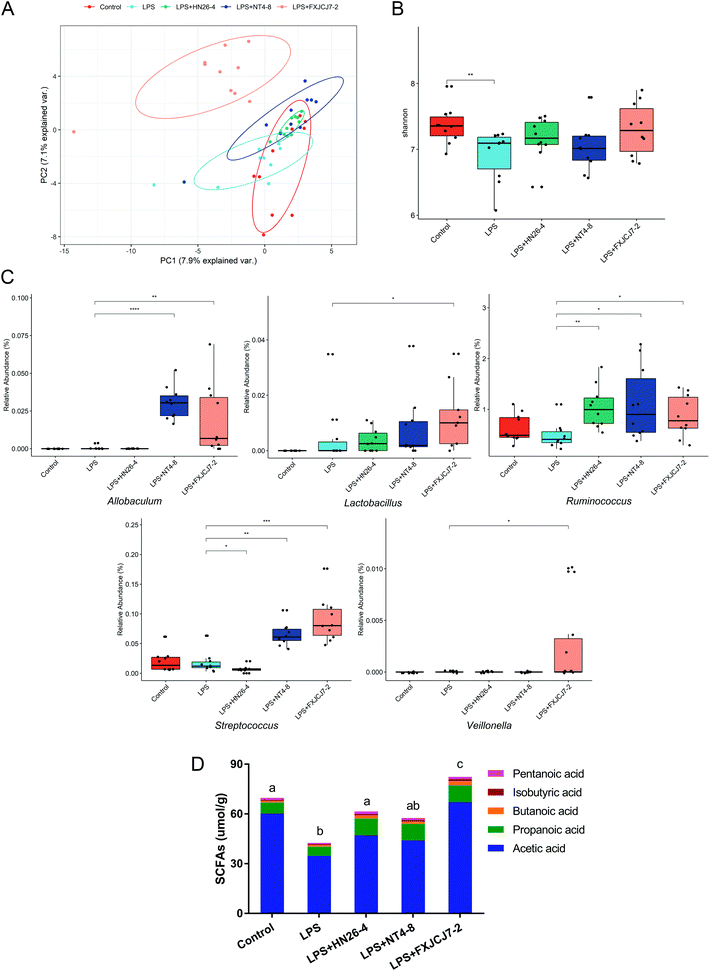
LPS treatment did not significantly affect the composition of the gut microbiota but induced a marked decrease in microbial diversity (Fig. 9A and B). Co-treatments with LPS and L. salivarius strains, especially L. salivarius FXJCJ7-2, significantly altered gut microbiota structure (Fig. 9A, C and S7†). Compared to LPS treatment alone, the abundances of Allobaculum, Lactobacillus, Streptococcus, Veillonella, and Ruminococcus were up-regulated after the co-treatment with LPS and L. salivarius FXJCJ7-2 (Fig. 9C, P < 0.05).LPS treatment significantly reduced the levels of acetic acid, isobutyric acid, pentanoic acid, and total SCFAs in the colonic contents of the mice (Fig. 9D and Table S2,†P < 0.05). These alterations were reversed by the co-treatment with LPS and L. salivarius. Among the tested strains, L. salivarius FXJCJ7-2 showed the most marked effects on the recovery of SCFAs in the colonic contents.
4. Discussion
The intestinal tract of humans is a vital habitat for L. salivarius strains.41 However, other enterogenous lactic acid bacteria may possess nutritional and physiological properties that are similar to those of L. salivarius and thus may interfere with the isolation of this species.42 Therefore, a selective medium for the isolation of L. salivarius strains from human fecal samples was first developed in the present study. Based on the carbohydrate utilization characteristics of different Lactobacillus species reported in Bergey's Manual,42 rhamnose, raffinose, mannitol, and sorbitol were selected as potential carbon sources for L. salivarius isolation, as these carbohydrates can only be utilized by a relatively small number of Lactobacillus species (including L. salivarius). The utilization of these four carbohydrates was further evaluated in 10 species of lactic acid bacteria that were commonly isolated from human fecal samples,43 and rhamnose was selected as the optimized carbon source for L. salivarius isolation (Tables 3 and 4). Different species of lactic acid bacteria have been reported to show varied resistances to antibiotics.44 As shown in Table 5, a dose of streptomycin at 256 μg mL−1 was simultaneously selected to further enhance the isolation efficiency of L. salivarius strains. The rate for L. salivarius isolation in the modified medium was six times higher than the basic LAMVAB medium, which significantly accelerated our work for the screening of L. salivarius strains from Chinese populations.A phylogenetic tree of L. salivarius showed that all isolates from Chinese human feces belonged to sub-types I, III, and IV, whereas all strains in South Korea isolated from pig feces clustered (as downloaded from the NCBI database) into sub-type II. Such genotype dissimilarity may be due to the geographical separation and/or niche difference of L. salivarius strains, as previously reported for L. reuteri.45 Notably, the Chinese isolates intermixed and cannot be clearly clustered along the tree in terms of provinces, indicating a geographical transmission of L. salivarius strains, which may correspond with the migration and mobility of human populations. Three L. salivarius strains (L. salivarius HN26-4, NT4-8, and FXJCJ7-2) that were clearly categorized in sub-type I, III, and IV were randomly selected for further studies.
