Effects of ornithine α-ketoglutarate on growth performance and gut microbiota in a chronic oxidative stress pig model induced by D-galactose
Yuying
Li
 ab,
Peng
Wang
ab,
Jie
Yin
ab,
Peng
Wang
ab,
Jie
Yin
 abc,
Shunshun
Jin
ab,
Wenxuan
Su
ab,
Junquan
Tian
ab,
Tiejun
Li
abd and
Kang
Yao
*abd
abc,
Shunshun
Jin
ab,
Wenxuan
Su
ab,
Junquan
Tian
ab,
Tiejun
Li
abd and
Kang
Yao
*abd
aHunan Provincial Key Laboratory of Animal Nutritional Physiology and Metabolic Process, Key Laboratory of Agro-ecological Processes in Subtropical Region, Institute of Subtropical Agriculture, Chinese Academy of Sciences, National Engineering Laboratory for Pollution Control and Waste Utilization in Livestock and Poultry Production, Hunan Provincial Engineering Research Center for Healthy Livestock and Poultry Production, Scientific Observing and Experimental Station of Animal Nutrition and Feed Science in South-Central, Ministry of Agriculture, Changsha, Hunan 410125, P. R. China. E-mail: yaokang@isa.ac.cn
bUniversity of Chinese Academy of Sciences, Beijing 100039, China
cDepartment of Animal Science, Hunan Agriculture University, Changsha, Hunan 410128, China
dHunan Co-Innovation Center of Animal Production Safety, Changsha, Hunan 410128, China
First published on 26th November 2019
Abstract
The aim of this study was to evaluate the protective effects and underlying mechanisms of ornithine α-ketoglutarate (OKG) on D-galactose (D-gal)-induced chronic oxidative stress in a pig model. A total of 40 castrated young pigs were randomly separated into five groups, including a control group, a model group treated with 5 mg per kg body weight (BW) D-gal, and three D-gal + OKG groups in which the pigs received 0.5%, 1%, and 2% OKG (n = 8). The experiment lasted for 28 days. The growth performance, serum oxidative stress index, expression of relative intestinal genes, gut microbiota, and serum amino acid pool were determined. The results demonstrated that administration of D-gal significantly affected growth performance and superoxide dismutase (SOD) and glutathione peroxidase (GSH-Px) levels including related mRNA expression suppression, malondialdehyde (MDA) levels enhancement, gut microbiota dysfunction, and serum amino acid alteration in pigs. However, treatment with 0.5% OKG markedly ameliorated the reduction in the growth performance, as evidenced by the reversed final body weight, average feed intake, and average body weight. Also, 0.5% OKG enhanced the SOD and GSH-Px levels including relative mRNA expression in the intestine and inhibited lipid oxidation subsequent to MDA generation. The intestinal abundances of Firmicutes were increased and those of Proteobacteria, Fusobacteria, Bacteriodetes, and Euryarchaeota were decreased in the pigs supplemented with 0.5% OKG. Meanwhile, 0.5% OKG increased the glutamate, proline, aspartate, threonine, valine, isoleucine and leucine levels in the serum. Collectively, these results indicate that D-gal induced chronic oxidative stress and also proved the positive effects of 0.5% OKG on altering the pig gut microbe, restoring serum amino acid and alleviating the growth-suppression induced by D-gal chronic oxidative stress.
1. Introduction
With the growing population and the changing environment, chronic oxidative stress is increasingly becoming a major public health concern worldwide, affecting the health of humans and animals.1 Chronic oxidative stress is an imbalance between reactive oxygen species (ROS) and antioxidant agents, and is implicated in the pathogenesis of chronic oxidative disease. D-Galactose (D-gal) is mainly derived from lactose in the milk of animals and can induce chronic oxidative stress and mild inflammation. For instance, D-gal readily reacts with free amino acid amines in proteins and peptides in vivo and in vitro to form advanced glycation end products2 that elevate ROS production, decrease the activity of mitochondrial respiration complexes and cause chronic oxidation stress damage.1,3,4 Previous studies have shown that D-gal in mice causes cognitive impairment,3 neurotoxicity,5 hepatic disorders,1 and chronic kidney injury.6 Feng et al. investigated whether chronic subcutaneous administration with D-gal(100 mg kg−1) induced the activation of oxidative stress and inflammation in the liver and kidneys.6 In addition, rats administered with D-gal(100 mg kg−1) had increased oxidative DNA damage and renal podocyte loss.2 In our previous study, treatment of weaned pigs for three weeks with 5 and 10 g kg−1 day−1D-gal induced oxidative stress and inhibited the growth performance (unpublished data).Ornithine α-ketoglutarate (OKG) is a nutriment salt composed of two molecules of ornithine and one molecule of α-ketoglutarate, and has been used for centuries owing to its therapeutic and anabolic properties,7,8 including: increasing bone mineral density,9 curing burns and trauma,10 and its anti-stress properties.11,12 For example, short-term pretreatment with OKG before induction of ischemia-reperfusion decreased tissue damage, and increased pyruvate utilization for energy production in the Krebs cycle.13 OKG (0.5, 1.5 and 4.5 g kg−1 day−1) increased tissue glutamine concentration and N accumulation.14 Notably, OKG is not simply generated by AKG and Orn, but also the proline, arginine and glutamine levels precursor in vivo.7,15 Previous studies have reported that OKG can more effectively induce insulin secretion and the growth hormone than either alone16,17 and increases pyruvate utilization for energy production in the Krebs cycle.13 In a pig oxidative stress model, diquat, LPS or H2O2 are usually used to induce oxidative stress.18–20 However, injection of diquat, LPS or H2O2 can cause severe stress that is difficult to control in the body. Moreover, the effect of OKG on the growth performance and gut microbiota in a chronic oxidative stress pig model remains to be elicited. Thus, this study aims to investigate whether OKG alleviates the damage caused by chronic oxidative stress in growing pigs challenged with D-gal.
2. Materials and methods
2.1. Animals and groups
A total of 40 castrated young pigs (Landrace × Yorkshire, 28 days old, 7.68 ± 0.56 kg) were randomly assigned to one of five groups (n = 8 per group): a control group in which pigs were fed with a basal diet (see Table 1) according to NRC (2012), a model group in which pigs were fed with a basal diet with 5 mg per kg body weight D-galactose and another three groups in which pigs were fed with a basal diet with 5 mg per kg body weight D-galactose and 0.5% OKG (low OKG, SOKG), 1% OKG (middle OKG, MOKG), or 2% OKG (high OKG, LOKG), respectively. The test was started after pre-feeding for three days. Each pig was housed in a single cage and had free access to drinking water and the respective diet for 28 days. At the end of the feeding experiment, six pigs were randomly selected from each group and killed for sample collection. This study was carried out in accordance with the recommendations of the Declaration of Helsinki. All animal procedures were approved by the Committee on Animal Care of the Institute of Subtropical Agriculture, Chinese Academy of Sciences.| Feed ingredients (%) | Basal diet |
|---|---|
a The premix provided the following per kg of diet: nicotinic acid 50 mg, pantothenic acid 5 mg, folic acid 2 mg, biotin 0.2 mg, VA 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 IU, VD3 4000 IU, VE 40 IU, VK 3 4 mg, VB1 6 mg, VB2 12 mg, VB6 6 mg, VB12 0.05 mg, Cu 45 mg, Fe 90 mg, Mn 37 mg, Zn 41 mg, and Se 0.3 mg. 800 IU, VD3 4000 IU, VE 40 IU, VK 3 4 mg, VB1 6 mg, VB2 12 mg, VB6 6 mg, VB12 0.05 mg, Cu 45 mg, Fe 90 mg, Mn 37 mg, Zn 41 mg, and Se 0.3 mg.
