Open Access Article
Open Access ArticlePreferential interactions of primary amine-terminated quantum dots with membrane domain boundaries and lipid rafts revealed with nanometer resolution†
Arielle C.
Mensch
 a,
Eric S.
Melby‡
a,
Eric S.
Melby‡
 a,
Elizabeth D.
Laudadio
b,
Isabel U.
Foreman-Ortiz
b,
Yongqian
Zhang
b,
Alice
Dohnalkova
a,
Dehong
Hu
a,
Elizabeth D.
Laudadio
b,
Isabel U.
Foreman-Ortiz
b,
Yongqian
Zhang
b,
Alice
Dohnalkova
a,
Dehong
Hu
 a,
Joel A.
Pedersen
a,
Joel A.
Pedersen
 bc,
Robert J.
Hamers
bc,
Robert J.
Hamers
 b and
Galya
Orr
b and
Galya
Orr
 *a
*a
aEnvironmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, P.O. Box 999 MSIN K8-88, Richland, WA 99354, USA. E-mail: Galya.Orr@pnnl.gov; Tel: +1 509 371 6127
bDepartment of Chemistry, University of Wisconsin, Madison, WI 53706, USA
cDepartments of Soil Science, and Civil & Environmental Engineering, University of Wisconsin, Madison, WI 53706, USA
First published on 23rd December 2019
Abstract
The initial interactions of engineered nanoparticles (NPs) with living cells are governed by physicochemical properties of the NP and the molecular composition and structure of the cell membrane. Eukaryotic cell membranes contain lipid rafts – liquid-ordered nanodomains involved in membrane trafficking and molecular signaling. However, the impact of these membrane structures on cellular interactions of NPs remains unclear. Here we investigate the role of membrane domains in the interactions of primary amine-terminated quantum dots (Qdots) with liquid-ordered domains or lipid rafts in model membranes and intact cells, respectively. Using correlative atomic force and fluorescence microscopy, we found that the Qdots preferentially localized to boundaries between liquid-ordered and liquid-disordered phases in supported bilayers. The Qdots also induced holes at these phase boundaries. Using super resolution fluorescence microscopy (STORM), we found that the Qdots preferentially co-localized with lipid rafts in the membrane of intact trout gill epithelial cells – a model cell type for environmental exposures. Our observations uncovered preferential interactions of amine-terminated Qdots with liquid-ordered domains and their boundaries, possibly due to membrane curvature at phase boundaries creating energetically favorable sites for NP interactions. The preferential interaction of the Qdots with lipid rafts supports their potential internalization via lipid raft-mediated endocytosis and interactions with raft-resident signaling molecules.
Environmental significanceThe initial interactions of engineered nanoparticles (NPs) at the cell membrane govern NP internalization and impact on cellular functions. These interactions depend on physicochemical properties of the NP and the molecular structure of the cell membrane. Liquid-ordered domains, or lipid rafts, are key structures in eukaryotic cell membranes but their role in NP interactions is unclear. We focus on quantum dots (Qdots) – technologically relevant NPs with broad applications and concerns over their environmental release. We show that primary amine-terminated Qdots preferentially interact with liquid-ordered domain boundaries in bilayers and with lipid rafts in intact cells. These findings shed new light on mechanisms underlying NP-cell interactions and ultimately contribute to predictive evaluations of environmental health and safety for the use of NPs. |
Introduction
The growing use of nanoparticles (NPs) in emerging technologies raises significant concerns about their environmental fate and biological impact upon disposal.1,2 Nanoparticle–cell interactions are therefore an important factor to consider when evaluating environmental health and safety for the use of NPs.3 Interactions that occur between NPs and cells depend on the physicochemical properties of the NPs, such as size, shape, core material, aggregation state, and surface functionalization.4,5 The variability in the types of NPs in use, as well as the biomolecular complexity of the cell present a challenge for achieving a mechanistic understanding of these interactions. NP interactions with cells can occur through multiple mechanisms, among them are direct interactions with the cell membrane6 or within the cytoplasm after passive transport7,8 or endocytic uptake.9One route of NP uptake into eukaryotic cells is through lipid raft-mediated endocytosis.10,11 Lipid rafts in eukaryotic cells are membrane nanodomains enriched in sphingolipids and cholesterol or other sterols,12 which play an important role in both membrane trafficking and signal transduction.11,13 To understand the complex interactions between NPs and cells, supported lipid bilayers (SLBs) are often used as a model for the plasma membrane.14–17 While SLBs lack the chemical complexity of intact cells, they enable controlled and systematic study of the chemical components that serve important roles in the function of the cell membrane.16,18,19 Membrane domains can be formed in SLBs by using lipid species that phase segregate into either liquid-ordered (Lo) or liquid-disordered (Ld) phases, where the Lo domains mimic lipid rafts. Phase-segregation in lipid bilayers has been shown previously to be an important factor in NP interactions. Anionic diamond NPs can alter the packing and shape of ordered gel domains.20 Positively-charged dendrimers and polymeric NPs have been shown to selectively disrupt the liquid phase of the bilayer over the gel phase.21,22 Amphiphilic dendrimers have been shown to interact distinctly with bilayers of different phases – inducing partial solubilization of fluid bilayers, lipid patches and local depressions in gel bilayers, and a ribbon-like network with spherical aggregates in bilayers with both fluid and gel phases.23 Polymeric quantum dots have shown increased attachment to two-phase bilayers as compared to the single-component phases, with preferential interaction occurring within the Ld phase.24 Recent work by our group showed that quaternary amine-terminated quantum dots (Qdots) caused the collapse of the Lo phase.25 Furthermore, experimental16 and computational26 studies suggested that primary amine-terminated gold NPs interact favorably at the Lo/Ld phase boundary, but direct observation of such interaction has not yet been achieved. While these unique interactions with phase-segregated bilayers have been explored for a very small subset of NP types, the generalizability of these results to NPs with other surface functionalizations remains elusive. Importantly, while a preferential interaction of primary amine-terminated NPs at the boundary of the Lo and Ld phases has been hypothesized, direct observations with the required nanometer-scale spatial resolution and detection selectivity have not been achieved.
Observation of the interactions between NPs and membrane nanodomains requires tools that can localize NPs in relation to important components within the plasma membranes with nanometer spatial resolution. Atomic force microscopy (AFM) has been used previously to study interactions between NPs and phase-segregated bilayers, providing Ångström resolution and a three-dimensional analysis of the surface.21,22,25 Fluorescence microscopy has also been used to examine interactions between NPs and phase-segregated model membranes.20,24,27 While not attaining the resolution of AFM, fluorescence microscopy provides high detection selectivity enabled by fluorescence tagging.28 Correlative AFM and fluorescence microscopy can localize NPs in relation to other fluorescently tagged biomolecules with very high spatial resolution and detection selectivity. The correlative approach has been used to characterize phase segregation in SLBs29,30 and membrane restructuring due to the addition of ceramide,31,32 as well as to study the impact of 2,4-dichlorophenoxyacetic acid on bacterial, fungal, and human cells33 and localize labeled fusion proteins within mammalian cells.34 Such a correlative approach holds promise for localizing the interactions of NPs with membrane molecules or structures with exceptional accuracy.
