Open Access Article
Open Access ArticleChemically directed structure evolution for crystal structure prediction
Paul M.
Sharp
,
Matthew S.
Dyer
 ,
George R.
Darling
*,
John B.
Claridge
and
Matthew J.
Rosseinsky
,
George R.
Darling
*,
John B.
Claridge
and
Matthew J.
Rosseinsky
Department of Chemistry, University of Liverpool, Crown Street, L69 7ZD Liverpool, UK. E-mail: darling@liverpool.ac.uk
First published on 6th August 2020
Abstract
We present a new method of evolving crystal structures for crystal structure prediction. The method of chemically directed structure evolution uses chemical models to quantify the environment of atoms and vacancy sites in a crystal structure, with that information used to inform how to modify the structure to make a move on the potential energy surface. We have developed a method of chemically directed swapping, where we swap atoms in the least favourable chemical environments. This method has been implemented in the crystal structure prediction code ChemDASH (Chemically Directed Atom Swap Hopping), which explores the potential energy surface using a basin-hopping approach, and evaluates chemical environments using either the bond valence sum or electrostatic site potential. ChemDASH has a variety of methods of initialising structures, optimising structures, and swapping atoms. This gives ChemDASH the flexibility to be applied to a wide range of systems. We used ChemDASH to examine the effectiveness of the directed swapping method. Directed swapping finds the ground states of TiO2 and SrTiO3 faster than random (non-directed) swapping, but is less effective than random swapping for Y2Ti2O7.
Introduction
When attempting to discover new materials, the large number of chemical elements to choose from, which can be arranged according to a wide variety of possible crystal structure types, necessitates searching a very large chemical space.2,3 Exploring the potential energy surface (PES) formed from all possible arrangements of a set of atoms is an extremely difficult task, especially given the exponential scaling of the number of minima on the PES with increasing number of atoms.2,4,5 Nevertheless, a large number of methods used for searching the PES have been developed,3,6 including: sampling random structures,7 simulated annealing,8 metadynamics,9,10 and genetic algorithms.11–15 All of these methods explore the PES by evolving an initial structure, or set of structures. Here, we present a new method of evolving structures, where the evolution is chemically directed. This method involves using chemical models to quantitatively assess the environment of each atom in the structure, and using this information to direct how the structure is evolved. In this work, we have chosen to implement a method of chemically directed swapping, where atoms in the most unfavourable chemical environments are swapped to generate new structures.We have implemented chemically directed swapping in the crystal structure prediction code ChemDASH (Chemically Directed Atom Swap Hopping). ChemDASH uses a simple basin-hopping algorithm16 to explore the PES, with chemically directed swapping used to generate new structures. The basin-hopping algorithm has previously been employed in approaches involving a guided search through the PES for cluster-based systems, such as the use of tailored basin-hopping moves,17,18 where atoms are prioritised for swapping based on their neighbours and their coordination numbers, and the generalised basin hopping method,19–21 which searches for biminima, which are local minima on the PES from which it is not possible to access a deeper local minimum by a pairwise atom swap and structure relaxation. The biminimum approach has been extended beyond clusters to biomolecules.22 In our approach we measure the chemical environment by analysing the current relaxed structure to determine which ions have a bond valence sum furthest from ideal, or by computing an electrostatic site potential for each ion to compare to an optimum value. Initial structures are generated by populating sites on intersecting orthorhombic grids of anions or cations, or by populating a close-packed grid of anion sites and placing cations in tetrahedral and octahedral interstitial sites. Structures are optimised using either force-fields (with GULP23) or Density Functional Theory (with VASP24). We have implemented an algorithm that can efficiently swap the positions of any number of ions and/or vacancies in a structure. ChemDASH can be used within the probe structure-based materials discovery approach,25 in which it has already played a role in discovering a new compound.26
In this work, we first describe the method of chemically directed swapping. We will show how the bond valence sum27 and electrostatic site potential23 can be used to quantitatively measure the chemical environment of each ion in a crystal structure. We then explain how we use these values to rank ions and vacancy sites according to their chemical environment, and how these rankings are used to select which ions are swapped. We introduce ChemDASH, detailing how structures are initialised, optimised, and accepted/rejected; as well as showing how ions are swapped to generate new structures. We then use ChemDASH to investigate how effective chemically directed swapping is at finding the ground states of titanium dioxide (TiO2), strontium titanate (SrTiO3) and yttrium titanate (Y2Ti2O7) compared to swapping atoms at random. The detailed explanation of the ChemDASH swapping algorithm is given in the Appendix.
