Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Subtoxic dose of lithium cobalt oxide nanosheets impacts critical molecular pathways in trout gill epithelial cells†
Arielle C.
Mensch‡
 a,
Hugh D.
Mitchell‡
a,
Hugh D.
Mitchell‡
 b,
Lye Meng
Markillie
a,
Elizabeth D.
Laudadio
c,
Jenny K.
Hedlund Orbeck
b,
Lye Meng
Markillie
a,
Elizabeth D.
Laudadio
c,
Jenny K.
Hedlund Orbeck
 c,
Alice
Dohnalkova
a,
Michael P.
Schwartz
c,
Robert J.
Hamers
c,
Alice
Dohnalkova
a,
Michael P.
Schwartz
c,
Robert J.
Hamers
 c and
Galya
Orr
c and
Galya
Orr
 *a
*a
aEnvironmental Molecular Science Laboratory, Pacific Northwest National Laboratory, Richland, Washington 99354, USA. E-mail: galya.orr@pnnl.gov
bBiological Sciences Division, Pacific Northwest National Laboratory, Richland, Washington 99354, USA
cDepartment of Chemistry, University of Wisconsin–Madison, Madison, Wisconsin 53706, USA
First published on 20th October 2020
Abstract
Increasing use of nanoscale lithium cobalt oxide (LCO) particles in nanotechnologies, including catalysis and energy storage, raises concerns over their release into the environment and subsequent biological impact. Here we study the impact of LCO nanosheets on trout gill epithelial cells – model cells for environmental exposures – by global transcriptomics using RNA-Seq. We identify molecular processes impacted by subtoxic and toxic nanoparticle (NP) doses as well as dissolved Li+ and Co2+ ions. We found that the ions, at concentrations released from the toxic NP dose, did not impact cell viability and downregulated the expression of few genes following 24 h exposure, which recovered to normal levels at 48 h. In contrast, the toxic NP dose upregulated the expression of over 1000 genes at each time point, indicating the intact NPs are responsible for perturbing gene expression and toxicity. Importantly, the subtoxic NP dose, despite having no impact on cell viability, upregulated the expression of over 500 genes at 24 h, and 150 genes at 48 h. Clustering analysis showed distinct gene expression profiles induced by the toxic and subtoxic NP doses, and functional enrichment identified pathways with distinct patterns of regulations. The impacted pathways fell into four main functional categories: metabolic and energy related processes, oxygen and hypoxia related processes, membrane binding and internalization processes, and developmental processes. Together, our observations indicate that LCO NP toxicity originates from the intact NP, not the dissolved ions, and even subtoxic NP dose impacts multiple pathways critical to the normal function of the cell.
Environmental significanceLithium cobalt oxide (LCO) nanoparticles are being pursued as cathode materials in lithium-ion batteries among other nanotechnologies. With little infrastructure and low economic incentive for recycling, the release of LCO nanoparticles into the environment is inevitable. Nanoparticles released into the environment are found at a wide range of local concentrations, including low concentrations that induce undetectable cellular responses by common toxicity assays, such as cell death. Using global transcriptomic analysis, we uncover key molecular processes impacted in trout gill epithelial cells – a model for aquatic exposures – by LCO nanoparticles, even at a subtoxic concentration that shows no impact on cell viability. The molecular processes point to disruption of energy and oxygen balance, and multiple cellular attempts to restore normal functions. |
Introduction
The inevitable release of engineered nanoparticles (NPs) from consumer products into the environment raises concerns over their environmental and health impacts.1–4 The environmental concentrations of released engineered NPs are expected to vary depending on the identity of the NP and the environment.5,6 Concentrations of NPs, such as TiO2, ZnO, CeO2 and Ag NPs, as well as carbon nanotubes and fullerenes, in surface waters are estimated to be in the range of 10−5 to 10 μg L−1, with higher estimates in wastewater treatment plant effluents and biosolids.6 Such variability highlights the need to study the impact of NPs especially at low, subtoxic concentrations. While many assays are focused on acute toxicity endpoints, such as cell viability and membrane integrity,7,8 less effort has been focused on molecular and cellular processes that might be induced by lower doses that are more environmentally relevant. Several studies have demonstrated the importance of investigating the impact of subtoxic nanoparticle concentrations. For example, gold nanoparticles can disrupt F-actin structure in human dermal fibroblasts9 and induce modifications at the genetic level.10 Additionally, ZnO NPs disrupt metal homeostasis in hepatocytes,11 and affect genes related to the immune system and inflammation in zebrafish.12 Exposure of three types of human cell lines to low concentrations of either TiO2 NPs or multi-walled carbon nanotubes elicited transcriptomic and proteomic expression patterns dependent on both NP and cell types.13 Understanding how nanomaterials impact molecular and cellular functions at low, subtoxic doses is critical for understanding the environmental implications of nanotechnology.Whole transcriptome analysis using RNA-sequencing (RNA-Seq) is an effective approach to uncover changes in gene expression and molecular processes that might be impacted by NP exposures,14–20 including exposures to low or subtoxic NP concentrations.14–16 Recent work found human lung cells exposed to a non-cytotoxic dose of TiO2 NPs for 24 h resulted in over 2000 differentially expressed genes as compared to unexposed cells.14 Using single cell RNA-Seq, alveolar epithelial cells were found to upregulate their responses to low doses of quantum dots, whereas downregulated responses were generally observed for higher doses.15 Exposure of adult zebrafish to a low dose of carboxymethyl cellulose stabilized FeS NPs resulted in changes in the expression level of genes related to oxidative stress, DNA damage/repair, inflammatory responses, and detoxification.18 Taken together these studies demonstrate that both the nanoparticle properties and concentration play a role in impacting molecular processes in cells and organisms.
Lithium cobalt oxide (LCO) is commonly used as a cathode material for Li-ion batteries.21,22 and efforts are in place to utilize nano-sized LCO particles due to the enhanced lithium and electron transport at the nanoscale.23 Furthermore, during electrochemical cycling, sintered aggregates of commercial LCO NPs can be fractured into smaller nanometer-sized flakes.24 With little infrastructure in place and low economic incentive to recycle Li-ion batteries,25 these nanomaterials and their constituents are likely to end in landfills, leachate, or as air emissions,26 thus highlighting the need to understand the environmental and health impact of Li-ion battery materials.27–30
We conducted global transcriptomic analysis using RNA-Seq to uncover key molecular processes in trout gill epithelial cells – a model cell type for aquatic environmental exposures29–33 – in response to LCO nanosheets, referred to as NPs throughout the text, at subtoxic (5 mg L−1) and toxic (25 mg L−1) doses, determined by monitoring cell viability. We also used this approach to determine the contribution of the dissolved Li+ and Co2+ ions to the observed impact. Our studies indicate that the intact NPs, not the dissolved ions, are responsible for perturbing gene expression and inducing toxicity. Furthermore, the subtoxic NP dose, despite having no impact on cell viability, induced significant changes in gene expression and pathways. Although most of the regulated pathways were shared between the subtoxic and toxic doses, the patterns, as a function of time and degree of regulation, were different. Our observations suggest that both subtoxic and toxic LCO NP doses induce energy imbalance and depletion, as well as hypoxia, driving the cells to respond by increasing glucose import, transport and homeostasis, and upregulating energy and oxygen related pathways to restore balance.