Here we use correlative atomic force and fluorescence microscopy to localize primary amine-terminated Qdots in SLBs containing phase-segregated domains with nanometer resolution and high selectivity. We further use super resolution fluorescence microscopy – stochastic optical reconstruction microscopy (STORM) – to localize Qdots with respect to lipid rafts at the membrane of intact cells with nanometer resolution. We chose CdSe/ZnS core/shell Qdots due to their technological relevance35 and the concern over their release into the environment,36 as well as their fluorescence, which can be detected by both conventional fluorescence microscopy and super resolution STORM. The primary amine surface termination was chosen as part of a systematic effort to decipher the impact of distinct positive functional groups at NP surfaces on the membrane structure.16,25 Further, primary amine-terminated NPs have been hypothesized previously to interact with the Lo/Ld phase boundary,16,26 but direct observations have not been achieved. We chose the trout gill epithelial cell line, an established model system for studying environmental exposures on aquatic organisms.37,38 Our studies provide direct observations of preferential interactions of primary amine-terminated Qdots at domain boundaries in model membranes and with lipid rafts at the cell surface with exceptional spatial resolution and detection selectivity. These observations provide a molecular level understanding of the initial interactions of primary amine-terminated NPs with the cell membrane, and a molecular context for further studies of their internalization mechanism and impact on the cellular function.
Experimental
Quantum dot characterization
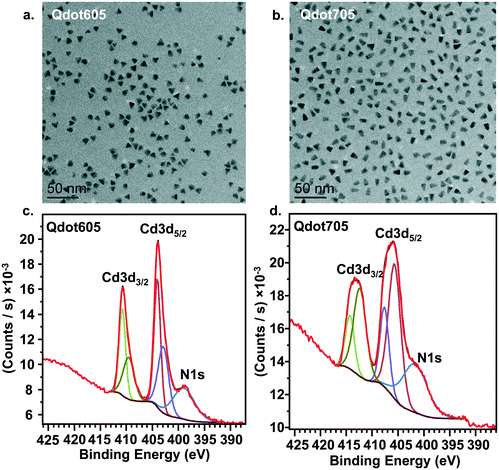
Quantum dots with a cadmium selenide core and zinc sulfide shell were procured from Invitrogen (QDot605 ITK™ or QDot705 ITK™). The Qdots were functionalized with amine-derivatized PEG, which conferred stability in aqueous solution. The company estimate for the number of amine groups per Qdot was 80–100. Using Nuclear magnetic resonance (NMR), we found 107.8 amines per Qdot (see Supplemental methods and Fig. S3†). Using Brunauer–Emmett–Teller (BET), we also found a surface amine density of 0.1 amine per nm2. Transmission electron microscopy (TEM), laser Doppler microelectrophoresis, and x-ray photoelectron spectroscopy (XPS) were conducted as follows on the particles.For size determination by TEM, 5 μL of stock Qdot solution (8000 nM) or a 1000-fold dilution in growth medium were dropcast onto a 100 Cu mesh grid coated with Formvar and carbon (Electron Microscopy Sciences). Excess solution was wicked away using a piece of filter paper, and the sample was left to air dry. Samples were imaged using a Tecnai T-12 TEM (FEI) operating at 120 kV with a LaB6 filament. A 2 × 2 k Ultrascan 1000 CCD (Gatan) was used to digitally collect the images. Size analysis was conducted by individually counting the width and length of 100 different triangular particles using Fiji.39 The apparent ζ-potential of the Qdots was determined by laser Doppler microelectrophoresis (Malvern Zetasizer Nano ZS) at a concentration of 5 nM in either 0.010 M NaCl buffered to pH 7.4 with 0.010 M HEPES or Leibotvitz's L-15 media (ATCC) supplemented with 10% fetal bovine serum (ThermoFisher) and 1% antibiotics.
To confirm the presence of amine groups on the surface of the Qdots, we acquired XPS measurements of electron emission from the N(1s) and Cd(3d) levels. Qdots from suspension (8000 nM) were dropcast onto a clean Si wafer. XPS spectra were measured using a Thermo Fisher Scientific K-alpha XPS. Spectra were collected at 45° photoelectron takeoff angles, and XPS spectra were fit using CasaXPS software.40 C 1s, N 1 s, O 1s, Cd 3d, S 2p, S 2s, Se 3d, and Se 3s peaks were monitored. Because Cd 3d5/2, Cd 3d3/2, and N 1s contributions occur at similar binding areas, we fit each peak to a single region between binding energies of 415 and 393 eV which was background corrected with a Shirley function. The Cd 3d5/2 and Cd 3d3/2 were each fit to two peaks with a 30% Gaussian, 70% Lorentzian lineshape. An area constraint of 2/3 the area of the Cd 3d5/2 peak was applied to each of the Cd 3d3/2 peaks to reflect the different contributions due to spin–orbit splitting. The N 1s peak was fit to a single peak with a 30% Gaussian, 70% Lorentzian lineshape. The peak fit yielded a residual χ2 value of 1.73 and 1.07 for Qdots605 and Qdots705, respectively.
Lipid vesicle preparation
We prepared small, unilamellar vesicles consisting of 60 mol% 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC, 850375C, Avanti Polar Lipids), 20 mol% plant-derived cholesterol (Chol, 700100P, Avanti Polar Lipids) and 20 mol% sphingomyelin (N-acyl-4-sphingenyl-1-O-phosphorylcholine) from chicken egg yolk (SM, S0756, Sigma Aldrich) with 0.1 mol% of the fluorescent phospholipid 1-palmitoyl-2-(dipyrrometheneboron difluoride)undecanoyl-sn-glycero-3-phosphocholine (TopFluor® PC, 810281C, Avanti Polar Lipids), as previously described.16 Briefly, the desired amounts of each bilayer component in chloroform were mixed, the chloroform was removed via a stream of N2 gas, and the dried lipid films were left under vacuum overnight to remove any trace chloroform. The film was rehydrated and sonicated for 30 min in 0.001 M NaCl buffered to pH 7.4 with 0.010 M HEPES to a stock concentration of 1.25 mg mL−1. The solution underwent three freeze (liquid N2)/thaw (bath sonication) cycles and was extruded 11 times (Avanti 610![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 extruder kit) through a 50 nm polycarbonate membrane filter (Whatman) to give small unilamellar vesicles.
000 extruder kit) through a 50 nm polycarbonate membrane filter (Whatman) to give small unilamellar vesicles.