Chemically directed swapping
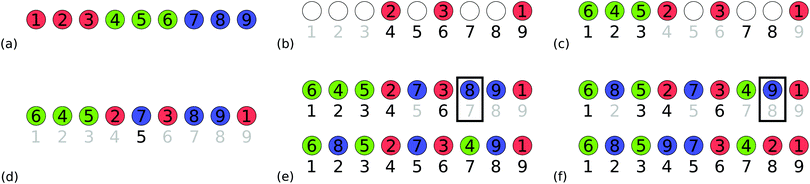
The method of chemically directed structure evolution is motivated by using chemistry to determine how the PES is explored. In an ideal crystal structure, we would expect each atom to be in an optimal chemical environment. Therefore, by quantitatively assessing the environment for each atom in structures generated whilst exploring a PES, we can determine the atoms in the least favourable chemical environments. We can then prioritise modifying these atoms when forming a new crystal structure to further explore the PES. Once we have a value for the chemical environment of each atom we take the difference between the value we find and the ideal value for each atom and rank the atoms of each species on that basis. It is also advantageous to consider moving atoms into vacancy sites when forming new structures, e.g., interstitial cation sites in close-packed lattices. Therefore, we also rank vacancies according to the chemical environment each atom in the structure would have if it occupied the vacancy. For atoms, the rankings run from the atom in the worst environment to the atom in the best environment. For vacancies the rankings are reversed, they run from the best environment for a particular atom to the worst. We have investigated two different measures of the chemical environment for each atom: the bond valence sum (BVS),27 and the electrostatic site potential.23 These models are examples of how we can quantify the chemical environment of atoms and vacancies in crystal structures but we are free to use alternative chemical models within our method of chemically directed structure evolution. We have chosen to use the environment rankings to perform atom swaps. Once we have chosen the species of the atoms to be swapped, the atom rankings list the order for choosing which particular atoms of each species are to be swapped. This allows us to swap atoms in bad environments with one another, or to move atoms from sites with a bad environment into vacancies with a good environment for the particular atom. The chemically directed swapping method is illustrated in Fig. 1.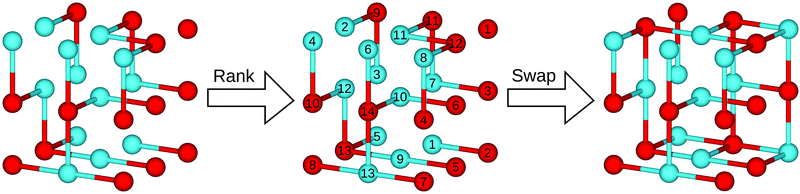 | ||
| Fig. 1 The chemically directed swapping method. We take a structure produced through any method of crystal structure prediction, then rank each species of atoms according to their chemical environment, from worst to best. The numbered labels on each atom represent these rankings, with a set of rankings each for the red and blue atoms. When swapping atoms, we prioritise atoms at the top of the rankings for each species. Hence, the red atom “1” is swapped with the blue atom “1”, and the red atom “2” is swapped with the blue atom “2”. Structure models are produced using VESTA.1 | ||
Ranking atoms and vacancies
We have considered three methods of measuring the chemical environment of atoms in a crystal structure: the bond valence sum, the electrostatic site potential, and a measure combining the bond valence sum and electrostatic site potential (referred to as “BVS+”). The bond valence sum and site potential can be simply calculated for all atoms, enabling us to direct atom–atom swaps. We also need to calculate bond valence sum and site potential values for vacancy sites. The electrostatic site potential can be calculated for any site regardless of whether or not an atom is present. However, a value for the bond valence sum cannot be calculated for a vacancy. Instead, we determine a set of BVS values for each vacancy, by assuming the vacancy is occupied by each species of atom present in the structure. Hence we have a set of rankings for vacancies with the bond valence sum, depending on the atom chosen to occupy the vacancy. This then enables us to use the bond valence sum to determine the best vacancy site for any given atom in the structure.Bond valence sum
The bond valence model is an empirical model that describes bipartite chemical bonding. For all of the bonds made by a particular atom, the valence sum rule states that the sum of the bond valences will equal the atomic valence of the atom in the ionic limit, Vi, i.e.,27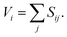 | (1) |