Experimental
Lithium cobalt oxide synthesis and characterization
LCO nanosheets were synthesized as described previously.34–36 Additional details about the synthesis and characterization (SEM, TEM, XPS, AFM, laser Doppler microelectrophoresis, and ion dissolution measurements), as well as quantification of ion dissolution are found in the ESI.†Cell culture
Oncorhynchus mykiss (rainbow trout) gill epithelial cells (RTgill-W1, ATCC CRL-2523) were routinely cultured in Leibovitz's L-15 growth medium (ATCC) supplemented with 1% antibiotics and 10% fetal bovine serum (ThermoFisher), referred to as “growth medium” throughout. Cells were incubated in ambient atmosphere at 19 °C with growth medium replaced twice weekly. Following 100% confluency, cells were harvested for experiments and passage. Growth medium was removed, cells were rinsed with phosphate buffered saline (PBS, ThermoFisher), and were detached from the surface using 0.25% trypsin-0.53 mM EDTA solution (ATCC).LCO preparation for exposure
A 1 mg mL−1 stock solution was prepared in growth medium. NP solutions were sonicated in ice water in a Branson Digital Sonifier 450 operated at 10 W for 4 × 2.5 min. Stock solution was immediately diluted into sonicated growth medium for cell exposure experiments. This same procedure was used for exposure to ions and unexposed control cells to account for any protein denaturation or reactive oxygen species generation that may have occurred due to sonication.37Cell viability assessment.
Cell viability was measured using the CellTiter96® AQueous One Solution Cell Proliferation Assay (MTS, Promega). Cells were seeded at 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells/well in 96-well plates. Following 100% confluence, cells were rinsed with fresh growth medium, and exposed to 100 μL of LCO NP solution at various concentrations (0–50 mg L−1) for 24 or 48 h. As a negative control, cells were exposed to lysis buffer (Promega) for 30 min. After 24 or 48 h, cells were rinsed 3× with phenol red- and FBS-free growth medium and incubated with 20 μL of MTS in 100 μL of phenol red- and FBS-free growth medium for 4 h at 19 °C. The absorbance at 490 nm was read using a Beckman Coulter DTX 880 multimode detector plate reader.
000 cells/well in 96-well plates. Following 100% confluence, cells were rinsed with fresh growth medium, and exposed to 100 μL of LCO NP solution at various concentrations (0–50 mg L−1) for 24 or 48 h. As a negative control, cells were exposed to lysis buffer (Promega) for 30 min. After 24 or 48 h, cells were rinsed 3× with phenol red- and FBS-free growth medium and incubated with 20 μL of MTS in 100 μL of phenol red- and FBS-free growth medium for 4 h at 19 °C. The absorbance at 490 nm was read using a Beckman Coulter DTX 880 multimode detector plate reader.
Sample preparation for RNA-sequencing
Trout gill epithelial cells were seeded at 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells/dish in 60 mm dishes (Falcon® 3004). At 80–90% confluency, cells were exposed to fresh growth medium (unexposed control), 5 mg L−1 LCO NPs (subtoxic), 25 mg L−1 LCO NPs (toxic), 125 or 139 μM LiCl (Li+ ions), or 47 or 53 μM CoCl2 (Co2+ ions) for 24 or 48 h. The subtoxic and toxic LCO NP doses were determined using cell viability assessments as described above. The LiCl or CoCl2 concentrations were determined by quantifying dissolution from the toxic dose in growth medium at 24 or 48 h. We separated the ions from intact particles using ultrafiltration and determined the concentrations of Li+ and Co2+ using ICP-OES, as described in the ESI.† Five replicates of each sample condition were prepared and analyzed. After the desired time point, the solution was aspirated, 600 μL of lysis buffer (RTL) + β-mercaptoethanol was added, cells were scrapped from the dish and placed into a 2 mL tube. Samples were put on dry ice until all samples were collected and then stored at −80 °C. RNA was extracted using a Qiagen RNeasy mini kit (cat# 74104) according to the manufacturer's instructions.
000 cells/dish in 60 mm dishes (Falcon® 3004). At 80–90% confluency, cells were exposed to fresh growth medium (unexposed control), 5 mg L−1 LCO NPs (subtoxic), 25 mg L−1 LCO NPs (toxic), 125 or 139 μM LiCl (Li+ ions), or 47 or 53 μM CoCl2 (Co2+ ions) for 24 or 48 h. The subtoxic and toxic LCO NP doses were determined using cell viability assessments as described above. The LiCl or CoCl2 concentrations were determined by quantifying dissolution from the toxic dose in growth medium at 24 or 48 h. We separated the ions from intact particles using ultrafiltration and determined the concentrations of Li+ and Co2+ using ICP-OES, as described in the ESI.† Five replicates of each sample condition were prepared and analyzed. After the desired time point, the solution was aspirated, 600 μL of lysis buffer (RTL) + β-mercaptoethanol was added, cells were scrapped from the dish and placed into a 2 mL tube. Samples were put on dry ice until all samples were collected and then stored at −80 °C. RNA was extracted using a Qiagen RNeasy mini kit (cat# 74104) according to the manufacturer's instructions.
RNA-sequencing
The quality of the RNA samples was assessed using an Agilent 2100 Bioanalyzer. TruSeq stranded mRNA (cat# 20020594) was used to generate the cDNA library for Illumina NextSeq550 platform according to the manufacturer's protocol. Single-read sequencing of the cDNA libraries with a read length of 150 was performed using NextSeq 500/550 High Output v2 kit 150 cycles (cat# 20024907).Bioinformatics and statistical analyses applied to the RNA-Seq data
Read-trimming was conducted using bbduk38 with the following parameters: trimq = 10, qtrim = “rl”, maq = 10. Read quality was assessed using FastQC.39 Reads were aligned to the Oncorhynchus mykiss genome (GenBank accession GCA_002163495.1) by Bowtie2 (ref. 40) using parameters -local, --sensitive-local. Reads were aligned to genes using HTSeq-count41 with parameters -a = 1, --mode = “union”. Differential gene expression was assessed using the R package DESeq2,42 with all subsequent analysis performed in R unless otherwise stated. MDS plots were also generated using DESeq2.Differentially expressed genes (DEG), defined as having a log2 fold change greater than 1 or less than −1 as compared to controls, and having an adjusted p-value less than 0.05, were taken as input for enrichment analysis, with the set of O. mykiss genes as the background and using a Fisher's exact test with the EASE adjustment.43 Gene ontology (GO) terms acquired from annotations from the EGGNoG web service44 were used as target gene sets in the analysis. Up- and down-regulated genes were taken separately as separate lists in the analysis. Results from each test were assigned an enrichment score, which was the −log10 of the p-value of the test result. Enrichment scores of 6 and above were included in heatmap displays. Heatmaps were colored according to enrichment scores for each experimental condition and GO term, with greater intensity indicating higher scores. Hue indicates whether up- or down-regulated gene lists were found to be significantly enriched within genes of each GO term (reds = up-regulation, blues = down-regulation). When both up-and down-regulated genes were significantly enriched (lower enrichment score > 2), the cell was split between the two colors. Venn diagrams showing set relationships of DEGs were generated using the BioVenn webservice.45
Distances between experimental samples in MDS plots were calculated using Euclidean distance after a variance stabilizing transformation (VST) of the data.46 For distances between GO terms, the kappa statistic test47 was used. GO term clusters were then determined using hierarchical clustering with the average setting.