Supported lipid bilayer formation
To conduct correlative AFM and fluorescence imaging on supported lipid bilayers, a modified approach to a previously established method,41 was utilized to enable fluorescence imaging on a thin layer of mica. Mica was chosen as the substrate as it enables an atomically flat surface for AFM imaging, allowing for subtle height changes due to Qdots to be detected. Briefly, a cyanoacrylate glue was used to adhere a mica disc (12 mm, highest grade V1, Ted Pella) to the center of a glass bottom dish (50 × 7 mm, PELCO®). The mica was cleaved using a razor blade and Scotch™ tape until it was thin enough to be transparent by eye. The dish was rinsed with ultrapure water (>18 MΩ) and dried with N2 gas. Supported lipid bilayers were formed on the mica surface, as previously described.25 Briefly, the mica was equilibrated in 3 mL of 0.150 M NaCl and 0.005 M CaCl2 buffered to pH 7.4 with 0.010 M HEPES, incubated in 3 mL of 0.03125 mg mL−1 of 60/20/20 mol% DOPC/SM/Chol + 0.1 mol% TopFluor® PC at 45 °C for 1 h, and allowed to cool to room temperature. The solution was exchanged with vesicle and calcium-free solution to remove loosely bound vesicles and calcium (15 mL in three 5 mL aliquots). The ionic strength of the solution was lowered to 0.010 M NaCl by subsequent rinses (15 mL in three 5 mL aliquots) with 0.150 M NaCl and 0.010 M NaCl both in 0.010 M HEPES solution.Correlative atomic force microscopy (AFM) and fluorescence imaging on supported lipid bilayers
Optimization of AFM imaging parameters and sample adherence was performed to render the sample optically transparent as well as able to withstand being submerged in aqueous buffer and the heating conditions required for bilayer preparation. AFM imaging was conducted in 0.010 M NaCl buffered to pH 7.4 with 0.010 M HEPES and the mica substrate with bilayer remained submerged in buffer throughout the duration of imaging. All AFM images presented are the highest quality images collected that are representative of images collected from many different spots across at least 7 distinct bilayer samples. Prior to the addition of Qdots, bilayers were screened for quality and only bilayers that contained no defects or holes over at least three areas, scanned by AFM microns away from one another, were used for subsequent exposure experiments.To conduct correlative measurements, fluorescence imaging was performed using a Zeiss inverted microscope (Axiovert 200) with a 100× oil immersion objective. The surface of the bilayer was first focused on using fluorescence microscopy by exciting the TopFluor PC with a 488 nm laser (Coherent Inc., Cellx). The AFM tip (Bruker, DNP-C, nominal k = 0.24 N m−1) was then lowered and centered on the sample using the optical microscope. The objective was lowered, to minimize noise in the AFM images, and representative “before images” were collected using AFM. AFM images were acquired in AC mode at a scan rate of 0.3 Hz using an MFP-3D AFM (Oxford Instruments). Optimization of set point and gain was monitored for each image to minimize the amount of force applied to the sample and maximize the tip tracking of the sample surface. Following “before image” collection, the AFM tip was raised straight up, the scanner removed, the sample remained on the sample stage, and 5 nM of Qdots were added. Following 20 min incubation with the Qdots, the sample was rinsed 3× with 5 mL of Qdot-free buffer and the AFM scanner was replaced. The 100× objective was moved back into place, and the bilayer surface was focused on and fluorescence images were collected. The TopFluor PC (ex. 495 nm, em. 503 nm) was excited with 488 nm laser at 0.5 s exposure, and emission was collected between 490 and 550 nm using a CCD camera (Andor iXon). Following bilayer image collection, the 488 nm laser was left on to completely photobleach the signal from the TopFluor PC. The Qdots605 were imaged using STORM. Excitation was done using 488 nm laser, and emission was acquired between 510 and 740 nm using the same CCD camera. 4000 exposures at 0.03 s each were acquired to capture the photoblinking events. ThunderSTORM,42 an open-source plug-in for Fiji was used, where single-molecule localization algorithm was applied to the fluorescence imaging data to localize the Qdots. STORM could not be applied to image the bilayer due to the rapid diffusion of the lipids. In addition, TopFluor PC, which does not blink, was chosen over other fluorescent lipids that blink as its tail group, rather than the head group, is labeled with the dye and thus would not interfere with the lipid-Qdot interaction. Overlay of the green and red emission channels was conducted using the open source Fiji software.39 Following fluorescence imaging, the objective was again moved out of place, the AFM tip was lowered, and “after Qdots” AFM images were collected. AFM images were processed and analyzed using Gwyddion.43 All correlated AFM-fluorescence images presented are the highest quality images that are representative of images collected from many spots across at least three distinct bilayer samples.
Trout gill epithelial cell culture
Rainbow trout (Oncorhynchus mykiss) gill epithelial cells (RTgill-W1, ATCC CRL-2523) were routinely cultured in Leibotvitz's L-15 media (ATCC) supplemented with 10% fetal bovine serum (ThermoFisher) and 1% antibiotics. Hereby referred to as “complete media.” Cells were incubated in ambient atmosphere at 19 °C. Cells were harvested for experiments and passage following 100% confluency, at which point media was removed, and cells were detached from the surface using 0.25% Trypsin-0.53 mM EDTA solution (ATCC). For fluorescence imaging experiments, cells were seeded onto glass coverslip-bottom dishes (50 × 7 mm, PELCO®) that had been incubated overnight in complete media to enhance cell adhesion on the glass surface. Fluorescence experiments were conducted with cells at 80–90% confluency.Cell exposure, fluorescence labeling and STORM imaging of lipid rafts within trout gill epithelial cells
Cells were exposed to 10 nM Qdots in complete media for 10 min and subsequently, the lipid rafts were labeled using a commercially available lipid raft labeling kit, Vybrant Alexa Fluor 488 Lipid Raft Labeling Kit (Invitrogen). Briefly, following Qdots exposure, the cells were rinsed 3× with a complete medium. The cells were incubated in 1 μg mL−1 of the Alexa Fluor 488 dye labeled cholera toxin subunit B (CT-B) for 10 min at 4 °C, which selectively binds a ganglioside (GM1) composed of a glycosphingolipid enriched in lipid rafts. The cells were rinsed 3× with PBS buffer (Gibco) and then exposed to a 200× dilution of the anti-CT-B antibody rabbit serum, which crosslinks the CT-B-labeled lipid rafts into patches on the cell membrane, for 15 min at 4 °C. The cells were then rinsed 3× with PBS, fixed with 4% paraformaldehyde for 10 min, rinsed 3× with PBS, counter-stained with DAPI to identify the cell nucleus, and rinsed 3× with PBS prior to imaging. To conduct STORM imaging, the Alexa Fluor 488 dye (ex. 495 nm, em. 503 nm) was excited with 488 nm laser at 0.03 s exposure for 4000 frames and emission between 490 and 550 nm was collected using CCD camera (Andor iXon). Following lipid raft image collection, the 488 nm laser was left on to completely photobleach the signal from the Alexa Fluor dye. To ensure separation between the dye and the Qdots, we chose to work with Qdots705 (ex. broad, em. 705 nm). The Qdot705 were excited with 488 nm laser and emission was acquired at 510–740 nm for 4000 exposures at 0.03 s each. All imaging was conducted in PBS buffer. Traditional STORM buffer contains both an oxygen scavenging system and a primary thiol, however, these components were found to quench the fluorescence of the Qdots. Therefore, we chose to use Alexa Fluor 488 in PBS as it has been demonstrated to have sufficient photoblinking under these conditions.44 Chromatic aberration was calibrated using fluorescent beads (Molecular Probes, ex. 488/em. 560) and corrected in post-processing. We used ThunderSTORM,42 an open-source plug-in for Fiji, to analyze the single-molecule and quantum dot data. Overlay of the two channels was conducted using the open source Fiji software.39Results and discussion