We use the following expression for the bond valence, Sij27
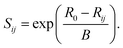 | (2) |
For each atom, the ideal value of the bond valence sum is the atomic valence. Therefore, we take the difference between the calculated value of the bond valence sum and the atomic valence for each atom, and rank atoms on that basis. Atoms with a large difference between the values are in a bad chemical environment, and hence are prioritised for swapping.
Electrostatic site potential
The electrostatic site potential is a measure of the coulomb interaction experienced by a unit charge at a given position in space.23 The site potential is given by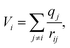 | (3) |
Combined bond valence sum and electrostatic site potential – BVS+
The final method used to rank the chemical environment of atom and vacancy sites combines the bond valence sum with the electrostatic site potential. In this method, bond valence sum values are calculated for atoms as above. We then rank atoms according to their bond valence sum values, however, for vacancies we first use the sign of the site potential value to determine whether the vacancy in question is a cation or an anion site (i.e., a positive site potential is an anion site, a negative site potential is a cation site). Ranking vacancies is then a two-step process; we first separate cation vacancies and anion vacancies and rank them according to their BVS values. We then combine the rankings such that, when considering a cation to occupy the vacancy, we first consider the cation vacancies, then the anion vacancies. For anion species occupying the vacancy, we first consider anion vacancies, then cation vacancies. This means then that the final sets of rankings for vacancies consist of vacancies with the correct sign for the site potential, ranked by BVS values, followed by vacancies with the incorrect sign for the site potential, ranked by BVS values. The idea behind this is to account for the repulsion between like ions when using the BVS to indicate the suitability of a vacancy for a particular ionic species.Swapping atoms
Whichever method has been chosen for ranking atoms and vacancies, we now have a set of rankings from worst to best environment for each atomic species, and a set of rankings for the vacancies from best to worst environment for each species of occupying atom. When it comes to swapping atoms, these rankings can now be used to inform the choice of atoms and vacancies either by working from the top of the rankings, choosing based on a distribution biased towards the top of each ranking list, or any other method. In this work, we choose to draw atoms and vacancies from the top of the appropriate ranking list to maximise the potential benefits of our directed swapping approach.ChemDASH
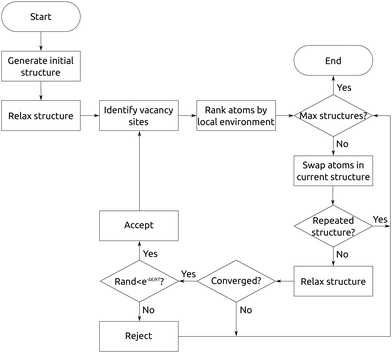
Our method of chemically directed swapping is implemented in the crystal structure prediction code ChemDASH. Fig. 2 shows the general procedure followed when running ChemDASH. ChemDASH implements a Monte-Carlo basin-hopping algorithm to search structures. We note however that the directed swapping method could in principle be implemented with any one of a range of search algorithms. There are two variants of the basin-hopping algorithm, which differ by the geometry in which we swap the atoms, we can either perform the swap in the structure after it has been optimised, or return to the unoptimised structure, and swap atoms in that geometry. In ChemDASH, swapping atoms in optimised and unoptimised geometries are both supported. ChemDASH has already played a significant role in the discovery of new compounds,26 with application to the method of probe structure based materials discovery.25 ChemDASH is written in python version 3.5+ and depends on the atomic simulation environment (ASE),30 spglib,31 and their subsequent dependencies. ChemDASH has been released under the terms of the GNU General Public Licence, and the source code is available at: https://github.com/lrcfmd/ChemDASH.Initialisation