|
|
| Soybean meal, 43 | 25.50 |
| Corn | 70.00 |
| L-Lysine | 0.10 |
| L-Cysteine | 0.02 |
| Choline | 0.07 |
| Betaine | 0.02 |
| Complex enzyme | 0.06 |
| Sweetener | 0.04 |
| Acidifier | 0.26 |
| Antioxidant | 0.03 |
| CaHPO4 | 0.63 |
| Limestone | 1.82 |
| NaCl | 0.45 |
| Premixa | 1.00 |
| Total | 100.00 |
| Nutritional level | |
| Digestible energy MJ kg−1 | 13.61 |
| Crude protein | 17.00 |
| Lysine | 1.56 |
| Methionine | 0.50 |
| Threonine | 0.91 |
| Tryptophan | 0.25 |
| Calcium | 0.94 |
| Phosphorus | 0.44 |
2.2. Growth performance
During the entire experimental period, the daily feed intake from each pig was recorded and all pigs were weighed individually at the beginning and at the end of the experiment. The average daily feed intake (ADFI), average daily gain (ADG), and the ratios of the feed intake to weight gain (F/G) were calculated according to the feed consumption and weight of each pig.2.3. Serum oxidative stress index
Blood samples were obtained from the vascular vein. Serum was prepared via centrifugation at 3000g and 4 °C for 10 min and stored at −80 °C until analysis.21 Malondialdehyde (MDA), superoxide dismutase (SOD), glutathione peroxidase (GSH-Px), and catalase (CAT) in serum were measured using assay kits in accordance with the manufacturer's instructions (Beyotime Biotechnology, China)192.4. Isolation, characterization and bioinformatics of microbiota
Total DNA samples from the terminal ileum were extracted using the cetyltrimethylammonium bromide/sodium dodecylsulfate CTAB/SDS method and DNA concentration and purity was determined using 1% agarose gels.22,23 16S rDNA genes of distinct regions were amplified using the specific primer with the barcode. Then, the products were purified with a GeneJETTM Gel Extraction Kit (Thermo Scientific).24 Sequencing libraries were generated using the Ion Plus Fragment Library Kit 48 rxns (Thermo Scientific) following the manufacturer's recommendations. The library quality was assessed on the Qubit@2.0 Fluorometer (Thermo Scientific). Lastly, the library was sequenced on an Ion S5TM XL platform and 400 bp/600 bp single-end reads were generated.25,26Single-end reads were assigned to samples based on their unique barcode and truncated by cutting off the barcode and primer sequence.27 Quality filtering on the raw reads was performed under specific filtering conditions to obtain the high-quality clean reads according to the Cutadapt28,29 (V1.9.1, http://cutadapt.readthedocs.io/en/stable/) quality controlled process. The reads were compared with the reference database (Silva database, https://www.arb-silva.de/)30 using the UCHIME algorithm (UCHIME Algorithm, http://www.drive5.com/usearch/manual/uchime_algo.html)31,32 to detect chimera sequences, and then the chimera sequences were removed.33 Then, the clean reads were finally obtained. Sequence analysis was performed using Uparse software (Uparse v7.0.1001, http://drive5.com/uparse/).34 Sequences with greater than or equal to 97% similarity were assigned to the same operational taxonomic units (OTUs). A representative sequence for each operational taxonomic unit (OTU) was screened for further annotation. For each representative sequence, the Silva Database (https://www.arb-silva.de/)30 was used based on the Mothur algorithm to annotate the taxonomic information. In order to study the phylogenetic relationship of different OTUs, and the difference between the dominant species in different samples (groups), multiple sequence alignments were conducted using the MUSCLE software (Version 3.8.31, http://www.drive5.com/muscle/).35 The abundance information of the OTUs was normalized using a standard sequence number corresponding to the sample with the least sequences. Subsequent analysis of the alpha diversity and beta diversity were all performed based on this output normalized data. The alpha diversity is applied to analyze the complexity of the species diversity in a sample. All of the indices in our samples were calculated using QIIME (Version1.7.0) and displayed with R software (Version 2.15.3).36
2.5. Amino acid determination
Eighteen amino acids (lysine, methionine, threonine, tryptophan, glutamate, aspartate, valine, isoleucine, leucine, phenylalanine, arginine, serine, histone, glycine, alanine, proline, cysteine, and tyrosine) in the serum were detected via a Hitachi L-8900 automatic amino acid analyzer according to the method used in our previous reports.192.6. Real-time quantitative polymerase chain reaction
The total RNA from intestine samples was isolated with TRIZOL reagent (Invitrogen, USA) and then treated with DNase I (Invitrogen, USA). Reverse transcription was performed at 37 °C for 15 min, and 95 °C for 5 s. The primers used in this study were designed using Primer 5.0 according to the pig gene sequence (Table 2). β-Actin was chosen as the house-keeping gene to normalize the target gene levels. The PCR cycling conditions were 36 cycles at 94 °C for 40 s, 60 °C for 30 s and 72 °C for 35 s.| Gene | Gene bank no. | Sequence (5′–3′) |