Nanoparticle characterization
Fig. 1a presents transmission electron microscopy (TEM) images of the two primary amine-terminated Qdots used in this study, showing their triangular morphology. The two Qdot types differ only by their emission wavelengths, which were chosen to accommodate the two experimental designs, one for the bilayers and one for the intact cells. The average width and length of the Qdots were determined by measuring 100 individual particles for each type. For the Qdots with an emission peak at 605 nm (Qdot605), which were used in the bilayer experiments, the average width was 8.9 ± 0.9 nm and the average length was 9.0 ± 1.0 nm. For the Qdots with an emission peak at 705 nm (Qdot705), which were used in the cell membrane experiments, the average width was 7.3 ± 1.1 nm and the average length was 12.1 ± 1.0 nm. The apparent ζ-potential of Qdot605 in the solution used for lipid bilayer experiments (0.010 M NaCl buffered to pH 7.4 with 0.010 M HEPES) was −10 ± 1 mV. The apparent ζ-potential of Qdot705 in complete cell culture medium was −4 ± 1 mV. The near neutral ζ-potential value of both types of Qdots suggests low levels of protonated amine groups. However, we confirmed the presence of amine groups on the surface of the Qdots using x-ray photoelectron spectroscopy (XPS) analysis. Fig. 1c and d show XPS spectra for both types of Qdots, which shows the nitrogen 1s binding energy peak between 405 and 395 eV, confirming the presence of amine groups on the surface of the Qdots.Primary amine-terminated Qdots preferentially interact with domain boundaries in phase-segregated lipid bilayers as observed by AFM
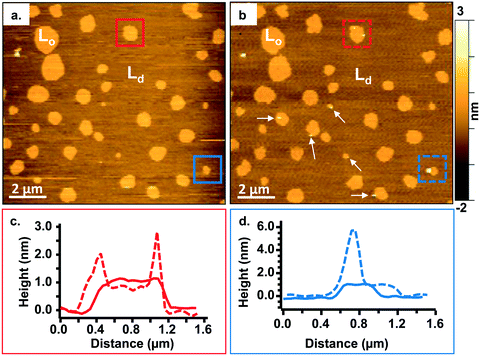
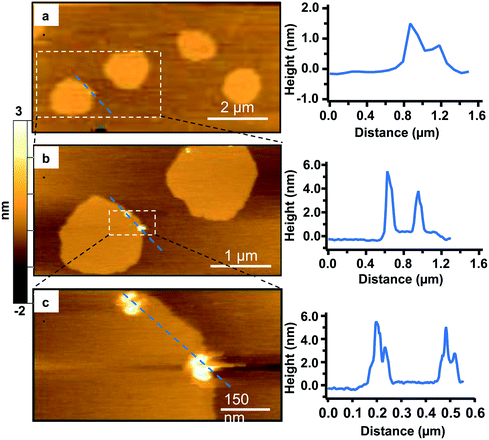
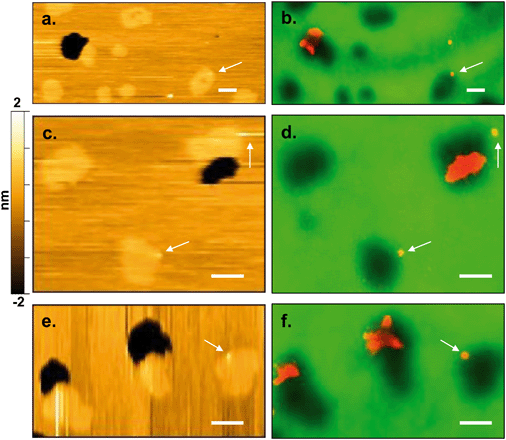
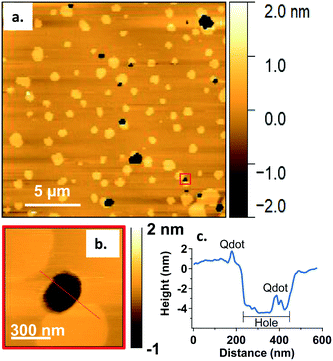
To study the interactions of the Qdots with distinct membrane structures, we used AFM to localize the Qdots in SLBs with high spatial resolution and 3D analysis. We used SLBs comprised of 60/20/20 mol% phosphatidylcholine (DOPC)/sphingomyelin (SM)/cholesterol (Chol) + 0.1 mol% of TopFluor PC, which has a phosphocholine headgroup and the TopFluor fluorescent dye covalently conjugated to one of the lipid acyl chains. At this bilayer composition, two distinct phases are observed, a liquid-ordered (Lo) phase and a liquid-disordered (Ld) phase, as shown in Fig. 2a in yellow and brown respectively. The Lo phase is ∼1 nm taller than the Ld phase, as shown by the solid red (Fig. 2c) and solid blue trace (Fig. 2d). The Lo domains are enriched in sphingomyelin and cholesterol, similar to lipid rafts in the cell membrane, whereas the Ld phase consists predominantly of DOPC and TopFluor PC. Previous work has shown that in the Lo domains, the cholesterol induces ordering of the unsaturated acyl chains of the sphingomyelin resulting in this ∼1 nm height change compared to the predominantly DOPC phase.16,45 Therefore, our starting bilayers are consistent with previous reports for this bilayer composition.To visualize the interaction between the Qdots and the two phases in the bilayers, we incubated the bilayers with Qdots for 20 min, rinsed with buffer, and took representative “after” AFM images of the membrane. Examples of these AFM images are shown in Fig. 2–4. Panels a. and b. in Fig. 2 show corresponding before and after AFM images of the same region of a bilayer following exposure to the Qdots and subsequent rinsing. Fig. 2b shows seven Lo domains (denoted by the white arrows or in the boxes), where small bright features were found near the domain boundaries after exposure to the Qdots (absent in Fig. 2a). These features, which appear only after adding the Qdots, are higher than the surrounding domains and are likely to be the Qdots. Furthermore, height profiles across the domains before and after addition of Qdots show a range of heights for these features between 2–9 nm. An example of such height profiles can be seen in Fig. 2c and d. While this range is consistent with the average values determined by TEM (8.9 ± 1.0 (width), 9.0 ± 1.0 (length), it includes smaller values than anticipated based on the TEM measurements. It has been shown previously,46 that even when imaging parameters are optimized, AFM may provide an apparent height that is lower than the true height. This could be due to sample deformation, high tip-sample forces, and/or as a result of local AFM probe-sample geometry, which is particularly relevant when measuring nanoscale objects.46,47 We also hypothesize that this range of sizes could be due to the particles intercalating within the bilayer. This would mean that some of the Qdots may be on top of the bilayer, while others may be embedded within the membrane up to ∼5 nm. This interpretation could account for the large variability in heights of the Qdots observed by AFM. Fig. 3 shows a second example of the co-localization of Qdots with domain boundaries. In this case, the same domain boundary is imaged at increasing spatial resolution by AFM. Two bright features (∼6 nm in height), likely Qdots, are found at the boundary. At the highest resolution (Fig. 3c), two peaks are resolved for each of the two bright features, suggesting the presence of more than one Qdot or an aggregate. Treating the aggregates as single entities and analyzing 37 of these entities across five different membranes we observed that 68% of particles or aggregates found on the bilayer were localized at Lo/Ld phase boundaries, and 22% were localized in the Lo phase.
In addition to co-localization of Qdots at domain boundaries, we also observed hole formation at these boundaries after adding the Qdots, as shown in Fig. 4a. A height profile (Fig. 4b) across the dark regions shows a height change of ∼4 nm, which is consistent with previous reports for bilayers comprised solely of DOPC.48 In addition, the presence of what seems to be a Qdot cluster is detected in the hole by the height profile (Fig. 4c). The presence of Qdot clusters in holes is further supported by the correlative imaging as later demonstrated in Fig. 6. This suggests that Qdots not only bind to the domain boundaries but also induce defects at these sites. Previous work using lipid bilayers representing the lung surfactant layer, which phase segregate into gel and fluid phase domains, showed that poly(amidoamine) dendrimers accumulated at defects sites and removed lipids preferentially from the fluid phase domains.21 The dendrimers were terminated with protonated primary amines, similar to the termination of the Qdots used in our study.