LPS has been reported to activate the Toll-like receptor 4 signal transduction pathway, disrupt intestinal tight junctions, and induce enterocolitis and inflammatory bowel disease.46 In the present study, oral administration of three L. salivarius strains showed protective effects against LPS-induced intestinal inflammation in mice, as they recovered the gut barrier function (Fig. 4), alleviated abnormal colonic histopathology (Fig. 5), and prevented LPS-induced alterations in the levels of colonic immunomodulatory indicators (Fig. 7). Th17 cells, a subset of CD4+ lymphocytes induced by IL-6 and transforming growth factor (TGF)-β, have been reported to trigger inflammation via secretion of the pro-inflammatory cytokine IL-17.47 The inflammatory activity of Th17 can be attenuated by Tregs that inhibit T-cell proliferation and autoimmune responses.48 In the present study, both Tregs in the spleen and IL-17 and IL-6 levels in the serum were increased after LPS treatment (Fig. 6). Similar results have been reported in a previous study focusing on the effects of LPS stimulation on chronic obstructive pulmonary disease.49 The authors proposed that LPS treatment induced more significant release of inflammatory cytokines (such as IL-17 and IL-6) than that of anti-inflammatory cytokines (such as IL-10), thus leading to a exacerbation of the inflammatory response.49 The LPS-induced disruption of the Treg/Th17 balance was recovered by administration of three L. salivarius strains (Fig. 6), which may explain the anti-inflammatory effects of these strains. The protective mode is in accordance with a previous study showing that the intestinal commensal microbe Bifidobacterium infantis 35624 can ameliorate LPS-induced intestinal inflammation by inhibiting the production of pro-inflammatory cytokines and activating the generation and function of Tregs.50
Taking all the above-mentioned in vivo biological indicators into consideration, L. salivarius FXJCJ7-2 showed more significant protective effects against LPS-induced intestinal inflammation in mice than the other tested L. salivarius strains. Analyses of NF-κB pathway activation and of gut microbiota were performed to further explain the underlying protective mechanism of L. salivarius FXJCJ7-2. Previous reports have indicated that LPS stimulation may trigger the phosphorylation of IκBα, thus activating the NF-κB pathway and inducing inflammatory responses.51,52 As shown in Fig. 8, co-treatment with LPS and L. salivarius FXJCJ7-2 significantly prevented the phosphorylation of IκBα, which can partly explain its anti-inflammatory property. Moreover, L. salivarius FXJCJ7-2 administration significantly increased the abundances of such SCFAs-producing intestinal microbes as Allobaculum, Lactobacillus, Streptococcus, Veillonella, and Ruminococcus (Fig. 9C).53,54 Consistently, the levels of SCFAs in the colonic contents were markedly increased after the co-treatment with LPS and L. salivarius FXJCJ7-2. It has been reported that SCFAs can regulate colonic Treg and Th1 cell homeostasis, induce the production of IL-10, and alleviate intestinal inflammation.55,56 This may be another protective mode of L. salivarius FXJCJ7-2 against LPS-induced inflammatory responses.