Results and discussion
Nanoparticle characterization
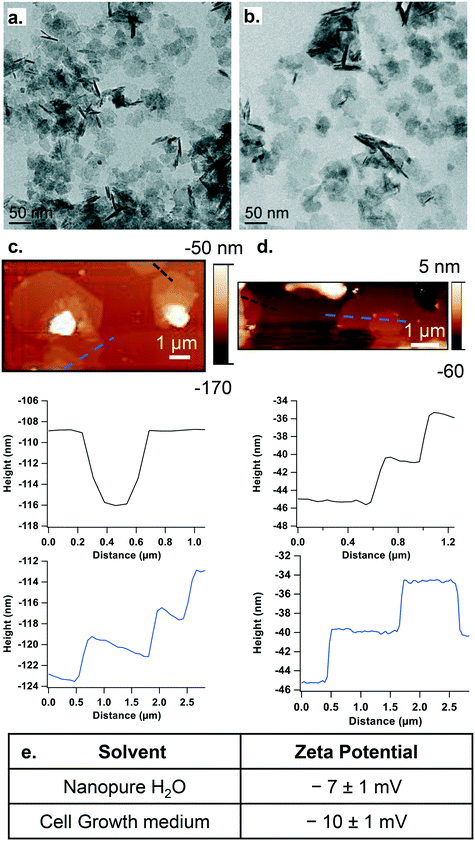
We focused on LixCoO2 NPs, which are used as cathode material in Li-ion batteries23 and are expected to be released in the environment with no infrastructure or incentive for recycling. The NPs used here have stoichiometry of Li0.87CoO2 determined by ICP-OES (referred to throughout as “LCO NPs”). XPS spectra for the Co 2p (Fig. S1b†) and Li 1s (Fig. S1c†) regions confirm the chemical composition of the LCO NPs. These NPs have a sheet-like morphology as shown by TEM (Fig. 1a and b), SEM (Fig. S1a†) and AFM (Fig. 1c and d), and a thickness of ∼5 nm in both nanopure H2O (Fig. 1a and c) and growth medium (Fig. 1b and d). The ζ-potential of the particles is −7 ± 1 mV in nanopure water and −10 ± 1 mV in growth medium (Fig. 1e). These results are in agreement with previous characterization of LCO NPs.29,35,36LCO NPs decrease cell viability independent of the dissolved ions
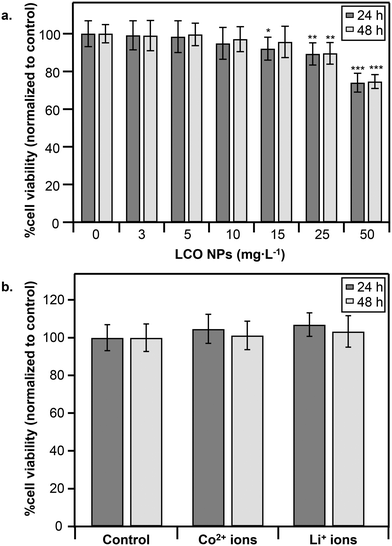
In the present study we aimed to determine the impact of LCO NPs on molecular pathways at doses above (toxic) and below (subtoxic) the threshold for detectable decrease in cell viability. It is important to examine effects at subtoxic doses since adverse effects can still occur in the absence of cell death. Fig. 2a shows dose-dependent responses of trout gill epithelial cells to LCO NPs following 24 h (dark gray) and 48 h (light gray) exposures at concentrations spanning 0–50 mg L−1. Following 24 h exposure, 15 mg L−1 was the lowest NP dose to induce a significant decrease in cell viability (92 ± 6% viability). Following 48 h exposure (Fig. 2a, light gray), a significant decrease in cell viability was observed in response to 25 mg L−1 (90 ± 6% viability), suggesting some recovery from the adverse response to NPs at 15 mg L−1 has occurred by 48 h. Based on these results, we used 5 mg L−1 (50 μM) as the subtoxic NP dose and 25 mg L−1 (260 μM) as the toxic NP dose for transcriptomic analysis.To determine the contribution of dissolved ions to the observed toxicity, we measured cell viability following exposure to Li+ or Co2+ ions at concentrations released into the growth medium by the toxic LCO NP dose (25 mg L−1) as determined by ICP-OES. Co2+ concentrations released at 24 h and 48 h were 47 ± 4 μM and 53 ± 5 μM (∼18% or 20% cobalt released from NPs), respectively. Li+ concentrations released at 24 h and 48 h were 125 ± 35 μM and 139 ± 38 μM (∼48% or 53% lithium released from NPs), respectively. Notably, no significant decrease in cell viability was observed following exposure to Li+ or Co2+ ions at their respective concentrations at 24 h (Fig. 2b, dark gray) or 48 h (Fig. 2b, light gray). While it is possible that a combination of Li+ and Co2+ ions could impact cell viability, our previous work studying the impact of LCO NPs in trout gill epithelial cells showed that a combination of Li+ and Co2+ ions at concentrations much greater than those released from the toxic NP dose in our study caused no impact on cell viability following 24 h exposure.29 These observations suggest that the decrease in cell viability following exposure to LCO NPs is not due to metal ions released into the culture medium, which is in agreement with previous observations in these cells exposed to LCO NPs for 3 h.29
Cells exposed to toxic or subtoxic LCO NP doses show distinct gene expression patterns, whereas cells exposed to ions show similar patterns as unexposed cells
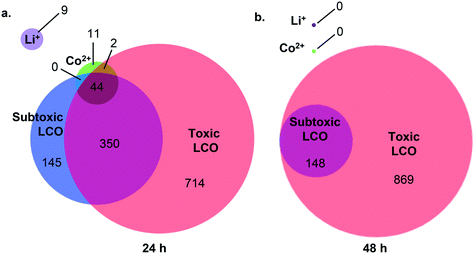
Using RNA-Seq, we quantified changes in gene expression and molecular pathways following exposure to LCO NPs at the subtoxic (5 mg L−1) and toxic (25 mg L−1) doses, as well as to Li+ or Co2+ ion at concentrations released from the toxic dose. Fig. 3 shows the total number of differentially expressed genes (DEGs) for all exposure conditions (p-value < 0.05 and a log2 fold change of >1 or <−1). At 24 h (Fig. 3a), all treatment groups showed changes in gene expression levels compared to unexposed control cells. Cells exposed to Co2+, Li+, LCO NPs at the subtoxic dose, and LCO NPs at the toxic dose resulted in 57, 9, 539, and 1110 DEGs, respectively. At 24 h, the number of DEGs that were unique to each of these exposure conditions were 11, 9, 145, and 714 DEGs, respectively. These results indicate a more pronounced impact on gene expression by the intact NPs compared to the ions. Furthermore, despite having no detectable impact on cell viability (Fig. 2), the subtoxic NP dose and, to a lesser extent, the ions impacted the expression of multiple genes. At 48 h (Fig. 3b), no difference in gene expression was detected in cells exposed to the ions compared to unexposed cells. Previous work