 | ||
| Fig. 4 a. Representative AFM image showing defects or holes (dark areas) at domain boundaries following 20 min incubation with 5 nM primary amine-terminated Qdots and subsequent rinsing. b. Zoomed-in image of the region marked by the red square in (a). c. Height profile along the red dashed line in (b), showing the presence of a hole ∼4 nm in height and a Qdot at the domain boundary, as well as what seems to be a Qdot cluster within the hole (further supported by the correlative AFM and fluorescence images (Fig. 6)). | ||
The observations of Qdot localization and hole formation at domain boundaries suggest the phase boundary may be a favorable site of interaction for primary amine-terminated Qdots. Our previous work showed an increase in attachment of primary amine-terminated gold NPs to bilayers containing both Ld and Lo phases relative to those comprised solely of a Ld phase.16 In that work, we hypothesized altered water structure at the phase boundary49 or localized electrostatic interactions between the exposed phosphate groups in the sphingomyelin and the primary amine-terminated NPs may be the cause for the increase in attachment.16 Recent computational studies have demonstrated a free energy minimum exists at the interface between the Lo phase and the domain boundary due to the intrinsic curvature at the phase boundary caused by the height difference between the two phases, providing opportunity for the NP to make multiple contacts with the bilayer without incurring an energetic penalty to inducing bilayer curvature.26 While the particles used in the current study exhibit slightly negative ζ-potentials, the primary amine groups are expected to be protonated at the experimental pH of 7.4 and thus localized positively charged groups are found at the surface of the Qdots. Therefore, the current study provides direct experimental evidence of favorable interactions at the phase boundary and helps to extend experimental results to primary amine-terminated NPs that have a slightly negative ζ-potential. This suggests that the chemistry of the surface ligand may play a larger role in dictating interactions with biological membranes, compared with the overall surface charge of the particles. Furthermore, the formation of holes by dendrimers has been rationalized previously by the energy barrier needed to remove lipids from the Ld phase being lower than that needed to remove lipids from the Lo phase.21 The lower bending modulus of the Ld phase relative to the Lo phase has also been postulated as a potential reason for stronger interactions between positively charged particles and the Ld phase.26 Finally, positively charged polymers have been shown to disrupt the electrostatic interactions between the bilayer and mica, potentially resulting in defect formation.50 A combination of these effects are likely in play in the case of the Qdot study presented here, as we observe Qdots mainly at the Ld/Lo boundary and no holes in the Lo phase. The presence of Qdot clusters on the mica surface (Fig. 4 and 6) supports the disruption of bilayer–mica electrostatic interactions. We hypothesize that, in our work, the negative curvature at the phase boundary between the Lo and Ld phases first promotes the attractive interactions with the Qdots. At a critical concentration of Qdots, hole formation at the boundary may begin to occur, likely due to a combination of favorable energetics to remove lipids from the Ld phase,21 a lower bending modulus of the Ld phase compared to the Lo phase,26 and a disruption of the electrostatic interactions between the bilayer and the mica substrate.50
Preferential interaction of Qdots with the Ld/Lo phase boundary confirmed by correlative atomic force and fluorescence microscopy
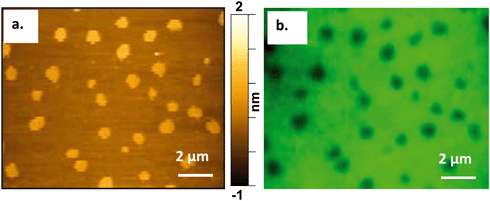
To verify our findings and ensure no spatial interference is introduced by the AFM probe, we conducted correlative fluorescence and atomic force microscopy to confirm the presence of the Qdots at the Ld/Lo boundaries before atomic force microscopy images were collected.Atomic force microscopy requires an ultraflat surface to ensure that the topography of the substrate does not interfere with the topography of the sample. For this reason, we used mica as the substrate for the bilayer. Mica has been demonstrated previously to be a suitable substrate for imaging phase-segregated domain containing SLBs.25,51 Mica also provides the optical transparency needed for fluorescence imaging, which was necessary for performing correlative AFM and fluorescence imaging on the same area of the bilayer. Modifying a previous approach,41Fig. 5 shows the successful correlation of a 60/20/20 mol% DOPC/SM/Chol + 0.1 mol% TopFluor PC supported lipid bilayer formed on a mica substrate and imaged by both AFM and fluorescence microscopy. The images in Fig. 5 were taken from the same area of the bilayer using (a) AFM and (b) fluorescence microscopy, respectively. TopFluor PC emits in the green region of the visible spectrum and preferentially segregates to the Ld phase. Thus, the Lo domains in the bilayer show up dark in color due to lack of the dye, whereas the Ld phase shows up bright green due to the dye's fluorescence emission following excitation at 488 nm. Holes in the membrane would also appear dark in the fluorescence image but would be distinguished from the Lo domains in the AFM image, as Lo domains show up ∼1 nm taller than the Ld phase, whereas holes would be ∼4–5 nm below the Ld phase.
To confirm the preferential interaction of the quantum dots at the Lo/Ld phase boundary we used the correlative AFM/fluorescence approach to localize the Qdots in relation to the Lo domains. Fig. 6 shows three pairs of correlative AFM and fluorescence images, acquired following addition of Qdots (shown in red in the fluorescence images). Taking both the AFM and fluorescence images together, the small and bright (taller) regions found at Lo/Ld phase boundaries in the AFM images were confirmed to be the Qdots (corresponding white arrows in both the AFM and fluorescence images in Fig. 6) by their red emission in the fluorescence images. Larger Qdot clusters were also found in the holes, as shown in the fluorescence images, which supports the interpretation that the taller feature observed in the hole in the high-resolution AFM scan in Fig. 4c is indeed a Qdot cluster. The presence of the Qdots in the holes supports our hypothesis that the Qdots induce defects at the phase boundaries and the idea that defect-formation in the Ld phase may be due to the disruption of the electrostatic interactions between the bilayer and the mica substrate.50 To our knowledge this is the first use of correlative atomic force and fluorescence microscopy to confirm the presence of nanoparticles on and characterize their interaction with a bilayer.
The Lo domains in the bilayer used in our study are designed to represent the sphingolipid- and cholesterol-rich lipid rafts present in the cytoplasmic membrane of eukaryotic cells, which are membrane domains that play important roles in membrane trafficking and signal transduction.11,13 Thus, the localization of Qdots with the Lo domains are likely to have important implications in the Qdot interactions, uptake, and impact in eukaryotic cells.
Primary amine-terminated Qdots preferentially co-localize with lipid rafts in intact cell membrane as revealed by super resolution fluorescence microscopy
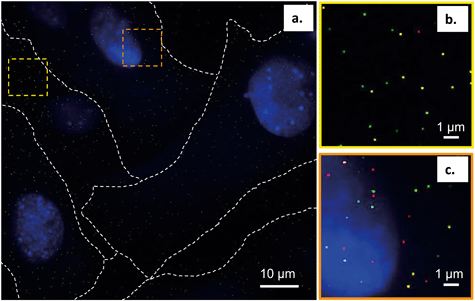
To further interpret our bilayer results in the context of the intact cell membrane, we characterized the spatial relationships between the primary amine-terminated Qdots and lipid rafts in the membrane of intact trout gill epithelial cells – a model cell type for environmental exposures of aquatic organisms. This was done using single molecule localization-based super resolution fluorescence imaging (STORM), which enables imaging intact hydrated cells with 20–30 nm resolution.Lipid rafts were labeled with Alexa 488-tagged cholera toxin subunit B (CT-B), which selectively binds a ganglioside (GM1) composed of a glycosphingolipid enriched specifically in lipid rafts. The use of (CT-B) to bind to GM1 as a marker for lipid rafts is well-established in the literature.52–54 Thus, when we refer to lipid rafts we are referring particularly to the nanodomains within the cellular membrane that contain GM1 that we were able to resolve experimentally using fluorescently labeled CT-B. Fig. 7 demonstrates the co-localization of the Qdots (red) and lipid rafts (green) at the membrane of intact gill epithelial cells grown in culture. Fig. 7b and c are zoomed-in images of the two, yellow (b) and orange (c), squared areas in the image on the left (a). Across eight cells from three different cell cultures, we determined that 59% of all Qdots found on the cells co-localized with lipid rafts (overlaid red and green shows as yellow). These observations are in line with those from the bilayer studies and suggest favorable interaction of the Qdots with lipid rafts. These observations also support the possibility that the Qdots enter the cell via lipid raft-mediated endocytosis. It is possible that the fluorescent signal from CT-B originates in some cases from the intracellular environment. In such cases, the clear colocalization of the signals coming from CT-B and the Qdots strongly supports their co-internalization and therefore their initial colocalization at the plasma membrane. The ζ-potential of quantum dots has been suggested to dictate their route of entry into cells.55 It has been shown that quantum dots with lower surface potentials (corresponding to ζ-potential between −10 and +10 mV), similar to those used in this work (apparent ζ-potential of −4 ± 1 mV in cell culture media), tend to enter the cell via lipid raft-mediated endocytosis.55
While the Qdots used in this study caused no discernable decrease in cell viability (Fig. S1†), it has been shown that NPs can induce significant changes in cell function, such as changes in gene expression, even without detectable changes in cell viability.56 Understanding the initial interactions of NPs at the cell membrane and the mechanisms of their internalization is critical to understanding such changes and consequent impacts on the normal function of the cell.