The variations in anti-inflammation properties among these three L. salivarius strains were further explained by comparative genomic analysis (Fig. 3A and Table 6). Among the 19 strain-specific genes for L. salivarius FXJCJ7-2, the gene LS_0660 may enable the strain to produce proline, a water-attracting osmolyte that offers protection against bile salt stress on L. salivarius cells.57 Genes LS_0665 and LS_0667 are responsible for selenocysteine synthesis and transportation. These genes have also been reported to enhance the tolerance of lactic acid bacteria to environmental stress.58 Supporting these analyses, L. salivarius FXJCJ7-2 exhibited superior tolerance to the simulated gastrointestinal tract conditions over the other two strains (Fig. 3B). This guarantees a higher number of viable L. salivarius FXJCJ7-2 cells in the colon, which could offer more marked protective effects against intestinal inflammation than the other two strains. Supporting these analyses, a species-specific quantitative real-time PCR approach confirmed that oral administration of L. salivarius FXJCJ7-2 induced a higher abundance of L. salivarius species than the other two tested strains in the feces of mice (unpublished data). Genes LS_0679 and LS_0681 are responsible for the metabolism of rare sugars. These genes have been reported to be related to the SCFA production abilities of probiotics in the colon and are protective against pathogenic infection in mice.59 Consistently, L. salivarius FXJCJ7-2 was more effective in recovering the levels of SCFAs in the colonic contents of LPS-treated mice than the other two strains (Fig. 9D and Table S2†). Moreover, gene LS_0621 encoding transketolase may be related to the immunomodulative effects of L. salivarius FXJCJ7-2, in that transketolase has been reported to be involved in the flagellation of bacteria60 and flagellin is important in the immune interaction between commensal bacteria and the host.61 This is supported by results from the RAW264.7 cell trial showing that L. salivarius FXJCJ7-2 treatment offered more significant effects on the modulation of anti-inflammatory cytokines (Fig. 3C) than the other two tested strains.
5. Conclusion
In this study, 73 L. salivarius strains were isolated from fecal samples of Chinese populations by a modified selective medium. Three strains that were clearly categorized in different sub-phylotypes of the phylogenetic tree were randomly selected for the evaluation of their effects against intestinal inflammation in mice. All tested strains reversed LPS-induced alterations in gut barrier function, colonic histopathology, Treg/Th-17 balance, colonic immunomodulatory indicators, and the intestinal microenvironment of the mice. L. salivarius FXJCJ7-2 offered a more significant protective effect than the other two strains, which may be related to its specific genes involved in the tolerance to a gastrointestinal environment, SCFA production, and host–bacterium interaction.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by the National Natural Science Foundation of China Program (No. 31601452 and 31871773), National First-Class Discipline Program of Food Science and Technology (JUFSTR20180102), Collaborative Innovation Center of Food Safety and Quality Control in Jiangsu Province and BBSRC Newton Fund Joint Centre Award (BB/J004529/1).References
- M. Rogosa, R. Wiseman, J. A. Mitchell, M. Disraely and A. Beaman, Species differentiation of oral Lactobacilli from man including descriptions of Lactobacillus salivarius, nov spec and Lactobacillus cellobiosus nov spec, J. Bacteriol., 1953, 65, 681–699 CrossRef CAS.
- Y. Li, E. Raftis, C. Canchaya, G. F. Fitzgerald, D. van Sinderen and P. W. O'Toole, Polyphasic analysis indicates that Lactobacillus salivarius subsp. salivarius and Lactobacillus salivarius subsp. salicinius do not merit separate subspecies status, Int. J. Syst. Evol. Microbiol., 2006, 56, 2397–2403 CrossRef CAS.
- H. Mandal and T. Bagchi, In vitro screening of indigenous Lactobacillus, isolates for selecting organisms with better health-promoting attributes, Appl. Biochem. Biotechnol., 2018, 185, 1060–1074 CrossRef CAS.
- X. Heng-Yi, T. Wan-Hong, W. Cui-Xiang, J. Li-Jun, W. Lan-Yin, Y. Jing, L. Chun-Mei, Z. Ming and W. Hua, Antagonistic potential against pathogenic microorganisms and hydrogen peroxide production of indigenous, Lactobacilli isolated from vagina of Chinese pregnant women, Biomed. Environ. Sci., 2008, 21, 365–371 CrossRef.
- P. Švec, I. Sedláček, L. Žáčková, D. Nováková and M. Kukletová, Lactobacillus spp. associated with early childhood caries, Folia Microbiol., 2009, 54, 53–58 CrossRef PubMed.