using microarray analysis of human lung epithelial cells exposed to silver NPs reported 1000 DEGs following AgNP exposure as compared to only 133 DEGs following exposure to silver ions.48 Furthermore, lung epithelial cells can tolerate eight folds higher concentrations of intracellular zinc ions generated by exposure to ZnSO4 (or Zn2+) compared to those generated by exposure to intact zinc oxide NPs.49 This study also showed that exposure to ZnSO4 led to the accumulation of the ions in large intracellular vacuoles, whereas exposure to the intact NPs led to ion accumulation in endosomes and lysosomes.49 These observations, along with the results presented here, indicate different internalization and accumulation mechanisms, as well as response pathways, for the intact NPs vs. the ions, with possibly more evolved and adaptive response to the ions in certain cell types.At 48 h, the number of DEGs markedly decreased in cells exposed to the subtoxic NP dose, from 539 to 148 DEGs, while staying roughly the same in cells exposed to the toxic NP dose (1110 at 24 h and 1017 at 48 h) (Fig. 3b). These observations suggest a partial recovery in response to the subtoxic dose over time. 148 DEGs were identified in response to the subtoxic NP dose at 48 h, all of which were shared with the DEGs identified in response to the toxic dose (Fig. 3b), likely representing the cells' ongoing effort to survive the NP insult. Such decrease in DEGs over time was also observed in response to silver NPs exposure of HeLa cells, where fewer DEGs were found at 48 h compared with 24 h.50
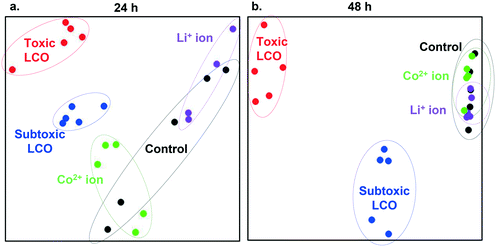
Fig. 4 utilizes multidimensional scaling (MDS) to visualize the variance in gene expression patterns between all samples generated in this study. In this approach, differences in gene expression behavior between samples are represented as distances between points on the grid. From these results we can conclude that cells exposed to ions show gene expression patterns similar to patterns found in control cells. However, distinct gene expression patterns are found in cells exposed to toxic and subtoxic doses. Despite the overlapping DEGs shown in Fig. 3, the individual samples under the toxic and subtoxic exposure conditions clustered into two distinct, toxic LCO and subtoxic LCO groups, indicating unique gene expression patterns under these two exposure conditions. These results hold across both time points with more clustered similarities arising between the unexposed control group and the ion exposure groups at 48 h.
Subtoxic and toxic NP doses upregulate pathways that fall into four main functional categories
To understand the impact of LCO NPs and ions on molecular processes or pathways we performed functional enrichment analysis. Fig. 5a shows the heatmap generated using this approach. Pathways (rows) with significant enrichment for up- or down-regulated genes in each condition (column) are represented. Higher color intensity indicates enrichment with greater significance, while color hue indicates the direction of change. Functional enrichment analysis in cells exposed to LCO NPs at both toxic and subtoxic doses predominantly resulted in up-regulated responses. The names of all pathways in the heatmap are listed in Table S1.†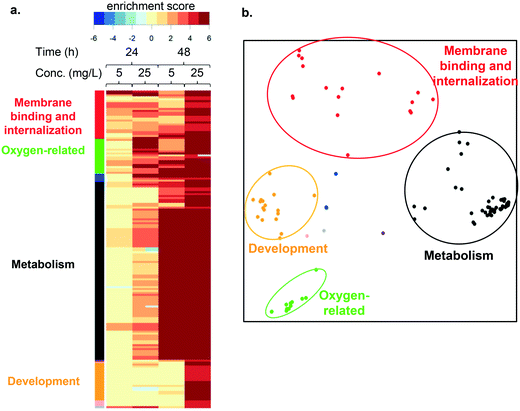 | ||
| Fig. 5 a. Heatmap showing results of the pathway enrichment analysis for cells exposed to the subtoxic or toxic LCO NP dose for 24 h or 48 h. A complete list of pathways can be found in Table S1.† The color bars on the left side of the heatmap indicate functional categories of the pathways, based on the clustering analysis described in b. Color intensity indicates significance level, while hue indicates direction: up- (red) or down- (blue) regulation. b. Pathways sharing similar groups of genes identified using multidimensional scaling, where distances between points (pathways) represent the degree of gene membership overlap between the pathways. Four major clusters (circled) were identified, which were named based on the key functions that they encompass. | ||
To understand how the enriched pathways are related to one another, we calculated distances between each pathway based on the overlap between the genes that comprise them. These distances were then used to graphically represent the relationships in an MDS plot (Fig. 5b), which uncovered 4 main clusters. We named these clusters according to the consensus functions within each. Cluster membership is indicated by the color-coded circles in Fig. 5b, and the color bars in Fig. 5a. These four functional categories include: membrane binding and internalization (red), oxygen-related processes (green), metabolism (black), and development (orange).