We begin by generating an initial structure containing the set of atoms supplied by the user. This can be done by supplying a CIF file containing a structure to use as a starting point, which is useful when focussing on a particular structure type or for considering dopants in known structures. Otherwise, we choose either an “orthorhombic” or a “close-packed” initialisation lattice and populate the appropriate sites in the chosen lattice with cations and anions. This ensures that the atoms start in positions that will encourage bipartite bonding during optimisation, and also ensures that atoms lie a sufficient distance apart from one another. This is necessary because it is known that a substantial fraction of the PES corresponds to structures in which some atoms are very close together, and this region contains almost no minima.7 For both initialisation grid types we first determine the number of grid points for the anions, and the spacing between each anion grid point along the a, b and c axes from user input. We then identify the available sites for cations and anions, and populate those sites randomly with the desired atoms, on an element-by-element basis. Once all of the atoms have been distributed onto the initialisation grid, all unoccupied grid points can be recorded in the structure as vacancies.Orthorhombic initialisation grids
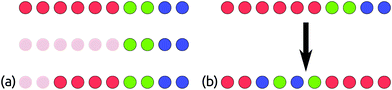
We build two interpenetrated primitive orthorhombic grids; one for cations and one for anions. Fig. 3 shows how orthorhombic initialisation grids are set up and populated. Each orthorhombic grid has a × b × c grid points, so there are a × b × c grid points available each for cations and anions. As shown in Fig. 3, once we have established the cation and anion sites, they are populated by the user-specified set of atoms. Any leftover sites are listed as vacancies.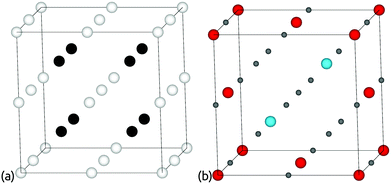 | ||
| Fig. 3 Populating orthorhombic initialisation grids in ChemDASH. The interpenetrating grids are set up in (a), with white sites available for anions and black sites available for cations. In (b), some of those sites are populated by atoms, and the unused sites shown in grey are listed as vacancies. Structure models are produced using VESTA.1 | ||
Close-packed initialisation grids
When creating an initialisation grid according to close-packing rules, we can stack anion layers in either the hexagonal close packed sequence, the cubic close packed sequence, or any specified sequence provided it does not contain any repeated layers. We can also let ChemDASH generate a random sequence of anion layers. With the anion stacking sequence established, we then identify the interstitial tetrahedral and octahedral sites available for cations. This means that there will be three times as many sites available for cations as there will be for anions. We can also use either oblique or centred rectangular lattices for the 2D layers of close-packed anions.Structure optimisation
Once we have a structure we must optimise the atomic positions to their local energy minimum.32–34 Structural optimisation and energy calculations in ChemDASH are handled by interfacing to existing materials modelling codes. ChemDASH currently supports optimisation using the force-field code GULP23 and the density functional theory (DFT) code VASP,24 which allows ChemDASH to be applied to a broad range of systems. In this work, we will focus exclusively on using GULP in ChemDASH. Use of the VASP mode has been reported previously.26ChemDASH has the ability to run structural optimisations consisting of multiple stages, where each stage is a run of the chosen optimisation software with a particular set of input parameters. For example, this can involve switching between conjugate gradient and quasi-Newton algorithms, or using different step sizes, etc. When optimising with DFT, we also enforce a stringent convergence condition for optimisations. Whilst VASP concludes a calculation after the forces on the atoms are sufficiently well converged, this will likely be after a number of self-consistent field (SCF) cycles, during which the basis functions accumulate history from the previous steps of the optimisation, which may influence the final values. We therefore run the final stage of any DFT optimisation until it converges within a single SCF cycle, or else a defined maximum number of convergence calculations are performed.