- D. I. Ayala, P. W. Cook, D. L. Campos, M. M. Brashears, H. den Bakker and K. K. Nightingale, Draft genome sequence of Lactobacillus salivarius, L28 isolated from ground beef, Genome Announce., 2017, 5, e00955–e00917 Search PubMed.
- A. A. Soro-Yao, P. Schumann, P. Thonart, K. M. Djè and R. Pukall, The use of MALDI-TOF mass spectrometry, ribotyping and phenotypic tests to identify lactic acid bacteria from fermented cereal foods in Abidjan (Côte d'Ivoire), Open Microbiol. J., 2014, 8, 78–86 CrossRef PubMed.
- X.-H. Guo, J.-M. Kim, H.-M. Nam, S.-Y. Park and J.-M. Kim, Screening lactic acid bacteria from swine origins for multistrain probiotics based on in vitro functional properties, Anaerobe, 2010, 16, 321–326 CrossRef.
- M. Thirabunyanon and P. Hongwittayakorn, Potential probiotic lactic acid bacteria of human origin induce antiproliferation of colon cancer cells via synergic actions in adhesion to cancer cells and short-chain fatty acid bioproduction, Appl. Biochem. Biotechnol., 2013, 169, 511–525 CrossRef CAS PubMed.
- E. A. Svetoch, B. V. Eruslanov, V. P. Levchuk, V. V. Perelygin, E. V. Mitsevich, I. P. Mitsevich, J. Stepanshin, I. Dyatlov, B. S. Seal and N. J. Stern, Isolation of Lactobacillus salivarius, 1077 (NRRL B-50053) and characterization of its bacteriocin, including the antimicrobial activity spectrum, Appl. Environ. Microbiol., 2011, 77, 2749–2754 CrossRef CAS.
- EFSA Panel on Biological Hazards (BIOHAZ), Update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 4: suitability of taxonomic units notified to EFSA until March 2016, EFSA J., 2016, 14, e04522 Search PubMed.
- E. J. Raftis, E. Salvetti, S. Torriani, G. E. Felis and P. W. O'Toole, Genomic diversity of Lactobacillus salivarius, Appl. Environ. Microbiol., 2011, 77, 954–965 CrossRef CAS PubMed.
- M. J. Claesson, Y. Li, S. Leahy, C. Canchaya, J. P. van Pijkeren, A. M. Cerdeño-Tárraga, J. Parkhill, S. Flynn, G. C. O'Sullivan and J. K. Collins, Multireplicon genome architecture of Lactobacilluss salivarius, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 6718–6723 CrossRef CAS.
- G. Zayed and Y. H. Roos, Influence of trehalose and moisture content on survival of Lactobacillus salivarius subjected to freeze-drying and storage, Process Biochem., 2004, 39, 1081–1086 CrossRef CAS.
- M. Ahamed and M. K. J. Siddiqui, Environmental lead toxicity and nutritional factors, Clin. Nutr., 2007, 26, 400–408 CrossRef CAS PubMed.
- S. Messaoudi, A. Madi, H. Prévost, M. Feuilloley, M. Manai, X. Dousset and N. Connil, In vitro evaluation of the probiotic potential of Lactobacillus salivarius SMXD51, Anaerobe, 2012, 18, 584–589 CrossRef CAS PubMed.
- M. Zhang, X. Fan, B. Fang, C. Zhu, J. Zhu and F. Ren, Effects of Lactobacillus salivarius Ren on cancer prevention and intestinal microbiota in 1, 2-dimethylhydrazine-induced rat model, J. Microbiol., 2015, 53, 398–405 CrossRef PubMed.
- D. Ren, C. Li, Y. Qin, R. Yin, S. Du, F. Ye, C. Liu, H. Liu, M. Wang and Y. Li, In vitro evaluation of the probiotic and functional potential of Lactobacillus, strains isolated from fermented food and human intestine, Anaerobe, 2014, 30, 1–10 CrossRef CAS PubMed.
- L. Peran, D. Camuesco, M. Comalada, A. Nieto, A. Concha, M. P. Diaz-Ropero, M. Olivares, J. Xaus, A. Zarzuelo and J. Galvez, Preventative effects of a probiotic, Lactobacillus salivarius ssp. salivarius, in the TNBS model of rat colitis, World J. Gastroenterol., 2005, 11, 5185–5192 Search PubMed.