|---|---|---|
| a F: Forward; R: reverse. | ||
| β-Actin | DQ845171.1 | F: CTGCGGCATCCACGAAACT |
| R: AGGGCCGTGATCTCCTTCTG | ||
| GPx1 | NM_214201 | F: TGGGGAGATCCTGAATTG |
| R: GATAAACTTGGGGTCGGT | ||
| GPx4 | NM_214407.1 | F: GATTCTGGCCTTCCCTTGC |
| R: TCCCCTTGGGCTGGACTTT | ||
| MnSOD | NM_214127 | F: GGACAAATCTGAGCCCTAACG |
| R: CCTTGTTGAAACCGAGCC | ||
| CuZnSOD | NM_001190422 | F: CAGGTCCTCACTTCAATCC |
| R: CCAAACGACTTCCASCAT | ||
The relative expression was expressed as the ratio of the target gene to the control gene using the formula 2−(ΔΔCt), in which ΔΔCt = (CtTarget − Ctβ-actin)treatment − (CtTarget − Ctβ-actin)control. The relative expression was normalized and expressed relative to the expression in the control group according to our previous report.19,37
2.7. Statistical analysis
All statistical analyses were performed using IBM SPSS 20 software. The data were subjected to one-way analysis of variance followed by Tukey's test. The values in the same row with different superscripts are significant (P < 0.05). Data were expressed as a mean ± standard error.3. Results
3.1. Growth performance
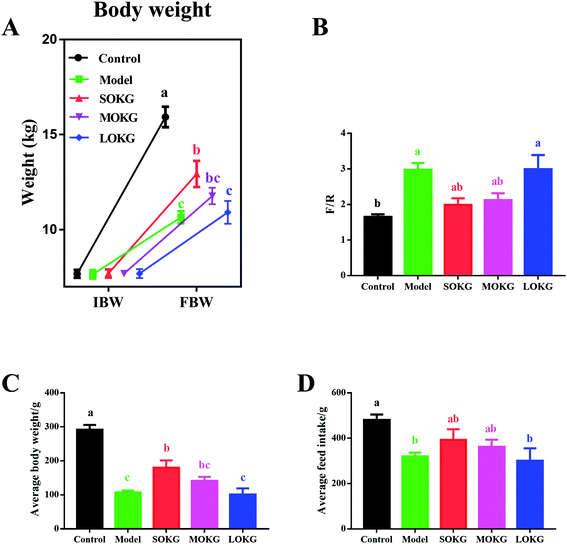
Compared with the control group, the D-gal treatment group showed a markedly reduced final body weight, ADG, ADFI and increased F/G (P < 0.05, Fig. 1). Interestingly, dietary supplementation with 0.5% OKG significantly reversed the final body weight, ADG, ADFI, and F/G compared with the model group (P < 0.05). However, we failed to notice any significant changes in the 1% OKG and 2% OKG group compared with the model group (P > 0.05).3.2. Serum oxidative stress index and intestine relative gene expressions
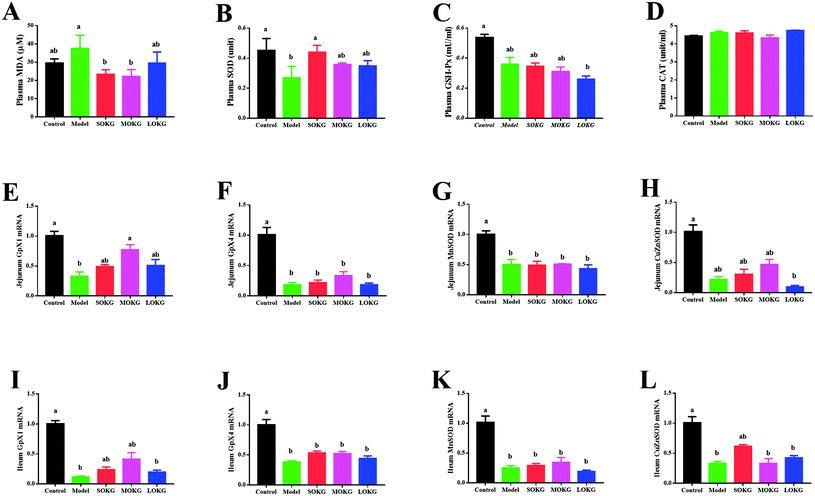
Data on the serum oxidative stress-related indexes, including the MDA, SOD, GSH-Px, CAT and intestine mRNA expression, including Gpx1, Gpx4, MnSOD, and CuZnSOD are summarized in Fig. 2. The results showed that the serum CAT activity was unchanged after dietary D-gal and OKG (P > 0.05). D-Gal treatment significantly increased the MDA content and reduced the serum SOD activity (P < 0.05), while the SOKG and MOKG group showed markedly reduced MDA levels (P < 0.05). Compared with the control group, the LOKG group showed markedly reduced serum GSH-Px activity (P < 0.05).We next preformed a real-time polymerase chain reaction (RT-PCR) to test the expression of Gpx1, Gpx4, MnSOD, and CuZnSOD. The results revealed that D-gal and LOKG treatment markedly decreased the gene expression of these in the intestine (P < 0.05). However, SOKG and MOKG ameliorated the reduction of the relative gene expression, including Gpx1 and CuZnSOD in the jejunum and ileum (P < 0.05).
3.3. Alpha-diversity of intestinal microbial communities
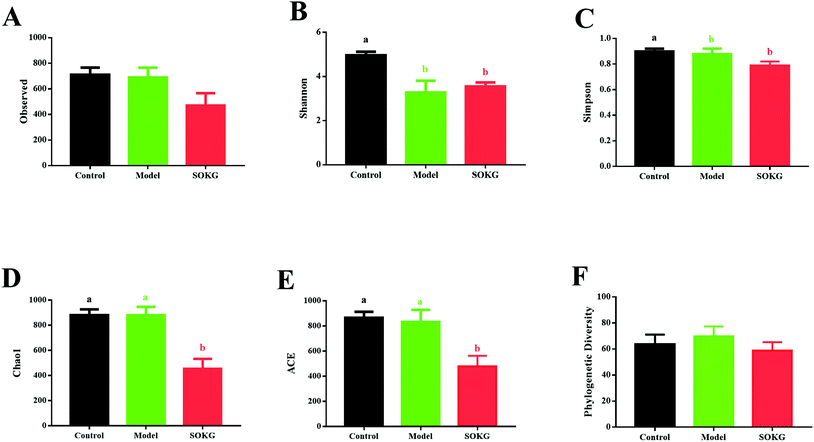
As mentioned above, SOKG alleviated the D-gal induced chronic oxidative stress via the growth performance and oxidative stress index. Thus, we further investigated the influence of 0.5% OKG and D-gal on the intestinal microbiota. The number of OTUs (observed) and the estimators of community evenness, richness, and diversity (chao1, Simpson, Shannon, and phylogenetic diversity) were examined (Fig. 3). Dietary 0.5% OKG significantly reduced the community evenness, richness, and diversity, as evidenced by the decreased Shannon, Simpson, chao1, and ACE indices compared with those of the control group (P < 0.05). However, treatment with D-gal only markedly reduced the Shannon and Simpson indices compared with those of the control group (P < 0.05).3.4. 16S rDNA bacterial sequences represented in terminal ileum samples
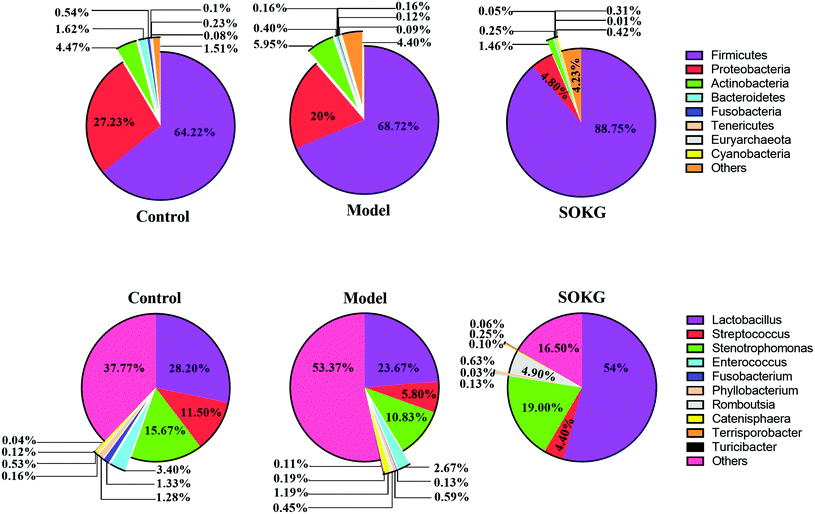
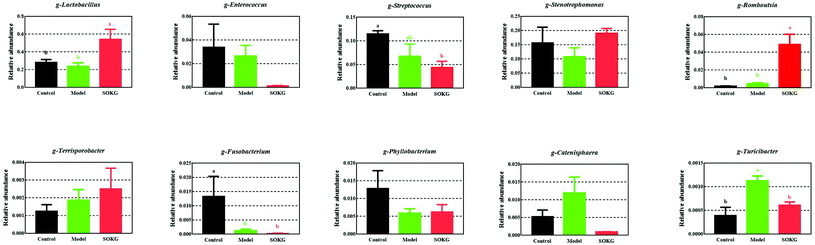
The overall microbial compositions from the three groups were altered at the phylum and genus levels (Fig. 4–6). The Firmicutes and Proteobacteria mainly accounted for the phylum level and the top 10 microbes changed in response to the dietary OKG and D-galactose (Fig. 4 and 5). Compared with the results for the control group, SOKG markedly enhanced the abundance of the Firmicutes and Cyanobacteria, while the abundance of the Proteobacteria, Fusobacteria, Bacteroidetes, and Euryachaeota decreased (P < 0.05). Lactobacillus, Streptococcus, and Stenotrophomonas mainly accounted for the genus level and the top 10 microbes changed in response to the dietary OKG and D-galactose (Fig. 4 and 6). The abundance of Lactobacillus and Romboutsia in the SOKG group was enhanced, while the abundance of Streptococcus and Fusobacterium was reduced (P < 0.05). In addition, the model group significantly decreased the abundance of Fusobacterium but increased the Turicibater (P < 0.05).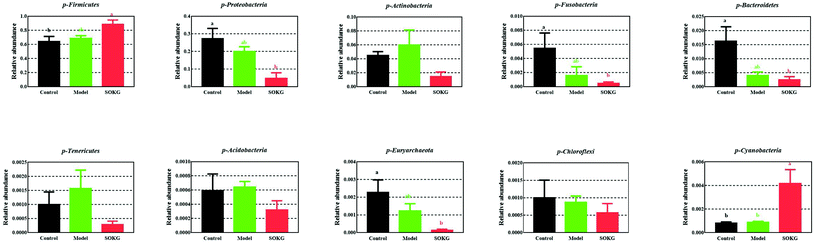 | ||