Conclusions
Our observations show that primary amine-terminated Qdots are preferentially found at the boundary between the Lo and Ld phases in supported lipid bilayers containing phase-segregated domains. We further show that the Qdots preferentially co-localize with lipid rafts at the membrane of intact cells, together supporting favorable interactions of the Qdots with the Lo domains and their boundaries. Previous studies have shown that the bending modulus of the membrane plays a large role in NP-membrane interactions.26,57,58 We hypothesize the observed favorable interactions are due to the intrinsic curvature at the phase boundary between the Ld and Lo domains, which lessens the need for large curvature deformations for NP-membrane interactions.26 The resulting free energy minimum at the phase boundary26 leads to NP attachment and subsequent attractive interactions between the amine groups on the particle and the negatively charged phosphate groups in the sphingomyelin exposed at the phase boundary.16Fig. 8 illustrates the potential mechanism underlying these observed favorable interactions. Previous work has shown that NPs will approach the membrane in a way that minimizes electrostatic repulsion,59 which is likely occurring in our studies as well. Computational studies have shown that the Lo/Ld phase boundaries are likely to be energetically favorable regions for interactions between lipid domains and external species,49 including NPs.26,60 The phase boundary has been shown to be a preferential site of attachment of HIV fusion peptides61,62 and toxins,63,64 and has been hypothesized to be a preferential site for the attachment of primary amine-terminated gold NPs.26 Here we provide direct visual evidence for the preferential interaction of primary amine-terminated quantum dots with the phase boundary using correlative atomic force and fluorescence microscopy with both high spatial resolution and chemical selectivity. Likewise, we determined the co-localization of the Qdots with lipid rafts at the intact cell membrane using two-color super resolution STORM imaging, achieving these observations with both high spatial resolution and detection specificity. The preferential co-localization with lipid rafts suggests that lipid raft-mediated endocytosis may play an important role in the uptake of these Qdots. The uptake of Qdots with similar ζ-potentials as those used in our study has been shown to occur via lipid raft-mediated endocytosis,55 although the use of an inhibitor of this uptake mechanism was used in that study, as opposed to the direct, high resolution observation of co-localization that was applied in our study.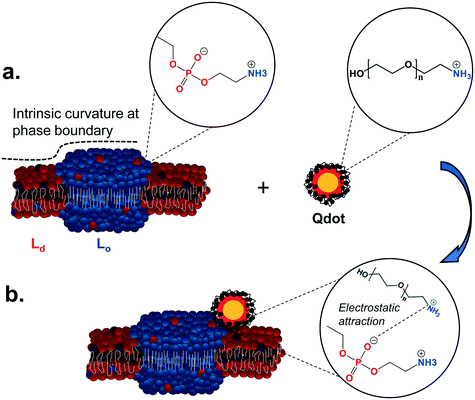 | ||
| Fig. 8 Illustration of the proposed mechanism underlying the interaction preference of primary amine-terminated Qdots with the boundary between liquid-ordered (Lo) and liquid-disordered (Ld) phases. a. The intrinsic curvature of the phase boundary (dashed line) leads to an energy minimum at this site.26 Furthermore, negatively charged phosphate groups in the sphingomyelin (blue balls) are exposed at this phase boundary, while positively charged primary amine groups are found on the surface of the Qdot. b. The intrinsic free energy minimum at the phase boundary potentially leads to favorable nanoparticle interactions at this site and subsequently favorable electrostatic interactions between the negatively charged phosphate groups and positively charged amine groups. | ||
To date, our group has demonstrated favorable interactions between positively-charged primary amine-terminated gold NPs,16 positively-charged quaternary amine-terminated Qdots,25 and now negatively charged primary amine-terminated Qdots with the Lo/Ld phase boundary, or in the case of the quaternary amine-terminated Qdots, the Lo phase, in phase-segregated supported lipid bilayers. The differences in interactions between primary and quaternary amine-terminated Qdots (preferential interactions at phase boundaries vs. collapse of the Lo domains) suggest the chemistry of the surface ligand plays a role in these interactions. Interestingly, our results extend these findings to negatively charged primary amine-terminated NPs, further suggesting that the surface ligand, more than the surface charge, may play a dominating role in controlling the interactions of nanomaterials at the cell membrane.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the NSF under the Center for Sustainable Nanotechnology, CHE-1503408. A portion of the research was performed using the Environmental Molecular Sciences Laboratory (EMSL), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research (BER) and located at Pacific Northwest National Laboratory (PNNL). The authors gratefully acknowledge use of facilities and instrumentation supported by NSF through the University of Wisconsin Materials Research Science and Engineering Center (DMR-1720415). E. D. L. was supported by the NSF Graduate Research Fellowship Grant, DGE-1747503. Support was also provided by the University of Wisconsin Alumni Research Foundation.References
- R. J. Hamers, Nanomaterials and global sustainability, Acc. Chem. Res., 2017, 50, 633–637 CrossRef CAS PubMed.
- A. Nel, T. Xia, L. Mädler and N. Li, Toxic potential of materials at the nanolevel, Science, 2006, 311, 622–627 CrossRef CAS.
- C. J. Murphy, A. M. Vartanian, F. M. Geiger, R. J. Hamers, J. Pedersen, Q. Cui, C. L. Haynes, E. E. Carlson, R. Hernandez, R. D. Klaper, G. Orr and Z. Rosenzweig, Biological responses to engineered nanomaterials: Needs for the next decade, ACS Cent. Sci., 2015, 1, 117–123 CrossRef CAS PubMed.
- M. Zhu, G. Nie, H. Meng, T. Xia, A. Nel and Y. Zhao, Physicochemical properties determine nanomaterial cellular uptake, transport, and fate, Acc. Chem. Res., 2013, 46, 622–631 CrossRef CAS PubMed.
- S. Behzadi, V. Serpooshan, W. Tao, M. A. Hamaly, M. Y. Alkawareek, E. C. Dreaden, D. Brown, A. M. Alkilany, O. C. Farokhzad and M. Mahmoudi, Cellular uptake of nanoparticles: Journey inside the cell, Chem. Soc. Rev., 2017, 46, 4218–4244 RSC.
- A. Lesniak, A. Salvati, M. J. Santos-Martinez, M. W. Radomski, K. A. Dawson and C. Aberg, Nanoparticle adhesion to the cell membrane and its effect on nanoparticle uptake efficiency, J. Am. Chem. Soc., 2013, 135, 1438–1444 CrossRef CAS PubMed.
- D. Bedrov, G. D. Smith, H. Davande and L. Li, Passive transport of C60 fullerenes through a lipid membrane: A molecular dynamics simulation study, J. Phys. Chem. B, 2008, 112, 2078–2084 CrossRef CAS.
- J. Lin and A. Alexander-Katz, Cell membranes open “doors” for cationic nanoparticles/biomolecules: Insights into uptake kinetics, ACS Nano, 2013, 7, 10799–10808 CrossRef CAS PubMed.