- E. M. Fernandez, V. Valenti, C. Rockel, C. Hermann, B. Pot, I. G. Boneca and C. Grangette, Anti-inflammatory capacity of selected Lactobacilli in experimental colitis is driven by NOD2-mediated recognition of a specific peptidoglycan-derived muropeptide, Gut, 2011, 60, 1050–1059 CrossRef CAS PubMed.
- S. Sierra, F. Lara-Villoslada, L. Sempere, M. Olivares, J. Boza and J. Xaus, Intestinal and immunological effects of daily oral administration of Lactobacillus salivarius CECT5713 to healthy adults, Anaerobe, 2010, 16, 195–200 CrossRef PubMed.
- I. Elawadli, J. Brisbin, B. Mallard, M. Griffiths, M. Corredig and S. Sharif, Differential effects of Lactobacilli on activation and maturation of mouse dendritic cells, Benefic. Microbes, 2014, 5, 323–334 CrossRef CAS PubMed.
- H. Kaplan and R. W. Hutkins, Fermentation of fructooligosaccharides by lactic acid bacteria and Bifidobacteria, Appl. Environ. Microbiol., 2000, 66, 2682–2684 CrossRef CAS PubMed.
- R. Abou-Shanab, P. Van Berkum and J. Angle, Heavy metal resistance and genotypic analysis of metal resistance genes in Gram-positive and Gram-negative bacteria present in Ni-rich serpentine soil and in the rhizosphere of Alyssum murale, Chemosphere, 2007, 68, 360–367 CrossRef CAS PubMed.
- R. Hartemink, V. Domenech and F. Rombouts, LAMVAB—a new selective medium for the isolation of Lactobacilli from faeces, J. Microbiol. Methods, 1997, 29, 77–84 CrossRef CAS.
- N. Tharmaraj and N. Shah, Selective enumeration of Lactobacillus delbrueckii, ssp. bulgaricus, Streptococcus thermophilus, Lactobacillus acidophilus, Bifidobacteria, Lactobacillus casei, Lactobacillus rhamnosus, and propionibacteria, J. Dairy Sci., 2003, 86, 2288–2296 CrossRef CAS PubMed.
- R. Luo, B. Liu, Y. Xie, Z. Li, W. Huang, J. Yuan, G. He, Y. Chen, Q. Pan and Y. Liu, SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler, GigaScience, 2012, 1, 18 CrossRef PubMed.
- T. Seemann, Prokka: rapid prokaryotic genome annotation, Bioinformatics, 2014, 30, 2068–2069 CrossRef CAS PubMed.
- A. J. Page, C. A. Cummins, M. Hunt, V. K. Wong, S. Reuter, M. T. Holden, M. Fookes, D. Falush, J. A. Keane and J. Parkhill, Roary: rapid large-scale prokaryote pan genome analysis, Bioinformatics, 2015, 31, 3691–3693 CrossRef CAS PubMed.
- Y. Cui, X. Yang, X. Didelot, C. Guo, D. Li, Y. Yan, Y. Zhang, Y. Yuan, H. Yang and J. Wang, Epidemic clones, oceanic gene pools, and eco-LD in the free living marine pathogen Vibrio parahaemolyticus, Mol. Biol. Evol., 2015, 32, 1396–1410 CrossRef CAS PubMed.
- A. L. Delcher, S. L. Salzberg and A. M. Phillippy, Using MUMmer to identify similar regions in large sequence sets, Curr. Protoc. Bioinf., 2003, 10, 10.3.1–10.3.18 CrossRef.
- R. L. Tatusov, M. Y. Galperin, D. A. Natale and E. V. Koonin, The COG database: a tool for genome-scale analysis of protein functions and evolution, Nucleic Acids Res., 2000, 28, 33–36 CrossRef CAS PubMed.
- Y. Huang and M. C. Adams, In vitro assessment of the upper gastrointestinal tolerance of potential probiotic dairy propionibacteria, Int. J. Food Microbiol., 2004, 91, 253–260 CrossRef PubMed.