| Fig. 5 Altered ileal microbiota at the phylum level in response to dietary D-gal and OKG in piglets. Values are expressed as the mean ± SEM (n = 8). | ||
 | ||
| Fig. 6 Altered ileal microbiota at the genus level in response to dietary D-gal and OKG in piglets. Values are expressed as the mean ± SEM (n = 8). | ||
3.5. Biofunction prediction of the intestinal microbial flora
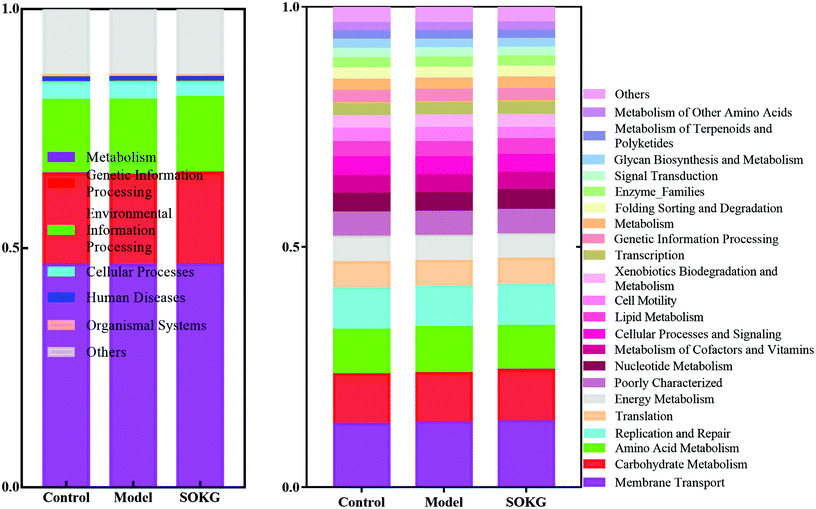
In this study, PICRUSt is based on the OTU tree in the Greengene database, and the genetic information on the OTU, inferring the gene function spectrum of their common ancestors, and inferring the gene function spectrum of other untested species in the Greengenes database to construct the archaeal and bacterial domain full spectrum of the gene function prediction spectrum, and finally “map” the sequenced microbial composition into the database to achieve prediction of the metabolic function of the flora.The predicted results can be enriched at two different levels of the KEGG pathways, in which the 1 and 2 level impressions are used for histograms (Fig. 7). The results showed that the metagenome has a highly regulated response to OKG and D-gal and can differentiate the microbiomes that mainly contribute to the metabolism (46%) at level 1. At the KEGG pathway level 2, the membrane transport, carbohydrate metabolism, amino acid metabolism, replication and repair, translation, and energy metabolism changed in response to OKG and D-gal.
 | ||
| Fig. 7 Predictive functional profiling of microbial communities by PICRUSt. KEGG pathway annotations in level 1 and level 2. | ||
3.6. Serum amino acid profiles
Compared with the results of the control group, the model group showed a significantly decreased serum glycine and proline content, but an increased phenylalanine content (P < 0.05). However, the SOKG group showed a markedly enhanced aspartate, glutamate, alanine, valine, leucine, tyrosine, and phenylalanine content, the MOKG group also showed an increased alanine, isoleucine, leucine, tyrosine, phenylalanine, lysine, and proline content (P < 0.05). In addition, the LOKG group showed a markedly enhanced phenylalanine, but decreased glycine, content (P < 0.05) (see Table 3).| Item | Control | Model | SOKG | MOKG | LOKG | P-Value |
|---|---|---|---|---|---|---|
| a Data are expressed as the mean ± SEM (n = 8). Values in the same row with different superscripts are significantly different. | ||||||
| Asp | 8.73 ± 0.70b | 10.99 ± 0.37ab | 12.06 ± 1.00a | 10.66 ± 0.73ab | 10.83 ± 0.75ab | 0.049 |
| Thr | 21.33 ± 2.28ab | 16.80 ± 2.64b | 28.73 ± 4.68a | 30.05 ± 1.30a | 16.51 ± 2.55b | 0.017 |
| Ser | 9.69 ± 0.85 | 11.61 ± 0.19 | 12.45 ± 0.71 | 10.95 ± 1.48 | 8.44 ± 0.11 | 0.056 |
| Glu | 39.50 ± 1.44b | 51.07 ± 1.72ab | 53.10 ± 5.64a | 51.85 ± 5.00ab | 43.09 ± 2.02ab | 0.037 |
| Gly | 54.48 ± 3.92a | 39.10 ± 1.53b | 49.61 ± 3.52ab | 40.66 ± 6.61ab | 35.46 ± 6.27b | 0.039 |
| Ala | 24.78 ± 1.13b | 26.71 ± 0.80b | 35.35 ± 4.31ab | 39.06 ± 5.52a | 33.43 ± 3.68ab | 0.049 |
| Cys | 9.56 ± 0.39 | 10.86 ± 0.06 | 9.77 ± 0.30 | 10.41 ± 0.25 | 9.73 ± 0.26 | 0.052 |
| Val | 11.75 ± 0.67b | 13.53 ± 0.96ab | 14.72 ± 0.82a | 13.34 ± 0.82ab | 11.23 ± 0.38b | 0.045 |
| Met | 4.52 ± 0.24 | 4.88 ± 0.26 | 4.52 ± 0.31 | 4.9 ± 0.35 | 4.03 ± 0.19 | 0.236 |
| Ile | 4.62 ± 0.27c | 6.04 ± 0.31bc | 8.06 ± 0.87ab | 9.57 ± 1.24a | 7.91 ± 0.78ab | 0.001 |
| Leu | 12.55 ± 1.17b | 15.95 ± 0.46ab | 17.12 ± 1.02a | 20.18 ± 1.40a | 16.69 ± 0.24ab | 0.001 |
| Tyr | 9.72 ± 2.42b | 13.64 ± 0.77ab | 17.14 ± 1.58a | 19.66 ± 1.14a | 16.48 ± 0.76ab | 0.005 |
| Phe | 7.68 ± 0.78b | 11.44 ± 0.44a | 12.57 ± 0.92a | 13.24 ± 0.69a | 11.51 ± 0.64a | 0.001 |
| Lys | 14.43 ± 0.54b | 13.33 ± 1.65b | 17.87 ± 1.51ab | 24.25 ± 2.37a | 14.50 ± 1.49b | 0.001 |
| His | 5.11 ± 0.34 | 6.08 ± 0.68 | 5.32 ± 0.53 | 6.23 ± 0.68 | 5.11 ± 0.39 | 0.454 |
| Arg | 11.80 ± 0.79 | 11.10 ± 0.34 | 13.88 ± 2.01 | 15.66 ± 1.76 | 12.6 ± 0.57 | 0.271 |
| Pro | 14.52 ± 0.32b | 16.42 ± 2.35a | 19.02 ± 1.81ab | 24.59 ± 3.04a | 20.37 ± 1.31ab | 0.023 |
4. Discussion
D-Galactose is a nutrient and reducing sugar that reacts with free amino acids in proteins to produce ROS, and thus causes chronic oxidative stress.3 Accumulating evidence has shown that OKG possesses beneficial and protective activities in animals and humans under chronic and pathological conditions, owing to its regulatory roles in oxidative stress, burns, injury and metabolism.7,15,38,39 However, evidence of the protective effects of OKG on chronic oxidative stress is still limited. Thus, in the current study, we further discovered that D-gal model pigs alleviated growth-suppression and restored serum amino acid profiles by altering the gut microbiome. It is of note that growth performance was inhibited in D-gal model pigs and 0.5% OKG alleviated growth-suppression, which is similar