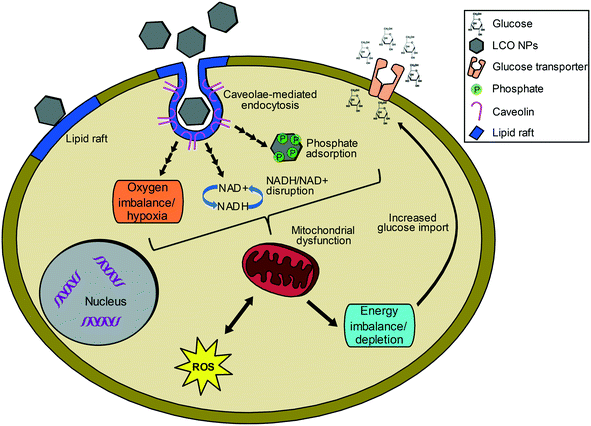
Engineered nanomaterials have been shown previously to disrupt the energy balance within cells, likely due to mitochondrial-related impacts.51,52 Impacts to mitochondrial function may account for the upregulation of metabolism related pathways observed here. Furthermore, LCO NPs have been shown previously to adsorb phosphate in solution.35,36 As phosphates are a crucial component in metabolism,53 among many other cellular functions, a depletion of phosphate due to sorption onto the NPs might disrupt metabolic balance and explain the upregulation of metabolic pathways observed here. Additionally, nanomaterials such as gold,54 platinum,55 and titanium dioxide56 have been shown previously to catalyze the oxidation of NADH to NAD+. An imbalance in NADH/NAD+ levels would have an impact on glycolysis. We hypothesize that the observed upregulation of NADH and glycolysis related pathways in our study indicates cellular attempts to restore or cope with the imbalance in NADH/NAD+ and/or energy levels. Overall, exposure to a toxic dose of LCO NPs for at least 24 h, as well as exposure to a subtoxic dose for 48 h, appeared to disrupt energy-related processes.
Fig. 6a shows the log2 fold change of the core genes (present in 90% of the pathways) in the metabolism cluster in response to the subtoxic and toxic NP doses at 24 and 48 h. These genes were strongly tied to glycolysis and were upregulated. The varying degrees of upregulation of both the genes (Fig. 6a) and the pathways (Fig. 5a) indicate dependency on both concentration and exposure time.
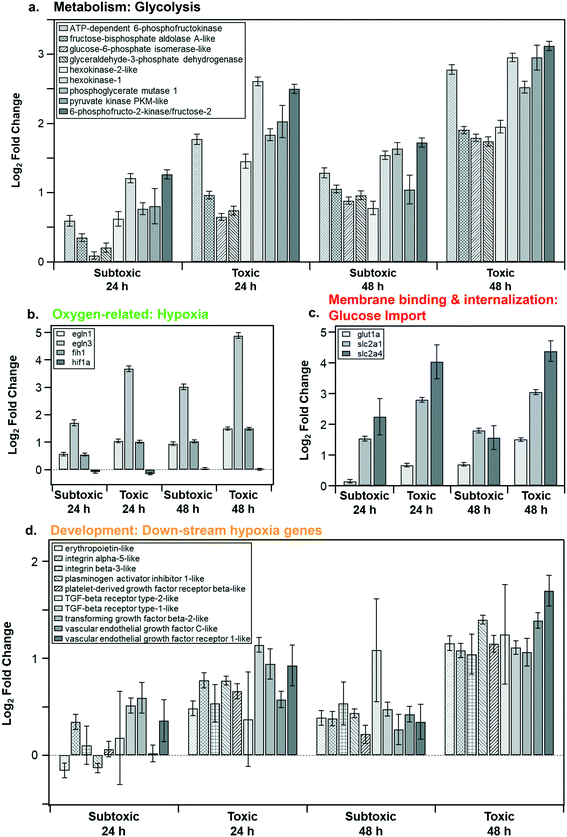 | ||
| Fig. 6 Expression levels of key genes (present in 90% of the pathways in their respective category) in response to subtoxic or toxic doses of LCO NPs for 24 or 48 h. The 4 main categories (Fig. 5) include: a. metabolism (glycolysis), b. oxygen-related (hypoxia), c. membrane binding and internalization (glucose import), d. development (down-stream hypoxia genes). | ||
Fig. 6b shows the log2 fold change for genes identified to play a key role in the oxygen-related category. We observed that egln1 and egln3 were upregulated, both of which are known to be responsive to hypoxia.59 The transcript expression of hif1a remains stable since its oxygen sensitivity is mediated at the protein level through continuous proteasome degradation during normoxia.59 The HIF-dependent hypoxia response is known to activate genes that control mitochondrial metabolism, angiogenesis, cellular oxygen homeostasis, and cell proliferation.60 Likely, the interplay between hypoxic conditions and energy depletion leads to our observed impacts on the enriched pathways.
To directly compare the responses of the toxic and subtoxic doses (rather than their degree of change compared to control), we first identified genes showing highly significant differences in their expression levels between the toxic and subtoxic doses at each of the time points. We then used these genes for pathway enrichment. The results are shown in Fig. S2.† Similar to pathways found in the metabolism category discussed above, the toxic dose caused increased expression of genes related to energy production and stress compared to the subtoxic dose at both time points.
Exposure to Li+ or Co2+ ions downregulated unique pathways, which recovered to normal levels at 48 h
24 h exposure to Li+ or Co2+ ions, at concentrations released by the toxic NPs dose, resulted in downregulation of unique responses, as indicated by the blue color in the heatmap in Fig. S3.† Membership in these pathways was encompassed by only 9 genes. Following 24 h exposure to Li+ ions, all the impacted pathways were related to metabolism, cellular respiration, or electron transport processes. Li+ ions have been shown previously to downregulate antioxidant genes,30,64 as well as inhibit Mg2+-dependent enzymes by competing for the binding site with Mg2+ ions, leading to a decrease in enzyme activity and in structural stability.65 The competition between Li+ and Mg2+ ions could lead to the observed downregulated pathways, although the exact mechanism of interaction remains largely unknown. 24 h exposure to Co2+ ions led to only two downregulated pathways, which are involved in collagen type VI trimer and collagen beaded filament. The downregulation of collagen-related genes and pathways by Co2+ ions has been reported previously in osteoblast cells,66,67 indicating such effects occur in different cell types. Importantly, cells exposed to Co2+ or Li+ ions for 48 h showed no DEGs compared to unexposed cells (Fig. 3b), indicating a full recovery of the cells to normal function. This is in stark contrast to the ∼1000 genes and ∼150 genes impacted by the 48 h exposure to the toxic and subtoxic NP doses, respectively. These observations highlight the NP-specific adverse effects of LCO NPs independent of the released ions.Conclusions
The main new findings that have emerged from our work include the identification of intact LCO NPs as the sole cause for perturbation of key molecular pathways and ultimately toxicity, with minimal contribution from the dissolved ions, and that a subtoxic dose of the NPs, which shows no impact on cell viability, induces significant changes in molecular processes critical to the normal function of the cells. While the subtoxic dose (5 mg L−1) used in our study might be higher than the expected NP concentrations in surface water,6 our findings highlight the need to study molecular level impacts of NPs at concentrations that show no impact on cell viability.We identified molecular pathways that were uniquely regulated by subtoxic and toxic NP doses. These pathways fell into four key functional categories: metabolism related, oxygen-related, membrane binding and internalization processes, and developmental processes. Drastic upregulation of metabolism related processes was observed at 48 h by both toxic and subtoxic NP doses, suggesting prolonged exposures disrupt energy balance. Multiple oxygen related processes, mainly hypoxia related, were upregulated at 24 h, with further upregulation by both subtoxic and toxic doses at 48 h. Membrane binding and internalization processes, mainly glucose import, transport and homeostasis related, were upregulated by both subtoxic and toxic doses at both 24 and 48 h, suggesting cellular attempts to increase glucose availability in response to energy depletion and hypoxia. Developmental processes were mainly upregulated by the toxic dose at 48 hours, including hypoxia-regulated genes. The regulation patterns of these pathways were found to be unique to the subtoxic and toxic doses, indicating unique physiological states of the cells.