In order to maximise the efficiency of the code, we screen our structures at each stage in order to eliminate those which are not likely to be successfully optimised. After each stage of the optimisation, we abandon the relaxation if GULP/VASP failed to perform the calculation, if the calculation times out (where the time limit is supplied by the user), or if the energy or the norm of the gradient (gnorm, the root mean square gradient per variable) are not representable as floating point numbers, i.e., they have blown up to an extremely large value. The gnorm provides a rough guide to convergence, where a small value indicates that the optimisation has entered the harmonic region.23 Therefore, for GULP we can also impose a maximum value of the final gnorm for any stage of the optimisation, where the optimisation is abandoned if this value is exceeded. In addition, if a swap results in a structure that has already been considered (i.e., the same atoms occupy the same absolute positions as in a previous structure), the structure is immediately rejected before relaxation. This improves performance by ensuring calculations are not repeated. Whilst some other structure prediction codes can impose symmetry constraints onto initial structures and/or during optimisation,7 ChemDASH does not do this at present. Therefore, after optimising each structure, we determine the symmetry and transform the unit cell to its standard setting.31 We have found that this can have the effect of improving the number of structures that are successfully optimised, particularly when swapping atoms in optimised geometries and optimising structures using force-fields.
Identifying vacancy sites
When a crystal structure is optimised, the atomic positions, unit cell dimensions and angles can all change considerably.34 Therefore, when swaps are based on the optimised geometries, we must re-examine the optimised structure to determine where vacancy sites lie. In ChemDASH, we do this by using a vacancy grid. This consists of a Cartesian grid of points placed onto the structure. We then need to ensure that vacancies do not lie too close to atoms, so we remove all points that lie within a user-specified exclusion radius of an atom, as shown in Fig. 4. In addition, as we swap atoms, we also remove vacancies within the exclusion radius of any vacancy that is chosen to be occupied by an atom. This is because it would otherwise be possible to place two atoms into nearby vacancies, resulting in those atoms being unphysically close to one another. This issue is particularly important for the directed swapping method, where it is likely that nearby vacancies will have similarly favourable environments for a particular atomic species.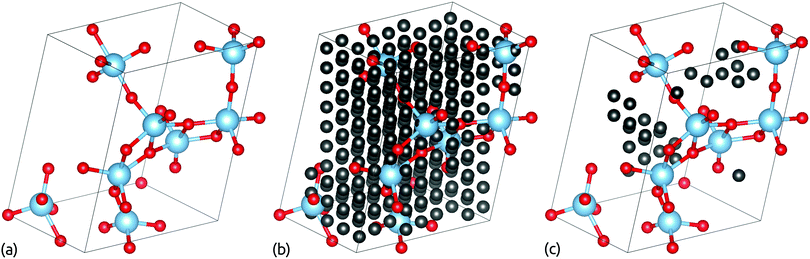 | ||
| Fig. 4 Schematic showing how vacancy sites are identified by use of a vacancy grid in ChemDASH. In (a) we take an optimised structure and then in (b) apply a Cartesian grid of vacancy sites. We then remove all vacancy sites that lie within the exclusion radius of the atoms in the optimised structure, reducing the set of vacancies to those shown in (c). Structure models are produced using VESTA.1 | ||
Swapping atoms
New structures are generated by swapping the positions of atoms and/or vacancies in the current structure. We always ensure that all swaps are non-trivial, that is to say, every atom and vacancy involved in the swap is replaced by an atom, or vacancy, of a different species. The procedure for swapping atoms is identical regardless of whether or not we use our directed swapping method. There are four stages involved in choosing the atoms to be swapped in ChemDASH:• choose which species of atoms in the structure are available to be swapped – the swap group,
• choose the total number of atoms to be swapped, m,
• choose how many atoms of each species in the swap group will be swapped,
• choose the specific atoms to swap.