to the effect of OKG on tumor rats40 and D-gal induced ageing mice, in which D-gal inhibited body weight in mice.41 Also, OKG alleviated D-gal induced chronic oxidative stress via enhancing the SOD and GSH-Px levels including the relative mRNA expression in the intestine and inhibiting lipid oxidation subsequent to MDA generation, similar to the results found in our previous study on glutamate restored diquat-induced oxidative stress in piglets.19,42Intestinal microbial communities play an important role in the status of the host, including the decomposition of food, metabolism, and defense against pathogen invasion.43–49 Diet rapidly and reproducibly influences gut microbial composition, structure, and metabolism.23,50–52 It was found that OKG can prevent bacterial translocation and dissemination and thus reduce gut-derived sepsis.53 Zhao et al. reported that D-gal markedly increased Firmicutes and decreased Bacteroidetes. In this study, we reported for the first time that 0.5% OKG suppressed alpha diversity, which refers to the diversity of species within a community including the Shannon, Simpson, Chao1, and ACE indices.54 We further discovered that OKG enhanced the intestinal abundance of Firmicutes and Cyanobacteria but decreased the Proteobacteria, Fusobacteria, Bacteriodetes, and Euryarchaeota at the phylum level. Vaughn et al. and Yin et al. reported that rats fed a high energy-dense diet and showed an increase in the Firmicutes/Bacteriodetes ratio may be associated with being overweight and with body fat accumulation.46,55Proteobacteria, Fusobacteria, and Euryarchaeota are harmful intestinal bacteria. Proteobacteria promotes chronic gut inflammation.56Fusobacteria are members of the oral and gastrointestinal flora and are important potential pathogens in human.57,58 We also observed an increase in the genus Lactobacillus and Romboutsia, but a decrease in the Streptococcus, Fusobacterium and Turicibacter in the 0.5% OKG group. Lactobacillus has a protective effect on the intestinal morphology, and the integrity of the intestinal epithelium of pigs and humans.59Romboutsia is commonly identified in the human gut and is often associated with a healthy status in patients, it is also a potential microbial indicator of disease.60 This result indicated that 0.5% OKG may alter bacteria and could further increase feed intake, body weight and the health of pigs.
PICRUSt was used to predict the metabolic function of the flora based on the OTU tree in the Greengene database, and by constructing the archaeal and bacterial domain full spectrum of the gene function prediction spectrum.61 The results showed that the altered microbes were mainly involved in membrane transport, carbohydrate metabolism, amino acid metabolism, replication and repair, translation and energy metabolism. Similarly, a previous study also indicated that OKG influenced amino acid metabolism.7,62 Thus, to explore the effect of D-gal and OKG on altering the gut microbiome, by modulating host physiological functions we further determined the serum amino acid pool to confirm the role of OKG on amino acid metabolism. The serum amino acid pool reflects the condition of the body and some diseases dynamically.63 Diquat or mycotoxin exposure reduced the level of most serum amino acids.19,64 Similarly, our results indicated that the concentration of most serum amino acids was reduced in the D-gal induced pigs. OKG can transform proline, arginine, glutamate, glutamine and other precursor which can respond to oxidative reactions and improve the immune status during stress.7,11,12,15,65 Meanwhile, aspartate, glutamate and threonine are the main sources of energy for the intestines and mainly metabolized in the intestines to synthesize intestinal mucosal proteins and improve the integrity of the intestinal epithelium.66,67 In our study dietary 0.5% and 1% OKG increased glutamate, proline, aspartate, and threonine levels, suggesting that OKG may convert glutamate, proline, aspartate, and threonine to alleviate D-gal induced intestinal oxidative stress. Branched-chain amino acids, including leucine, isoleucine, and valine, are essential dietary nutrients for the normal growth of pigs and can improve intestinal morphology and cell proliferation.68 In our study, 0.5% and 1% OKG markedly increased the concentration of the serum branched-chain amino acids.
In conclusion, dietary supplementation with 0.5% OKG restores serum amino acids and alleviates the growth-suppression induced by D-gal chronic oxidative stress and alters the composition of gut microbiota, especially for the Firmicutes and Bacteriodetes at the phylum level. Therefore, OKG may decreased the damage caused by D-gal in pigs and could be used as a nutritional additive. Further studies with prolonged supplementation of OKG in healthy animals and humans should be performed.