Fig. 7 illustrates hypothesized connections and possible explanations for the observed regulated processes, which represent potential areas for future studies. The upregulation of metabolism related processes by both subtoxic and toxic NP doses suggests energy imbalance or depletion, possibly due to mitochondrial disfunction,51,52 and cellular attempt to restore balance. We hypothesize this energy imbalance could result from NP-induced hypoxia, as suggested by the observed upregulation of pathways related to decreased oxygen levels and hypoxia by both the toxic and subtoxic doses at 24 and 48 hours. Furthermore, the energy imbalance could result from depletion of phosphate from the growth medium,35,36 and/or interference with NADH/NAD+ balance.54–56 The cellular attempt to restore energy balance by upregulating energy related machineries can also explain the observed upregulation of glucose import, transport, and homeostasis processes, among other key nutrients, by both the subtoxic and toxic doses at 24 and 48 h. Developmental processes, which include hypoxia-responsive genes, were mainly upregulated by the toxic dose at 48 h and further suggest mitochondrial dysfunction and energy depletion. Although the exact mechanism is unclear, both subtoxic and toxic NP doses impacted critical molecular processes governing cellular metabolism and energy production as well as oxygen homeostasis. While global transcriptional analysis provides insights to even slight deviations from the normal state of the cell, studying responses at the protein level will provide further insights to the functional state and deeper understanding of NP impacts on cellular functions.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Science Foundation under Grant No. CHE-2001611, the NSF Center for Sustainable Nanotechnology (CSN). The CSN is part of the Centers for Chemical Innovation Program. A portion of the research was performed using the Environmental Molecular Sciences Laboratory (EMSL), a DOE Scientific User Facility sponsored by the Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory (PNNL). E. D. L. is supported by the NSF Graduate Research Fellowship grant DGE-1747503, and by the University of Wisconsin Alumni Research Foundation. Authors acknowledge the use of facilities supported by NSF through the University of Wisconsin Materials Research Science and Engineering Center (DMR-1720415).References
- M. A. Maurer-Jones, I. L. Gunsolus, C. J. Murphy and C. L. Haynes, Toxicity of engineered nanoparticles in the environment, Anal. Chem., 2013, 85, 3036–3049 CrossRef CAS.
- A. Nel, T. Xia, L. Mädler and N. Li, Toxic potential of materials at the nanolevel, Science, 2006, 311, 622–627 CrossRef CAS.
- C. J. Murphy, A. M. Vartanian, F. M. Geiger, R. J. Hamers, J. Pedersen, Q. Cui, C. L. Haynes, E. E. Carlson, R. Hernandez, R. D. Klaper, G. Orr and Z. Rosenzweig, Biological responses to engineered nanomaterials: Needs for the next decade, ACS Cent. Sci., 2015, 1, 117–123 CrossRef CAS.
- R. J. Hamers, Nanomaterials and global sustainability, Acc. Chem. Res., 2017, 50, 633–637 CrossRef CAS.
- F. Gottschalk, T. Sonderer, R. W. Scholz and B. Nowack, Modeled environmental concentrations of engineered nanomaterials (TiO2, ZnO, Ag, CNT, fullerenes) for different regions, Environ. Sci. Technol., 2009, 43, 9216–9222 CrossRef CAS.
- F. Gottschalk, T. Sun and B. Nowack, Environmental concentrations of engineered nanomaterials: Review of modeling and analytical studies, Environ. Pollut., 2013, 181, 287–300 CrossRef CAS.
- S. A. Love, M. A. Maurer-Jones, J. W. Thompson, Y. S. Lin and C. L. Haynes, Assessing nanoparticle toxicity, Annu. Rev. Anal. Chem., 2012, 5, 181–205 CrossRef CAS.
- B. J. Marquis, S. A. Love, K. L. Braun and C. L. Haynes, Analytical methods to assess nanoparticle toxicity, Analyst, 2009, 134, 425–439 RSC.
- J. A. Yang, S. E. Lohse and C. J. Murphy, Tuning cellular response to nanoparticles via surface chemistry and aggregation, Small, 2014, 10, 1642–1651 CrossRef CAS.
- P. Falagan-Lotsch, E. M. Grzincic and C. J. Murphy, One low-dose exposure of gold nanoparticles induces long-term changes in human cells, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 13318–13323 CrossRef CAS.
- M. Chevallet, B. Gallet, A. Fuchs, P. H. Jouneau, K. Um, E. Mintz and I. Michaud-Soret, Metal homeostasis disruption and mitochondrial dysfunction in hepatocytes exposed to sub-toxic doses of zinc oxide nanoparticles, Nanoscale, 2016, 8, 18495–18506 RSC.
- J. S. Choi, R. O. Kim, S. Yoon and W. K. Kim, Developmental toxicity of zinc oxide nanoparticles to zebrafish (Danio rerio): A transcriptomic analysis, PLoS One, 2016, 11, e0160763 CrossRef.
- S. C. Tilton, N. J. Karin, A. Tolic, Y. Xie, X. Lai, R. F. Hamilton, Jr., K. M. Waters, A. Holian, F. A. Witzmann and G. Orr, Three human cell types respond to multi-walled carbon nanotubes and titanium dioxide nanobelts with cell-specific transcriptomic and proteomic expression patterns, Nanotoxicology, 2014, 8, 533–548 CrossRef CAS.
- D. T. Jayaram, A. Kumar, L. E. Kippner, P.-Y. Ho, M. L. Kemp, Y. Fan and C. K. Payne, TiO2 nanoparticles generate superoxide and alter gene expression in human lung cells, RSC Adv., 2019, 9, 25039–25047 RSC.
- H. D. Mitchell, L. M. Markillie, W. B. Chrisler, M. J. Gaffrey, D. Hu, C. J. Szymanski, Y. Xie, E. S. Melby, A. Dohnalkova, R. C. Taylor, E. K. Grate, S. K. Cooley, J. E. McDermott, A. Heredia-Langner and G. Orr, Cells respond to distinct nanoparticle properties with multiple strategies as revealed by single-cell RNA-Seq, ACS Nano, 2016, 10, 10173–10185 CrossRef CAS.