- Q. Zhai, R. Yin, L. Yu, G. Wang, F. Tian, R. Yu, J. Zhao, X. Liu, Y. Q. Chen and H. Zhang, Screening of lactic acid bacteria with potential protective effects against cadmium toxicity, Food Control, 2015, 54, 23–30 CrossRef CAS.
- B. R. Beck, G. S. Park, Y. H. Lee, S. Im, D. Y. Jeong and J. Kang, Whole genome analysis of Lactobacillus plantarum Strains isolated from kimchi and determination of probiotic properties to treat mucosal infections by Candida albicans and Gardnerella vaginalis, Front. Microbiol., 2019, 10, 433 CrossRef PubMed.
- H. Tan, J. Zhao, H. Zhang, Q. Zhai and W. Chen, Novel strains of Bacteroides fragilis and Bacteroides ovatus alleviate the LPS-induced inflammation in mice, Appl. Microbiol. Biotechnol., 2019, 103, 2353–2365 CrossRef CAS PubMed.
- Q. Zhai, F. Tian, J. Zhao, H. Zhang, A. Narbad and W. Chen, Oral administration of probiotics inhibits absorption of the heavy metal cadmium by protecting the intestinal barrier, Appl. Environ. Microbiol., 2016, 82, 4429–4440 CrossRef CAS PubMed.
- Q. Zhai, G. Wang, J. Zhao, X. Liu, F. Tian, H. Zhang and W. Chen, Protective effects of Lactobacillus plantarum CCFM8610 against acute cadmium toxicity in mice, Appl. Environ. Microbiol., 2013, 79, 1508–1515 CrossRef CAS PubMed.
- Q. Zhai, S. Cen, P. Li, F. Tian, J. Zhao, H. Zhang and W. Chen, Effects of dietary selenium supplementation on intestinal barrier and immune responses associated with its modulation of gut microbiota, Environ. Sci. Technol. Lett., 2018, 5, 724–730 CrossRef CAS.
- B. Mao, D. Li, C. Ai, J. Zhao, H. Zhang and W. Chen, Lactulose differently modulates the composition of luminal and mucosal microbiota in C57BL/6J mice, J. Agric. Food Chem., 2016, 64, 6240–6247 CrossRef CAS PubMed.
- F. Dal Bello, J. Walter, W. Hammes and C. Hertel, Increased complexity of the species composition of lactic acid bacteria in human feces revealed by alternative incubation condition, Microb. Ecol., 2003, 45, 455–463 CrossRef CAS PubMed.
- W. P. Hammes and C. Hertel, Lactobacillus, Bergey's manual of systematics of archaea and bacteria, John Wiley & Sons, Inc., in association with Bergey's Manual Trust, 2015, pp. 1–76 Search PubMed.
- H. G. Heilig, E. G. Zoetendal, E. E. Vaughan, P. Marteau, A. D. Akkermans and W. M. de Vos, Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA, Appl. Environ. Microbiol., 2002, 68, 114–123 CrossRef CAS PubMed.
- M. S. Ammor, A. B. Flórez and B. Mayo, Antibiotic resistance in non-enterococcal lactic acid bacteria and Bifidobacteria, Food Microbiol., 2007, 24, 559–570 CrossRef CAS PubMed.
- P. L. Oh, A. K. Benson, D. A. Peterson, P. B. Patil, E. N. Moriyama, S. Roos and J. Walter, Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution, ISME J., 2010, 4, 377–387 CrossRef PubMed.
- S. Guo, M. Nighot, R. Al-Sadi, T. Alhmoud, P. Nighot and T. Y. Ma, Lipopolysaccharide regulation of intestinal tight junction permeability is mediated by TLR4 signal transduction pathway activation of FAK and MyD88, J. Immunol., 2015, 195, 4999–5010 CrossRef CAS PubMed.
- I. I. Ivanov, B. S. McKenzie, L. Zhou, C. E. Tadokoro, A. Lepelley, J. J. Lafaille, D. J. Cua and D. R. Littman, The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells, Cell, 2006, 126, 1121–1133 CrossRef CAS PubMed.
- E. V. Dang, J. Barbi, H.-Y. Yang, D. Jinasena, H. Yu, Y. Zheng, Z. Bordman, J. Fu, Y. Kim and H.-R. Yen, Control of TH17/Treg balance by hypoxia-inducible factor 1, Cell, 2011, 146, 772–784 CrossRef CAS PubMed.