- S. Zhang, H. Gao and G. Bao, Physical properties of nanoparticle cellular endocytosis, ACS Nano, 2015, 9, 8655–8671 CrossRef CAS PubMed.
- L. W. Zhang and N. A. Monteiro-Riviere, Mechanisms of quantum dot nanoparticle cellular uptake, Toxicol. Sci., 2009, 110, 138–155 CrossRef CAS PubMed.
- D. W. Hilgemann, M. J. Lin, M. Fine and C. Deisl, On the existence of endocytosis driven by membrane phase separations, Biochim. Biophys. Acta, Biomembr., 2019, 183007 Search PubMed.
- K. Simons and E. Ikonen, Functional rafts in cell membranes, Nature, 1997, 387, 569–572 CrossRef CAS PubMed.
- E. Sezgin, I. Levental, S. Mayor and C. Eggeling, The mystery of membrane organization: Composition, regulation and roles of lipid rafts, Nat. Rev. Mol. Cell Biol., 2017, 18, 361–374 CrossRef CAS PubMed.
- K. L. Chen and G. D. Bothun, Nanoparticles meet cell membranes: Probing nonspecific interactions using model membranes, Environ. Sci. Technol., 2014, 48, 873–880 CrossRef CAS PubMed.
- P. R. Leroueil, S. A. Berry, K. Duthie, G. Han, V. M. Rotello, D. Q. McNerny, J. R. Baker, B. G. Orr and M. M. B. Holl, Wide varieties of cationic nanoparticles induce defects in supported lipid bilayers, Nano Lett., 2008, 8, 420–424 CrossRef CAS PubMed.
- E. S. Melby, A. C. Mensch, S. E. Lohse, D. Hu, G. Orr, C. J. Murphy, R. J. Hamers and J. A. Pedersen, Formation of supported lipid bilayers containing phase-segregated domains and their interaction with gold nanoparticles, Environ. Sci.: Nano, 2016, 3, 45–55 RSC.
- A. M. Farnoud and S. Nazemidashtarjandi, Emerging investigator series: Interactions of engineered nanomaterials with the cell plasma membrane; what have we learned from membrane models?, Environ. Sci.: Nano, 2019, 6, 13–40 RSC.
- E. S. Melby, C. Allen, I. U. Foreman-Ortiz, E. R. Caudill, T. R. Kuech, A. M. Vartanian, X. Zhang, C. J. Murphy, R. Hernandez and J. A. Pedersen, Peripheral membrane proteins facilitate nanoparticle binding at lipid bilayer interfaces, Langmuir, 2018, 34, 10793–10805 CrossRef CAS PubMed.
- K. H. Jacobson, I. L. Gunsolus, T. R. Kuech, J. M. Troiano, E. S. Melby, S. E. Lohse, D. Hu, W. B. Chrisler, C. J. Murphy, G. Orr, F. M. Geiger, C. L. Haynes and J. A. Pedersen, Lipopolysaccharide density and structure govern the extent and distance of nanoparticle interaction with actual and model bacterial outer membranes, Environ. Sci. Technol., 2015, 49, 10642–10650 CrossRef CAS PubMed.
- A. Chakraborty, N. J. Mucci, M. L. Tan, A. Steckley, T. Zhang, M. L. Forrest and P. Dhar, Phospholipid composition modulates carbon nanodiamond-induced alterations in phospholipid domain formation, Langmuir, 2015, 31, 5093–5104 CrossRef CAS PubMed.
- B. Erickson, S. C. DiMaggio, D. G. Mullen, C. V. Kelly, P. R. Leroueil, S. A. Berry, J. R. Baker, B. G. Orr and M. M. B. Holl, Interactions of poly(amidoamine) dendrimers with Survanta lung: The importance of lipid domains, Langmuir, 2008, 24, 11003–11008 CrossRef CAS PubMed.
- A. Mecke, D.-K. Lee, A. Ramamoorthy, B. G. Orr and M. M. B. Holl, Synthetic and natural polycationic polymer nanoparticles interact selectively with fluid phase domains of DMPC lipid bilayers, Langmuir, 2005, 21, 8588–8590 CrossRef CAS.
- T. K. Lind, P. Zielińska, H. P. Wacklin, Z. Urbańczyk-Lipkowska and M. Cárdenas, Continuous flow atomic force microscopy imaging reveals fluidity and time-dependent interactions of antimicrobial dendrimer with model lipid membranes, ACS Nano, 2014, 8, 396–408 CrossRef CAS.
- R. Chelladurai, K. Debnath, N. R. Jana and J. K. Basu, Nanoscale heterogeneities drive enhanced binding and anomalous diffusion of nanoparticles in model biomembranes, Langmuir, 2018, 34, 1691–1699 CrossRef CAS PubMed.
- A. C. Mensch, J. T. Buchman, C. L. Haynes, J. A. Pedersen and R. J. Hamers, Quaternary amine-terminated quantum dots induce structural changes to supported lipid bilayers, Langmuir, 2018, 34, 12369–12378 CrossRef CAS PubMed.
- J. K. Sheavly, J. A. Pedersen and R. C. Van Lehn, Curvature-driven adsorption of cationic nanoparticles to phase boundaries in multicomponent lipid bilayers, Nanoscale, 2019, 11, 2767–2778 RSC.
- J. Sot, S. A. Mendanha-Neto, J. V. Busto, A. B. Garcia-Arribas, S. Li, S. W. Burgess, W. A. Shaw, D. Gil-Carton, F. M. Goni and A. Alonso, The interaction of lipid-liganded gold clusters (Aurora ™) with lipid bilayers, Chem. Phys. Lipids, 2019, 218, 40–46 CrossRef CAS PubMed.
- L. Zhou, M. Cai, T. Tong and H. Wang, Progress in the correlative atomic force microscopy and optical microscopy, Sensors, 2017, 17(4), 938 CrossRef PubMed.
- J. E. Shaw, R. F. Epand, R. M. Epand, Z. Li, R. Bittman and C. M. Yip, Correlated fluorescence-atomic force microscopy of membrane domains: Structure of fluorescence probes determines lipid localization, Biophys. J., 2006, 90, 2170–2178 CrossRef CAS PubMed.
- A. R. Burns, Domain structure in model membrane bilayers investigated by simultaneous atomic force microscopy and fluorescence imaging, Langmuir, 2003, 19, 8358–8363 CrossRef CAS.
- Ira, S. Zou, D. M. Ramirez, S. Vanderlip, W. Ogilvie, Z. J. Jakubek and L. J. Johnston, Enzymatic generation of ceramide induces membrane restructuring: Correlated AFM and fluorescence imaging of supported bilayers, J. Struct. Biol., 2009, 168, 78–89 CrossRef CAS.
- S. Chiantia, N. Kahya, J. Ries and P. Schwille, Effects of ceramide on liquid-ordered domains investigated by simultaneous AFM and FCS, Biophys. J., 2006, 90, 4500–4508 CrossRef CAS PubMed.
- S. V. Bhat, T. Sultana, A. Kornig, S. McGrath, Z. Shahina and T. E. S. Dahms, Correlative atomic force microscopy quantitative imaging-laser scanning confocal microscopy quantifies the impact of stressors on live cells in real-time, Sci. Rep., 2018, 8, 8305 CrossRef PubMed.
- P. D. Odermatt, A. Shivanandan, H. Deschout, R. Jankele, A. P. Nievergelt, L. Feletti, M. W. Davidson, A. Radenovic and G. E. Fantner, High-resolution correlative microscopy: Bridging the gap between single molecule localization microscopy and atomic force microscopy, Nano Lett., 2015, 15, 4896–4904 CrossRef CAS PubMed.