Conflicts of interest
The authors have declared no conflict of interest.Funding
This study was supported by the National Science Foundation for Distinguished Young Scholars of Hunan Province (2016JJ1015), the National Natural Science Foundation of China (31872371, 31470132, 31772642), the Chinese Academy of Sciences “Hundred Talent” award, and the Open Foundation of Key Laboratory of Agro-ecological Processes in Subtropical Region, Institute of Subtropical Agriculture, Chinese Academy of Sciences (ISA2016101).Acknowledgements
We would like to thank Special Fund for Taishan Industry Leading Talent for the funding support. We are grateful to the Public Service Technology Center, Institute of Subtropical Agriculture, Chinese Academy of Sciences for technical support.References
- W. Yan, D. Li, T. Chen, G. Tian, P. Zhou and X. Ju, Umbilical Cord MSCs Reverse D-Galactose-Induced Hepatic Mitochondrial Dysfunction via Activation of Nrf2/HO-1 Pathway, Biol. Pharm. Bull., 2017, 40, 1174–1182 CrossRef CAS PubMed.
- S. Park, C. S. Kim, J. Lee, J. S. Kim and J. Kim, Effect of Regular Exercise on the Histochemical Changes of d-Galactose-Induced Oxidative Renal Injury in High-Fat Diet-Fed Rats, Acta Histochem. Cytochem., 2013, 46, 111–119 CrossRef PubMed.
- J. Zhong, F. Wang, Z. Wang, C. Shen, Y. Zheng, F. Ma, T. Zhu, L. Chen, Q. Tang and J. Zhu, Aloin attenuates cognitive impairment and inflammation induced by d-galactose via down-regulating ERK, p38 and NF-kappaB signaling pathway, Int. Immunopharmacol., 2019, 72, 48–54 CrossRef CAS PubMed.
- C. M. Liu, J. Q. Ma and Y. Lou, Chronic administration of troxerutin protects mouse kidney against D-galactose-induced oxidative DNA damage, Food Chem. Toxicol., 2010, 48, 2809–2817 CrossRef CAS PubMed.
- J. Zhang, W. Lin, R. Wu, Y. Liu, K. Zhu, J. Ren, S. Zhang and X. Ling, Mechanisms of the active components from Korean pine nut preventing and treating d-galactose-induced aging rats, Biomed. Pharmacother., 2018, 103, 680–690 CrossRef CAS.
- Y. Feng, Y. H. Yu, S. T. Wang, J. Ren, D. Camer, Y. Z. Hua, Q. Zhang, J. Huang, D. L. Xue, X. F. Zhang, X. F. Huang and Y. Liu, Chlorogenic acid protects D-galactose-induced liver and kidney injury via antioxidation and anti-inflammation effects in mice, Pharm. Biol., 2016, 54, 1027–1034 CrossRef CAS PubMed.
- C. Loi, D. Hamani, C. Moinard, L. Bishoff, N. Neveux, C. Garbay and L. Cynober, Does the ornithine-alpha-ketoglutarate ratio influence ornithine alpha-ketoglutarate metabolism in healthy rats?, Metab., Clin. Exp., 2007, 56, 105–114 CrossRef CAS.
- P. Segaud, B. Lardeux, M. C. Alexandre-Gouabau, F. Bleiberg-Daniel, S. Nakib, L. Cynober and C. Moinard, Pretreatment of starved rats with ornithine alpha-ketoglutarate: effects on hepatic mRNA levels and plasma concentrations of three liver-secreted proteins, Nutrition, 2005, 21, 732–739 CrossRef PubMed.
- M. R. Tatara, E. Sliwa, W. Krupski, A. Brodzki and K. Pasternak, Ornithine alpha-ketoglutarate increases mineralization and mechanical properties of tibia in turkeys, Bone, 2006, 39, 100–105 CrossRef CAS.
- M. Jeevanandam and S. R. Petersen, Substrate fuel kinetics in enterally fed trauma patients supplemented with ornithine alpha ketoglutarate, Clin. Nutr., 1999, 18, 209–217 CrossRef CAS.
- C. Moinard, B. Chauveau, S. Walrand, C. Felgines, J. Chassagne, F. Caldefie, L. A. Cynober and M.-P. Vasson, Phagocyte functions in stressed rats: comparison of modulation by glutamine, arginine and ornithine 2-oxoglutarate, Clin. Sci., 1999, 97, 59 CrossRef CAS.
- H. Schuster, M. C. Blanc, C. Genthon, P. Therond, D. Bonnefont-Rousselot, A. Le Tourneau, J. P. De Bandt and L. Cynober, Does dietary ornithine alpha-ketoglutarate supplementation protect the liver against ischemia-reperfusion injury?, Clin. Nutr., 2005, 24, 375–384 CrossRef CAS.
- E. S. Goncalves, C. M. Rabelo, P. Neto, J. H. Garcia, S. B. Guimaraes and P. R. Vasconcelos, Effect of short-term ornithine alpha-ketoglutarate pretreatment on intestinal ischemia-reperfusion in rats, Acta Cir. Bras., 2011, 26(Suppl 1), 2–7 CrossRef PubMed.
- P. Pernet, C. Coudray-Lucas, C. Schneid, A. Jardel and L. Cynober, Dose dependency of the effect of ornithine α-ketoglutarate on tissue glutamine concentrations and hypercatabolic response in endotoxaemic rats, Br. J. Nutr., 2007, 92, 627 CrossRef PubMed.
- C. Moinard, F. Caldefie, S. Walrand, A. Tridon, J. Chassagne, M.-P. Vasson and L. Cynober, Effects of ornithine 2-oxoglutarate on neutrophils in stressed rats: evidence for the involvement of nitric oxide and polyamines, Clin. Sci., 2002, 102, 287 CrossRef CAS PubMed.
- C. Schneid, J. P. De Bandt, L. Cynober, E. Torres, G. Reach and S. Darquy, In vivo induction of insulin secretion by ornithine alpha-ketoglutarate: involvement of nitric oxide and glutamine, Metab., Clin. Exp., 2003, 52, 344–350 CrossRef CAS PubMed.
- M. Jeevanandam, N. J. Holaday and S. R. Petersen, Ornithine-alpha-ketoglutarate (OKG) supplementation is more effective than its component salts in traumatized rats, J. Nutr., 1996, 126, 2141–2150 CrossRef CAS PubMed.
- J. Duan, J. Yin, W. Ren, T. Liu, Z. Cui, X. Huang, L. Wu, S. W. Kim, G. Liu, X. Wu, G. Wu, T. Li and Y. Yin, Dietary supplementation with L-glutamate and L-aspartate alleviates oxidative stress in weaned piglets challenged with hydrogen peroxide, Amino Acids, 2016, 48, 53–64 CrossRef CAS.
- J. Yin, M. Liu, W. Ren, J. Duan, G. Yang, Y. Zhao, R. Fang, L. Chen, T. Li and Y. Yin, Effects of dietary supplementation with glutamate and aspartate on diquat-induced oxidative stress in piglets, PLoS One, 2015, 10, e0122893 CrossRef.
- L. Wang, Y. Hou, D. Yi, Y. Li, B. Ding, H. Zhu, J. Liu, H. Xiao and G. Wu, Dietary supplementation with glutamate precursor alpha-ketoglutarate attenuates lipopolysaccharide-induced liver injury in young pigs, Amino Acids, 2015, 47, 1309–1318 CrossRef CAS.
- Y. Li, H. Han, J. Yin, J. Zheng, X. Zhu, T. Li and Y. Yin, Effects of glutamate and aspartate on growth performance, serum amino acids, and amino acid transporters in piglets, Food Agric. Immunol., 2018, 1–13 Search PubMed.