This procedure ensures that a non-trivial swap is guaranteed to be performed.
We first randomly choose the group of atoms that will be involved in the swap. This swap group is a set of species from which we will choose the atoms to swap, immediately neglecting all atoms of the remaining species. This enables swapping to be targeted to the most appropriate atoms. The possible groups are:
• cations – non-trivially swap a set of cations,
• anions – non-trivially swap a set of anions,
• atoms – non-trivially swap any atoms, but not vacancies,
• all – non-trivially swap any atoms and vacancies,
• atoms-vacancies – choose a set of atoms and swap each one with a vacancy.
Note that the “all” group differs from the “atoms-vacancies” group in that the “all” group consists of atom–atom swaps and/or atom–vacancy swaps, whereas the “atoms-vacancies” group is restricted to atom–vacancy swaps. In addition, custom swap groups can be specified that enable swaps to be restricted to atoms of particular species, for example, “Sr–O” would restrict swaps to Sr and O atoms. In this specification, vacancies are denoted as “X”.30 Custom swap groups can be constructed from any combination of elements, provided there are at least two elements present in the swap group and all of the elements are present in the structure. The choice of which swap groups to use, and how much each one is weighted, can be specified by the user.
Once a swap group is chosen it, together with the structure under consideration, defines a maximum number of atoms that can be swapped non-trivially, N. Fig. 5 shows how this maximum value is determined. We first consider all of the atoms in the chosen swap group and identify the species with the largest number of atoms. We then neglect this species and determine the total number of remaining atoms. Fig. 5(a) shows that the maximum number of atoms that can be swapped is then twice this total, corresponding to a swap where every atom from the less abundant species is swapped with one from the most abundant species. This maximum is capped by the total number of atoms of the chosen swap group in the structure.
ChemDASH then chooses the actual number of atoms to swap, m, by a weighted choice. The weights wm are determined such that it is more likely to swap a smaller number of atoms than it is to swap a larger number. The default set of weights are set according to the arithmetic sequence:
| wm = wm−1 − 1, where w2 = N − 1 | (4) |
Repeated structures
With our implementation of the directed swapping method, there is a far greater likelihood of revisiting structures that have already been considered compared to swapping atoms that are chosen at random. This is by virtue of the fact that the atoms involved in each swap are drawn from the top of the ranking list for each species. In order to combat this, we have taken inspiration from the minima hopping algorithm35 and related methods16,36 to ensure an efficient exploration of the PES. The minima hopping algorithm states that if a move across the PES (a molecular dynamics (MD) trajectory in the case of the original algorithm) results in a minimum that has already been visited, then the kinetic energy should be increased for the next MD trajectory. Conversely, if the move results in a new minimum, then the kinetic energy for the next MD trajectory should be reduced.We have applied these ideas to our directed swapping method by modifying the number of atoms available from each ranking list as we encounter repeated structures. Initially, if p atoms of a particular species are to be swapped, we take the first p atoms from the ranked list for that species. However, if this swap results in a structure that has been considered previously, then we make another atom available for the next swap, i.e., we choose p atoms from the set of p + 1 atoms at the top of each ranked list. If further repeated structures are generated, then we make a further atom available each time. However, when a new minimum is visited, i.e., we accept a new structure, we reset the number of extra atoms considered to zero.