- D. A. Cervilha, J. T. Ito, J. D. Lourenço, C. R. Olivo, B. M. Saraiva-Romanholo, R. A. Volpini, M. C. Oliveira-Junior, T. Mauad, M. A. Martins and I. F. Tibério, The Th17/Treg Cytokine Imbalance in Chronic obstructive pulmonary disease exacerbation in an animal model of cigarette smoke exposure and lipopolysaccharide challenge association, Sci. Rep., 2019, 9, 1921 CrossRef PubMed.
- C. O'Mahony, P. Scully, D. O'Mahony, S. Murphy, F. O'Brien, A. Lyons, G. Sherlock, J. MacSharry, B. Kiely and F. Shanahan, Commensal-induced regulatory T cells mediate protection against pathogen-stimulated NF-κB activation, PLoS Pathog., 2008, 4, e1000112 CrossRef PubMed.
- J. López-Moreno, S. García-Carpintero, R. Jimenez-Lucena, C. Haro, O. A. Rangel-Zúñiga, R. Blanco-Rojo, E. M. Yubero-Serrano, F. J. Tinahones, J. Delgado-Lista and P. Pérez-Martínez, Effect of dietary lipids on endotoxemia influences postprandial inflammatory response, J. Agric. Food Chem., 2017, 65, 7756–7763 CrossRef PubMed.
- E. A. El-Shoura, B. A. Messiha, S. M. Sharkawi and R. A. Hemeida, Perindopril ameliorates lipopolysaccharide-induced brain injury through modulation of angiotensin-II/angiotensin-1-7 and related signaling pathways, Eur. J. Pharmacol., 2018, 834, 305–317 CrossRef CAS PubMed.
- A. Koh, F. De Vadder, P. Kovatcheva-Datchary and F. Bäckhed, From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites, Cell, 2016, 165, 1332–1345 CrossRef CAS PubMed.
- X. Zhang, Y. Zhao, M. Zhang, X. Pang, J. Xu, C. Kang, M. Li, C. Zhang, Z. Zhang and Y. Zhang, Structural changes of gut microbiota during berberine-mediated prevention of obesity and insulin resistance in high-fat diet-fed rats, PLoS One, 2012, 7, e42529 CrossRef CAS PubMed.
- M. Sun, W. Wu, L. Chen, W. Yang, X. Huang, C. Ma, F. Chen, Y. Xiao, Y. Zhao and C. Ma, Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis, Nat. Commun., 2018, 9, 3555 CrossRef PubMed.
- P. M. Smith, M. R. Howitt, N. Panikov, M. Michaud, C. A. Gallini, M. Bohlooly-y, J. N. Glickman and W. S. Garrett, The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis, Science, 2013, 341, 569–573 CrossRef CAS PubMed.
- G. Wang, D. Li, X. Ma, H. An, Z. Zhai, F. Ren and Y. Hao, Functional role of oppA encoding an oligopeptide-binding protein from Lactobacillus salivarius Ren in bile tolerance, J. Ind. Microbiol. Biotechnol., 2015, 42, 1167–1174 CrossRef CAS PubMed.
- E. Galano, E. Mangiapane, J. Bianga, A. Palmese, E. Pessione, J. Szpunar, R. Lobinski and A. Amoresano, Privileged incorporation of selenium as selenocysteine in Lactobacillus reuteri, proteins demonstrated by selenium-specific imaging and proteomics, Mol. Cell. Proteomics, 2013, 12, 2196–2204 CrossRef CAS PubMed.
- K. Hase, Bifidobacteria can protect from enteropathogenic infection through production of acetate, Nature, 2012, 469, 543–547 Search PubMed.
- K.-I. Sasajima and T. Kumada, Deficiency of flagellation in a Bacillus subtilis, pleiotropic mutant lacking transketolase, Agric. Biol. Chem., 1983, 47, 1375–1376 CAS.
- F. Hayashi, K. D. Smith, A. Ozinsky, T. R. Hawn, C. Y. Eugene, D. R. Goodlett, J. K. Eng, S. Akira, D. M. Underhill and A. Aderem, The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5, Nature, 2001, 410, 1099–1103 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9fo02116g |
| This journal is © The Royal Society of Chemistry 2020 |