- A. R. Gliga, S. Di Bucchianico, J. Lindvall, B. Fadeel and H. L. Karlsson, RNA-sequencing reveals long-term effects of silver nanoparticles on human lung cells, Sci. Rep., 2018, 8, 6668 CrossRef.
- J. Ndika, U. Seemab, W.-L. Poon, V. Fortino, H. El-Nezami, P. Karisola and H. Alenius, Silver, titanium dioxide, and zinc oxide nanoparticles trigger miRNA/isomiR expression changes in THP-1 cells that are proportional to their health hazard potential, Nanotoxicology, 2019, 13, 1380–1395 CrossRef CAS.
- M. Zheng, J. Lu and D. Zhao, Toxicity and transcriptome sequencing (RNA-seq) analyses of adult zebrafish in response to exposure carboxymethyl cellulose stabilized iron sulfide nanoparticles, Sci. Rep., 2018, 8, 8083 CrossRef.
- D. Sun, L. Gong, J. Xie, X. He, S. Chen, L. A. Luodan, Q. Li, Z. Gu and H. Xu, Evaluating the toxicity of silicon dioxide nanoparticles on neural stem cells using RNA-Seq, RSC Adv., 2017, 7, 47552–47564 RSC.
- J. K. Carrow, L. M. Cross, R. W. Reese, M. K. Jaiswal, C. A. Gregory, R. Kaunas, I. Singh and A. K. Gaharwar, Widespread changes in transcriptome profile of human mesenchymal stem cells induced by two-dimensional nanosilicates, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E3905–E3913 CrossRef CAS.
- M. S. Whittingham, Lithium batteries and cathode materials, Chem. Rev., 2004, 104, 4271–4301 CrossRef CAS.
- J. B. Goodenough, Evolution of strategies for modern rechargeable batteries, Acc. Chem. Res., 2013, 46, 1053–1061 CrossRef CAS.
- N. Nitta, F. Wu, J. T. Lee and G. Yushin, Li-ion battery materials: Present and future, Mater. Today, 2015, 18, 252–264 CrossRef CAS.
- H. Wang, Y. I. Jang, B. Huang, D. R. Sadoway and Y. M. Chiang, TEM study of electrochemical cycling-induced damage and disorder in LiCoO2 cathodes for rechargeable lithium batteries, J. Electrochem. Soc., 1999, 146, 473–480 CrossRef CAS.
- J. B. Dunn, L. Gaines, J. C. Kelly, C. James and K. G. Gallagher, The significance of Li-ion batteries in electric vehicle life-cycle energy and emissions and recycling's role in its reduction, Energy Environ. Sci., 2015, 8, 158–168 RSC.
- K. M. Winslow, S. J. Laux and T. G. Townsend, A review on the growing concern and potential management strategies of waste lithium-ion batteries, Resour., Conserv. Recycl., 2018, 129, 263–277 CrossRef.
- M. N. Hang, I. L. Gunsolus, H. Wayland, E. S. Melby, A. C. Mensch, K. R. Hurley, J. A. Pedersen, C. L. Haynes and R. J. Hamers, Impact of nanoscale lithium nickel manganese cobalt oxide (NMC) on the bacterium Shewanella oneidensis MR-1, Chem. Mater., 2016, 28, 1092–1100 CrossRef CAS.
- J. Bozich, M. Hang, R. Hamers and R. Klaper, Core chemistry influences the toxicity of multicomponent metal oxide nanomaterials, lithium nickel manganese cobalt oxide, and lithium cobalt oxide to Daphnia magna, Environ. Toxicol. Chem., 2017, 36, 2493–2502 CrossRef CAS.
- E. S. Melby, Y. Cui, J. Borgatta, A. C. Mensch, M. N. Hang, W. B. Chrisler, A. Dohnalkova, J. M. Van Gilder, C. M. Alvarez, J. N. Smith, R. J. Hamers and G. Orr, Impact of lithiated cobalt oxide and phosphate nanoparticles on rainbow trout gill epithelial cells, Nanotoxicology, 2018, 12, 1166–1181 CrossRef CAS.
- Y. Cui, E. S. Melby, A. C. Mensch, E. D. Laudadio, M. N. Hang, A. Dohnalkova, D. Hu, R. J. Hamers and G. Orr, Quantitative mapping of oxidative stress response to lithium cobalt oxide nanoparticles in single cells using multiplexed in situ gene expression analysis, Nano Lett., 2019, 19, 1990–1997 CrossRef CAS.
- N. R. Bury, S. Schnell and C. Hogstrand, Gill cell culture systems as models for aquatic environmental monitoring, J. Exp. Biol., 2014, 217, 639–650 CrossRef CAS.
- L. C. Felix, V. A. Ortega and G. G. Goss, Cellular uptake and intracellular localization of poly (acrylic acid) nanoparticles in a rainbow trout (Oncorhynchus mykiss) gill epithelial cell line, RTgill-W1, Aquat. Toxicol., 2017, 192, 58–68 CrossRef CAS.
- A. C. Mensch, E. S. Melby, E. D. Laudadio, I. U. Foreman-Ortiz, Y. Zhang, A. Dohnalkova, D. Hu, J. A. Pedersen, R. J. Hamers and G. Orr, Preferential interactions of primary amine-terminated quantum dots with membrane domain boundaries and lipid rafts revealed with nanometer resolution, Environ. Sci.: Nano, 2020, 7, 149–161 RSC.
- H. Chen and C. P. Grey, Molten salt synthesis and high rate performance of the “desert-rose” form of LiCoO2, Adv. Mater., 2008, 20, 2206–2210 CrossRef CAS.
- E. D. Laudadio, P. Ilani-Kashkouli, C. M. Green, N. J. Kabengi and R. J. Hamers, Interaction of phosphate with lithium cobalt oxide nanoparticles: A combined spectroscopic and calorimetric study, Langmuir, 2019, 35, 16640–16649 CrossRef CAS.
- E. D. Laudadio, J. W. Bennett, C. M. Green, S. E. Mason and R. J. Hamers, Impact of phosphate adsorption on complex cobalt oxide nanoparticle dispersibility in aqueous media, Environ. Sci. Technol., 2018, 52, 10186–10195 CrossRef CAS.
- J. Cohen, G. Deloid, G. Pyrgiotakis and P. Demokritou, Interactions of engineered nanomaterials in physiological media and implications for in vitro dosimetry, Nanotoxicology, 2013, 7, 417–431 CrossRef CAS.
- Bbtools, https://jgi.doe.gov/data-and-tools/bbtools/, (accessed February, 2020) Search PubMed.
- Fastqc, https://www.bioinformatics.babraham.ac.uk/projects/fastqc/, (accessed February, 2020) Search PubMed.
- Bowtie 2, http://bowtie-bio.sourceforge.net/bowtie2/index.shtml, (accessed February, 2020) Search PubMed.
- S. Anders, P. T. Pyl and W. Huber, HTSeq--a Python framework to work with high-throughput sequencing data, Bioinformatics, 2015, 31, 166–169 CrossRef CAS.