- J. Yin, Y. Li, H. Han, Z. J. Liu, X. F. Zeng, T. Li and Y. Yin, Long-Term Effects of Lysine Concentration on Growth Performance, Intestinal Microbiome, and Metabolic Profiles in a Pig model, Food Funct., 2018, 9, 4153–4163 RSC.
- Y. Li, H. Han, J. Yin, X. He, Z. Tang, T. Li, K. Yao, Y. J. F. Yin and Function, D- and L-Aspartate regulates growth performance, inflammation and intestinal microbial community in young pigs, Food Funct., 2019, 10, 1028–1037 RSC.
- J. Chen, H. Yang, L. Long, Y. Zhao, J. Qian, W. Fei, B. Kang, S. Liu, T. O. Adebowale and K. Yao, The effects of dietary supplementation with α-ketoglutarate on the intestinal microbiota, metabolic profiles, and ammonia levels in growing pigs, Anim. Feed Sci. Tech., 2017, 234, S0377840117300834 CrossRef.
- J. Yin, Y. Li, H. Han, S. Chen, J. Gao, G. Liu, X. Wu, J. Deng, Q. Yu, X. Huang, R. Fang, T. Li, R. J. Reiter, D. Zhang, C. Zhu, G. Zhu, W. Ren and Y. Yin, Melatonin reprogramming of gut microbiota improves lipid dysmetabolism in high-fat diet-fed mice, J. Pineal Res., 2018, 65, e12524 CrossRef PubMed.
- W. Ren, J. Yin, H. Xiao, S. Chen, G. Liu, B. Tan, N. Li, Y. Peng, T. Li, B. Zeng, W. Li, H. Wei, Z. Yin, G. Wu, P. R. Hardwidge and Y. Yin, Intestinal Microbiota-Derived GABA Mediates Interleukin-17 Expression during Enterotoxigenic Escherichia coli Infection, Front. Immunol., 2016, 7, 685 Search PubMed.
- S. Liu, L. He, Q. Jiang, V. Duraipandiyan, N. A. Al-Dhabi, G. Liu, K. Yao and Y. Yin, Effect of dietary alpha-ketoglutarate and allicin supplementation on the composition and diversity of the cecal microbial community in growing pigs, J. Sci. Food Agric., 2018, 98, 5816–5821 CrossRef CAS PubMed.
- M. Martin, Cutadapt removes adapter sequences from high-throughput sequencing reads, EMBnet. J., 2011, 17, 10–12 CrossRef.
- X. Gao, Q. Ma and H. Zhu, Distribution, industrial applications, and enzymatic synthesis of D-amino acids, Appl. Microbiol. Biotechnol., 2015, 99, 3341–3349 CrossRef CAS PubMed.
- C. Quast, E. Pruesse, P. Yilmaz, J. Gerken, T. Schweer, P. Yarza, J. Peplies and F. O. Glöckner, The SILVA ribosomal RNA gene database project: improved data processing and web-based tools, Nucleic Acids Res., 2013, 41, 590–596 CrossRef PubMed.
- R. C. Edgar, B. J. Haas, J. C. Clemente, Q. Christopher and R. Knight, UCHIME improves sensitivity and speed of chimera detection, Bionformatics, 2011, 27, 2194 CrossRef CAS PubMed.
- Z. Yu, C. Shi, W. Cheng, Z. Lu, F. Wang, F. Jie, Y. Wang and A. Immunology, Effect of soybean meal fermented with Bacillus subtilis BS12 on growth performance and small intestinal immune status of piglets, Food Agric. Immunol., 2017, 1–4 Search PubMed.
- B. J. Haas, D. Gevers, A. M. Earl, M. Feldgarden, D. V. Ward, G. Giannoukos, D. Ciulla, D. Tabbaa, S. K. Highlander and E. J. G. r. Sodergren, Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons, 2011, vol. 21, pp. 494–504 Search PubMed.
- R. C. Edgar, UPARSE: highly accurate OTU sequences from microbial amplicon reads, Nat. Methods, 2013, 10, 996 CrossRef CAS PubMed.
- R. C. Edgar, MUSCLE: multiple sequence alignment with high accuracy and high throughput, Nucleic Acids, 2004, 32, 1792–1797 CrossRef CAS PubMed.
- A. D. Radkov and L. A. Moe, Bacterial synthesis of D-amino acids, Appl. Microbiol. Biotechnol., 2014, 98, 5363–5374 CrossRef CAS PubMed.
- Y. Jie, Y. Li, H. Hui, Z. Jie, L. Wang, W. Ren, C. Shuai, W. Fei, R. Fang, X. Huang and T. Li, Effects of Lysine deficiency and Lys-Lys dipeptide on cellular apoptosis and amino acids metabolism, Mol. Nutr., 2017, 61, 600754 Search PubMed.
- V. L. Albaugh, K. Mukherjee and A. Barbul, Proline Precursors and Collagen Synthesis: Biochemical Challenges of Nutrient Supplementation and Wound Healing, J. Nutr., 2017, 147, 2011–2017 CAS.
- T. Le Bricon, C. Coudray-Lucas, N. Lioret, S. K. Lim, F. Plassart, L. Schlegel, J. P. De Bandt, R. Saizy, J. Giboudeau and L. Cynober, Ornithine alpha-ketoglutarate metabolism after enteral administration in burn patients: bolus compared with continuous infusion, Am. J. Clin. Nutr., 1997, 65, 512–518 CrossRef CAS PubMed.
- T. Le Bricon, L. Cynober and V. J. M. Baracos, Ornithine α-ketoglutarate limits muscle protein breakdown without stimulating tumor growth in rats bearing Yoshida ascites hepatoma, Metabolism, 1994, 43, 899–905 CrossRef CAS.
- H. Zhang, Y. Li, C. Cui, T. Sun, J. Han, D. Zhang, C. Lu, J. Zhou, L. Cheong, Y. Li and X. Su, Modulation of gut microbiota by dietary supplementation with tuna oil and algae oil alleviates the effects of D-galactose-induced ageing, Appl. Microbiol. Biotechnol., 2018, 102, 2791–2801 CrossRef CAS PubMed.
- W. Ren, P. Wang, J. Yan, G. Liu, B. Zeng, T. Hussain, C. Peng, J. Yin, B. Tan and G. Zhu, Melatonin alleviates weanling stress in mice: involvement of intestinal microbiota, J. Pineal. Res., 2018, 64 CrossRef.
- P. D. Cani and N. M. Delzenne, The role of the gut microbiota in energy metabolism and metabolic disease, Curr. Pharm. Des., 2009, 15, 1546–1558 CrossRef CAS PubMed.
- B. Stecher and W. D. Hardt, The role of microbiota in infectious disease, Trends Microbiol., 2008, 16, 107–114 CrossRef CAS PubMed.
- H. H. Li, Y. P. Li, Q. Zhu, J. Y. Qiao and W. J. Wang, Dietary supplementation with Clostridium butyricum helps to improve the intestinal barrier function of weaned piglets challenged with enterotoxigenic Escherichia coli K88, J. Appl. Microbiol., 2018, 125, 964–957 CrossRef CAS PubMed.