Accepting structures
Once the final energy of an optimised structure has been obtained, we must decide whether or not to accept this new structure to replace the current structure. To do so, we use the Metropolis criterion:37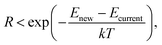 | (5) |
Results and discussion
In order to investigate the performance of the directed swapping algorithm in ChemDASH, we have run the code for three systems: 24 atoms of titanium dioxide (TiO2), 20 atoms of strontium titanate (SrTiO3), and 22 atoms of yttrium titanate (Y2Ti2O7). Each of these systems crystallises in a different structure type, and the oxidation states of each species are well defined in the atomic limit, i.e., Sr2+, Ti4+, Y3+, O2−. Hence, this will show how ChemDASH performs for a variety of systems. For each system we have tested the three methods of directed swapping alongside random swapping (i.e., no directed swapping), with swaps performed in optimised geometries. We measured the number of basin hopping (BH) moves, which are the number of times we swap atoms to generate a new structure (i.e., going around the loop in Fig. 2) that are required until we achieve the ground state structure. The fewer BH moves required, the faster ChemDASH finds the ground state. For each system, we have run ChemDASH thirty times for a given number of BH moves, and averaged the results over the thirty runs. This average gives the expected number of BH moves required to find the ground state for a particular system with either random swapping, or a particular method of directed swapping.However, quoting errors on this average is not straightforward. Firstly, the number of BH moves has a strict lower bound of zero, so the distribution is intrinsically positively skewed. Secondly, the Monte-Carlo method of exploring the PES means that the set of BH moves are not independent events. This is because, regardless of the method of swapping used, we are evolving a structure as we progress, and therefore expect to reach a lower-energy regime as the run progresses. Indeed, when considering all of the structures that relax to the ground state, we find that 50% of them have initial energies within 4.5 eV per atom of the ground state and relatively few have initial energies far from the ground state, with the largest relative initial energy being 100 eV per atom above the ground state. Hence, the trials we have conducted do not form a Gaussian distribution. We have therefore chosen to characterise our results by quoting the first four moments of the distributions of BH moves. These moments are the: mean, variance, skewness and kurtosis of the distribution.
For all of the distributions of BH moves that we obtain, we expect that the value of the skew will be positive due to the fact that each value in the distribution has a lower bound of zero. For a Gaussian distribution, the kurtosis has a value of three. A kurtosis smaller than this signifies a distribution with thinner tails and fewer and less extreme outliers than a Gaussian distribution. If the kurtosis is larger than three, then the distribution has wider tails, and hence produces more outliers than the normal distribution.
Each of the ChemDASH runs were initialised on orthorhombic initialisation grids, with anion grid points separated by 3.46 Å – so the minimum cation–anion distance is 3 Å, whilst like ions are separated by at least 3.46 Å. These values should help to promote bipartite bonding, without forcing cations and anions to be too close together. For TiO2 there were 3 × 3 × 3 cation and anion grid points, for SrTiO3 we used 2 × 2 × 3 grid points each for cations and anions, and cation and anion initialisation grids with 2 × 3 × 3 points were used for Y2Ti2O7. These values were chosen to accommodate each composition on the smallest possible initialisation grid. This should ensure that we do not have an initial structure consisting of separate fragments that are unlikely to join up during relaxation.7 When swapping atoms, we chose from the full set of swap groups involving two species of atoms. For example, for SrTiO3 we considered the swap groups: Sr–Ti, Sr–O, Ti–O, Sr–X, Ti–X, O–X. The value of kT was 0.025 eV per atom. We have optimised the structures using force-fields in GULP, which consisted of Buckingham potentials with a cut-off of 12 Å. The parameters used for each interaction are taken from the literature38,39 and given in Table 1. We employed a three-stage procedure for each optimisation:
| Interaction | A (eV) | ρ (Å) | C (eV Å−6) |
|---|---|---|---|
| O2−–O2− | 1388.77 | 0.36262 | 175 |
| Sr2+–O2− | 1952.39 | 0.33685 | 19.22 |
| Ti4+–O2− | 4590.7279 | 0.261 | 0 |
| Y3+–O2− | 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
0.24203 | 0 |
(1) Optimise the unit cell only (with conjugate gradient algorithm),
(2) Optimise atoms and cell (with conjugate gradient algorithm),
(3) Optimise atoms and cell (with quasi-Newton algorithm, BFGS update method).