- J. Yao, L. Li, P. Li and M. Yang, Quantum dots: From fluorescence to chemiluminescence, bioluminescence, electrochemiluminescence, and electrochemistry, Nanoscale, 2017, 9, 13364–13383 RSC.
- R. Hardman, A toxicologic review of quantum dots: Toxicity depends on physicochemical and environmental factors, Environ. Health Perspect., 2006, 114, 165–172 CrossRef.
- N. R. Bury, S. Schnell and C. Hogstrand, Gill cell culture systems as models for aquatic environmental monitoring, J. Exp. Biol., 2014, 217, 639–650 CrossRef CAS PubMed.
- L. C. Felix, V. A. Ortega and G. G. Goss, Cellular uptake and intracellular localization of poly (acrylic acid) nanoparticles in a rainbow trout (Oncorhynchus mykiss) gill epithelial cell line, RTgill-W1, Aquat. Toxicol., 2017, 192, 58–68 CrossRef CAS.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J. Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Fiji: An open-source platform for biological-image analysis, Nat. Methods, 2012, 9, 676–682 CrossRef CAS PubMed.
- N. Fairley, CasaXPS, 2.3.16, 2018 Search PubMed.
- A. Matysik and R. S. Kraut, Preparation of mica supported lipid bilayers for high resolution optical microscopy imaging, J. Visualized Exp., 2014,(88), e52054 Search PubMed.
- M. Ovesny, P. Krizek, J. Borkovec, Z. Svindrych and G. M. Hagen, Thunderstorm: A comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging, Bioinformatics, 2014, 30, 2389–2390 CrossRef CAS PubMed.
- D. Nečas and P. Klapetek, Gwyddion: An open-source software for SPM data analysis, Cent. Eur. J. Phys., 2012, 10, 181–188 Search PubMed.
- G. T. Dempsey, J. C. Vaughan, K. H. Chen, M. Bates and X. Zhuang, Evaluation of fluorophores for optimal performance in localization based super resolution imaging, Nat. Methods, 2011, 8, 1027–1036 CrossRef CAS.
- J. C. Lawrence, D. E. Saslowsky, M. Edwardson and R. M. Henderson, Real-time analysis of the effects of cholesterol on lipid raft behavior using atomic force microscopy, Biophys. J., 2003, 84, 1827–1832 CrossRef CAS.
- S. Santos, V. Barcons, H. K. Christenson, J. Font and N. H. Thomson, The intrinsic resolution limit in the atomic force microscope: Implications for heights of nano-scale features, PLoS One, 2011, 6, e23821 CrossRef CAS PubMed.
- R. García and R. Pérez, Dynamic atomic force microscopy methods, Surf. Sci. Rep., 2002, 47, 197–301 CrossRef.
- S. J. Attwood, Y. Choi and Z. Leonenko, Preparation of DOPC and DPPC supported planar lipid bilayers for atomic force microscopy and atomic force spectroscopy, Int. J. Mol. Sci., 2013, 14, 3514–3539 CrossRef CAS.
- K. H. Sheikh and S. P. Jarvis, Crystalline hydration structure at the membrane-fluid interface of model lipid rafts indicates a highly reactive boundary region, J. Am. Chem. Soc., 2011, 133, 18296–18303 CrossRef CAS PubMed.
- T. A. Spurlin and A. A. Gewirth, Poly-l-lysine-induced morphology changes in mixed anionic/zwitterionic and neat zwitterionic-supported phospholipid bilayers, Biophys. J., 2006, 91, 2919–2927 CrossRef CAS.
- R. M. Sullan, J. K. Li, C. Hao, G. C. Walker and S. Zou, Cholesterol-dependent nanomechanical stability of phase-segregated multicomponent lipid bilayers, Biophys. J., 2010, 99, 507–516 CrossRef CAS.
- M. M. Moreno-Altamirano, I. Aguilar-Carmona and F. J. Sanchez-Garcia, Expression of gm1, a marker of lipid rafts, defines two subsets of human monocytes with differential endocytic capacity and lipopolysaccharide responsiveness, Immunology, 2007, 120, 536–543 CrossRef CAS PubMed.
- C. A. Day and A. K. Kenworthy, Tracking microdomain dynamics in cell membranes, Biochim. Biophys. Acta, 2009, 1788, 245–253 CrossRef CAS PubMed.
- R. W. Ledeen and G. Wu, The multi-tasked life of gm1 ganglioside, a true factotum of nature, Trends Biochem. Sci., 2015, 40, 407–418 CrossRef CAS PubMed.
- A. Chakraborty and N. R. Jana, Clathrin to lipid raft-endocytosis via controlled surface chemistry and efficient perinuclear targeting of nanoparticle, J. Phys. Chem. Lett., 2015, 6, 3688–3697 CrossRef CAS PubMed.
- E. S. Melby, Y. Cui, J. Borgatta, A. C. Mensch, M. N. Hang, W. B. Chrisler, A. Dohnalkova, J. M. Van Gilder, C. M. Alvarez, J. N. Smith, R. J. Hamers and G. Orr, Impact of lithiated cobalt oxide and phosphate nanoparticles on rainbow trout gill epithelial cells, Nanotoxicology, 2018, 12, 1166–1181 CrossRef CAS PubMed.
- X. Chen, D. P. Tieleman and Q. Liang, Modulating interactions between ligand-coated nanoparticles and phase-separated lipid bilayers by varying the ligand density and the surface charge, Nanoscale, 2018, 10, 2481–2491 RSC.
- K. Yang, R. Yang, X. Tian, K. He, S. L. Filbrun, N. Fang, Y. Ma and B. Yuan, Partitioning of nanoscale particles on a heterogeneous multicomponent lipid bilayer, Phys. Chem. Chem. Phys., 2018, 20, 28241–28248 RSC.
- H. Ding, J. Li, N. Chen, X. Hu, X. Yang, L. Guo, Q. Li, X. Zuo, L. Wang, Y. Ma and C. Fan, DNA nanostructure-programmed like-charge attraction at the cell-membrane interface, ACS Cent. Sci., 2018, 4, 1344–1351 CrossRef CAS PubMed.
- D. L. Cheung, Aggregation of nanoparticles on one and two-component bilayer membranes, J. Chem. Phys., 2014, 141, 194908 CrossRef PubMed.
- S.-T. Yang, V. Kiessling, J. A. Simmons, J. M. White and L. K. Tamm, HIV gp41-mediated membrane fusion occurs at edges of cholesterol-rich lipid domains, Nat. Chem. Biol., 2015, 11, 424–431 CrossRef CAS PubMed.
- S.-T. Yang, V. Kiessling and L. K. Tamm, Line tension at lipid phase boundaries as driving force for HIV fusion peptide-mediated fusion, Nat. Commun., 2016, 7, 11401 CrossRef CAS PubMed.
- S. Tharad, B. Promdonkoy and J. L. Toca-Herrera, Lipid phase influences the binding of Bacillus thuringiensis Cyt2Aa2 toxin on model lipid membranes, Biochem. Biophys. Res. Commun., 2019, 511, 409–415 CrossRef CAS PubMed.
- S. M. Mate, R. F. Vazquez, V. S. Herlax, M. A. Daza Millone, M. L. Fanani, B. Maggio, M. E. Vela and L. S. Bakas, Boundary region between coexisting lipid phases as initial binding sites for Escherichia coli alpha-hemolysin: A real-time study, Biochim. Biophys. Acta, 2014, 1838, 1832–1841 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: The supporting information presents the cell viability assay in gill epithelial cells exposed to primary amine-terminated Qdots, including methods and results. See DOI: 10.1039/c9en00996e |
| ‡ Current Address: Department of Chemistry, Columbia Basin College, Pasco, WA 99301. |
| This journal is © The Royal Society of Chemistry 2020 |