- J. Yin, Y. Li, H. Han, S. Chen, J. Gao, G. Liu, X. Wu, J. Deng, Q. Yu, X. Huang, R. Fang, T. Li, R. J. Reiter, D. Zhang, C. Zhu, G. Zhu, W. Ren and Y. Yin, Melatonin reprogramming of gut microbiota improves lipid dysmetabolism in high-fat diet-fed mice, J. Pineal Res., 2018, 65(4), e12524 CrossRef PubMed.
- Z. Juan, S. Zhao-Ling, Z. Ming-Hua, W. Chun, W. Hai-Xia, L. Meng-Yun, H. Jian-Qiong, Z. Yue-Jie and S. J. R. Xin, Oral administration of Clostridium butyricum CGMCC0313-1 reduces ovalbumin-induced allergic airway inflammation in mice, Respirology, 2017, 22, 898 CrossRef PubMed.
- M. Goudarzi, M. J. Khodayar, H. T. Smt, H. Ghaznavi, I. Fatemi and S. Mehrzadi, Pretreatment with melatonin protects against cyclophosphamide-induced oxidative stress and renal damage in mice, Fundam. Clin. Pharmacol., 2017, 31 CAS.
- H. Han, Y. Li, J. Fang, G. Liu, J. Yin, T. Li and Y. Yin, Gut Microbiota and Type 1 Diabetes, Int. J. Mol. Sci., 2018, 19, 995 CrossRef PubMed.
- J. Gao, K. Xu, H. Liu, G. Liu, M. Bai, C. Peng, T. Li and Y. Yin, Impact of the Gut Microbiota on Intestinal Immunity Mediated by Tryptophan Metabolism, Front. Cell. Infect. Microbiol., 2018, 8, 13 CrossRef.
- C. Wang, C. Shi, Y. Zhang, D. Song, Z. Lu and Y. Wang, Microbiota in fermented feed and swine gut, Appl. Microbiol. Biotechnol., 2018, 102, 1–8 CrossRef PubMed.
- W. Ren, S. Chen, J. Yin, J. Duan, T. Li, G. Liu, Z. Feng, B. Tan, Y. Yin and G. Wu, Dietary arginine supplementation of mice alters the microbial population and activates intestinal innate immunity, J. Nutr., 2014, 144, 988–995 CrossRef CAS PubMed.
- L. Schlegel, C. Coudray-Lucas, F. d. r. Barbut, J. Le Boucher, A. Jardel, S. Zarrabian and L. Cynober, Bacterial dissemination and metabolic changes in rats induced by endotoxemia following intestinal E. coli overgrowth are reduced by ornithine α-ketoglutarate administration, J. Nutri., 2000, 130, 2897–2902 CrossRef CAS PubMed.
- A. Agrawal and K. Gopal, Application of Diversity Index in Measurement of Species Diversity, Springer India, 2013 Search PubMed.
- A. C. Vaughn, E. M. Cooper, P. M. Dilorenzo, L. J. O'Loughlin, M. E. Konkel, J. H. Peters, A. Hajnal, T. Sen, S. H. Lee and K. Czaja, Energy-dense diet triggers changes in gut microbiota, reorganization of gut–brain vagal communication and increases body fat accumulation, Acta Neurobiol. Exp., 2017, 77, 18–30 Search PubMed.
- F. A. Carvalho, O. Koren, J. K. Goodrich, M. E. Johansson, I. Nalbantoglu, J. D. Aitken, Y. Su, B. Chassaing, W. A. Walters, A. Gonzalez, J. C. Clemente, T. C. Cullender, N. Barnich, A. Darfeuille-Michaud, M. Vijay-Kumar, R. Knight, R. E. Ley and A. T. Gewirtz, Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice, Cell Host Microbe, 2012, 12, 139–152 CrossRef CAS PubMed.
- M. L. Santoru, C. Piras, A. Murgia, V. Palmas, T. Camboni, S. Liggi, I. Ibba, M. A. Lai, S. Orrù and L. Loizedda, Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients, Sci. Rep., 2018, 8, 9523 CrossRef PubMed.
- G. L. Roberts, Fusobacterial infections: an underestimated threat, Br. J. Biomed. Sci., 2000, 57, 156 CAS.
- D. Zhang, T. Shang, Y. Huang, S. Wang, H. Liu, J. Wang, Y. Wang, H. Ji and R. Zhang, Gene expression profile changes in the jejunum of weaned piglets after oral administration of Lactobacillus or an antibiotic, Sci. Rep., 2017, 7, 15816 CrossRef PubMed.
- M. Mangifesta, L. Mancabelli, C. Milani, F. Gaiani, N. de'Angelis, G. L. de'Angelis, D. van Sinderen, M. Ventura and F. Turroni, Mucosal microbiota of intestinal polyps reveals putative biomarkers of colorectal cancer, Sci. Rep., 2018, 8, 9 CrossRef PubMed.
- M. G. I. Langille, J. Zaneveld, J. G. Caporaso, D. McDonald, D. Knights, J. A. Reyes, J. C. Clemente, D. E. Burkepile, R. L. V. Thurber, R. Knight, R. G. Beiko and C. Huttenhower, Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences, Nat. Biotechnol., 2013, 31, 814–821 CrossRef CAS PubMed.
- L. Cynober, E. Lasnier, J. Le Boucher, A. Jardel and C. Coudray-Lucas, Effect of ornithine alpha-ketoglutarate on glutamine pools in burn injury: evidence of component interaction, Intensive Care Med., 2007, 33, 538–541 CrossRef CAS PubMed.
- G. Wu, Functional amino acids in nutrition and health, Amino Acids, 2013, 45, 407–411 CrossRef CAS PubMed.
- J. Duan, J. Yin, M. Wu, P. Liao, D. Deng, G. Liu, Q. Wen, Y. Wang, W. Qiu, Y. Liu, X. Wu, W. Ren, B. Tan, M. Chen, H. Xiao, L. Wu, T. Li, C. M. Nyachoti, O. Adeola and Y. Yin, Dietary glutamate supplementation ameliorates mycotoxin-induced abnormalities in the intestinal structure and expression of amino acid transporters in young pigs, PLoS One, 2014, 9, e112357 CrossRef PubMed.
- L. Cynober, C. Coudray-Lucas, J. P. de Bandt, J. Guechot, C. Aussel, M. Salvucci and J. Giboudeau, Action of ornithine alpha-ketoglutarate, ornithine hydrochloride, and calcium alpha-ketoglutarate on plasma amino acid and hormonal patterns in healthy subjects, J. Am. Coll. Nutr., 1990, 9, 2–12 CrossRef CAS PubMed.
- G. Wu, Functional amino acids in growth, reproduction, and health, Adv. Nutr., 2010, 1, 31–37 CrossRef CAS PubMed.
- G. Wu, Z. Wu, Z. Dai, Y. Yang, W. Wang, C. Liu, B. Wang, J. Wang and Y. Yin, Dietary requirements of “nutritionally non-essential amino acids” by animals and humans, Amino Acids, 2013, 44, 1107–1113 CrossRef CAS PubMed.
- Y. Duan, B. Tan, J. Li, P. Liao, B. Huang, F. Li, H. Xiao, Y. Liu and Y. Yin, Optimal branched-chain amino acid ratio improves cell proliferation and protein metabolism of porcine enterocytesin in vivo and in vitro, Nutrition, 2018, 54, 173–181 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2020 |