- M. I. Love, W. Huber and S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol., 2014, 15, 550 CrossRef.
- D. A. Hosack, G. D. Jr, B. T. Sherman, H. C. Lane and R. A. Lempicki, Identifying biological themes within lists of genes with ease, Genome Biol., 2003, 4, R70 CrossRef.
- Eggnog, http://eggnogdb.embl.de/#/app/emapper, (accessed February, 2020) Search PubMed.
- T. Hulsen, Venn diagram, http://biovenn.nl/venndiagram.tk/create.php, (accessed February, 2020) Search PubMed.
- S. Anders and W. Huber, Differential expression analysis for sequence count data, Genome Biol., 2010, 11, R106 CrossRef CAS.
- J. Cohen, A coefficient of agreement for nominal scales, Educ. Psychol. Meas., 1960, 20, 37–48 CrossRef.
- R. Foldbjerg, E. S. Irving, Y. Hayashi, D. S. Sutherland, K. Thorsen, H. Autrup and C. Beer, Global gene expression profiling of human lung epithelial cells after exposure to nanosilver, Toxicol. Sci., 2012, 130, 145–157 CrossRef CAS.
- C. Mihai, W. B. Chrisler, Y. Xie, D. Hu, C. J. Szymanski, A. Tolic, J. A. Klein, J. N. Smith, B. J. Tarasevich and G. Orr, Intracellular accumulation dynamics and fate of zinc ions in alveolar epithelial cells exposed to airborne ZnO nanoparticles at the air-liquid interface, Nanotoxicology, 2015, 9, 9–22 CrossRef CAS.
- L. Xu, T. Takemura, M. Xu and N. Hanagata, Toxicity of silver nanoparticles as assessed by global gene expression analysis, Mater. Express, 2011, 1, 74–79 CrossRef CAS.
- L. Pagano, E. Maestri, M. Caldara, J. C. White, N. Marmiroli and M. Marmiroli, Engineered nanomaterial activity at the organelle level: Impacts on the chloroplasts and mitochondria, ACS Sustainable Chem. Eng., 2018, 6, 12562–12579 CrossRef CAS.
- M. Burkard, A. Betz, K. Schirmer and A. Zupanic, Common gene expression patterns in environmental model organisms exposed to engineered nanomaterials: A meta-analysis, Environ. Sci. Technol., 2020, 54(1), 335–344 CAS.
- T. Tian, X. Y. Chu, Y. Yang, X. Zhang, Y. M. Liu, J. Gao, B. G. Ma and H. Y. Zhang, Phosphates as energy sources to expand metabolic networks, Life, 2019, 9(2), 43 CrossRef CAS.
- X. Huang, I. H. El-Sayed, X. Yi and M. A. El-Sayed, Gold nanoparticles: Catalyst for the oxidation of NADH to NAD+, J. Photochem. Photobiol., B, 2005, 81, 76–83 CrossRef CAS.
- K. Hikosaka, J. Kim, M. Kajita, A. Kanayama and Y. Miyamoto, Platinum nanoparticles have an activity similar to mitochondrial NADH:Ubiquinone oxidoreductase, Colloids Surf., B, 2008, 66, 195–200 CrossRef CAS.
- M. R. Majidi, N. Bagheri and J. Hassanzadeh, Nano TiO2 modified carbon-ceramic electrode and its application for electrocatalytic oxidation of NADH, J. Chin. Chem. Soc., 2015, 62, 632–639 CrossRef CAS.
- M. P. Murphy, How mitochondria produce reactive oxygen species, Biochem. J., 2009, 417, 1–13 CrossRef CAS.
- K. L. Eales, K. E. Hollinshead and D. A. Tennant, Hypoxia and metabolic adaptation of cancer cells, Oncogenesis, 2016, 5, e190 CrossRef CAS.
- L. del Peso, M. C. Castellanos, E. Temes, S. Martin-Puig, Y. Cuevas, G. Olmos and M. O. Landazuri, The von Hippel Lindau/hypoxia-inducible factor (HIF) pathway regulates the transcription of the HIF-proline hydroxylase genes in response to low oxygen, J. Biol. Chem., 2003, 278, 48690–48695 CrossRef CAS.
- H. Choudhry and A. L. Harris, Advances in hypoxia-inducible factor biology, Cell Metab., 2018, 27, 281–298 CrossRef CAS.
- A. Ouiddir, C. Planès, I. Fernandes, A. VanHesse and C. Clerici, Hypoxia upregulates activity and expression of the glucose transporter GLUT1 in alveolar epithelial cells, Am. J. Respir. Cell Mol. Biol., 1999, 21, 710–718 CrossRef CAS.
- C. Furuta, T. Miyamoto, T. Takagi, Y. Noguchi, J. Kaneko, S. Itoh, T. Watanabe and F. Itoh, Transforming growth factor-β signaling enhancement by long-term exposure to hypoxia in a tumor microenvironment composed of Lewis lung carcinoma cells, Cancer Sci., 2015, 106, 1524–1533 CrossRef CAS.
- X. Mingyuan, P. Qianqian, X. Shengquan, Y. Chenyi, L. Rui, S. Yichen and X. Jinghong, Hypoxia-inducible factor-1α activates transforming growth factor-β1/Smad signaling and increases collagen deposition in dermal fibroblasts, Oncotarget, 2018, 9, 3188–3197 CrossRef.
- M. S. Allagui, C. Vincent, A. El feki, Y. Gaubin and F. Croute, Lithium toxicity and expression of stress-related genes or proteins in A549 cells, Biochim. Biophys. Acta, 2007, 1773, 1107–1115 CrossRef CAS.
- E. Jakobsson, O. Arguello-Miranda, S. W. Chiu, Z. Fazal, J. Kruczek, S. Nunez-Corrales, S. Pandit and L. Pritchet, Towards a unified understanding of lithium action in basic biology and its significance for applied biology, J. Membr. Biol., 2017, 250, 587–604 CrossRef CAS.
- L. Anissian, A. Stark, H. Dahlstrand, B. Granberg, V. Good and E. Bucht, Cobalt ions influence proliferation and function of human osteoblast-like cells, Acta Orthop. Scand., 2009, 73, 369–374 CrossRef.
- A. Jonitz-Heincke, M. L. Sellin, A. Seyfarth, K. Peters, B. Mueller-Hilke, T. Fiedler, R. Bader and A. Klinder, Analysis of cellular activity short-term exposure to cobalt and chromium ions in mature human osteoblasts, Materials, 2019, 12(17), 2771 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Additional nanoparticle characterization, pathway enrichment following exposure to ion controls, and a complete list of pathways and associated GO IDs from Fig. 5. See DOI: 10.1039/d0en00844c |
| ‡ Equally contributed authors. |
| This journal is © The Royal Society of Chemistry 2020 |