Both the conjugate gradient and quasi-Newton algorithms evaluate the energy at a point x + dx by expanding the energy as a Taylor series.40 In the case of the conjugate gradient method, the Taylor expansion is truncated at the first order term, with each new search direction constructed orthogonally to all previous search directions.41 The quasi-Newton method includes terms up to second order. Given that the PES is quadratic around minima, it is totally determined by the Hessian matrix of second derivatives of the energy. However, the Hessian is not known exactly, so in quasi-Newton schemes an initial approximation of the Hessian (or its inverse) is improved as the optimisation progresses.34 Here, we use the BFGS method to update the Hessian matrix during the optimisation.33
This three-stage procedure is necessary because, given that ChemDASH does not impose constraints on symmetry, our starting structure is likely to lie far from its local minimum on the PES. We have found that using this procedure maximises the number of structures successfully optimised by GULP. The unit cell can change in all stages, but fractional atomic positions are fixed for the first stage of the relaxation. We use the low-memory BFGS algorithm in GULP, as this algorithm is written in parallel. We used a step size of 0.1 in the first two stages of the relaxation, increasing to 0.5 in the final stage. Cell vectors and atomic positions were optimized until the gnorm fell below 0.075.
Titanium dioxide
For TiO2, the ground state takes on the rutile structure, but the anatase and brookite polymorphs are metastable. Any individual run of ChemDASH can find more than one of these polymorphs, even by hopping out of the rutile ground state. Therefore, we have considered both the number of basin hopping moves required to find the rutile structure, and the number of moves to find any one of the three polymorphs. The results for TiO2 are shown in Table 2. As expected, in all cases the distributions of BH moves are positively skewed. Focussing first on the rutile ground state of TiO2, random swapping gives an average of 425 basin hopping moves to achieve the rutile structure. For all of the directed swapping methods, there is a considerable improvement, BVS directed swapping gives a factor of 2.4 improvement in the number of basin hopping moves compared to random swapping, the site potential shows a factor of 1.5 improvement, and the BVS+ method has a factor of 1.4 improvement over random swapping. Looking further to consider the three stable polymorphs, we firstly see that regardless of the method of swapping, ChemDASH achieves at least one of the polymorphs in each run of the code. With directed swapping, there is an improvement over random swapping by a factor of 1.22–1.47. Our results show that for TiO2 directed swapping improves the performance of ChemDASH in finding any of the three stable polymorphs, and in finding the rutile ground state in particular.| Random swapping | BVS swapping | Site potential swapping | BVS+ swapping | |
|---|---|---|---|---|
| Finding the rutile ground state of TiO 2 | ||||
| No. of runs that achieve rutile TiO2 | 30/30 | 28/30 | 28/30 | 29/30 |
| Average no. of BH moves to find rutile TiO2 | 426 | 178 | 278 | 252 |
| Variance of the distribution of BH moves | 1.38 × 105 | 1.29 × 104 | 3.13 × 104 | 5.23 × 104 |
| Skewness of the distribution of BH moves | 1.20 | 1.37 | 0.341 | 1.24 |
| Kurtosis of the distribution of BH moves | 0.408 | 2.67 | −0.944 | 0.809 |
| Finding any of the stable polymorphs of TiO 2 | ||||
| No. of runs that achieve rutile, anatase or brookite TiO2 | 30/30 | 30/30 | 30/30 | 30/30 |
| Average no. of BH moves to find rutile, anatase or brookite | 138 | 95 | 94 | 113 |
| Variance of the distribution of BH moves | 7.82 × 103 | 4.53 × 103 | 6.50 × 103 | 1.22 × 104 |
| Skewness of the distribution of BH moves | 0.769 | 0.914 | 1.79 | 1.28 |
| Kurtosis of the distribution of BH moves | 0.212 | 0.360 | 3.33 | 1.26 |
Strontium titanate
Focussing first on swapping atoms in optimised geometries, Table 3 shows that when running ChemDASH with SrTiO3, the perovskite ground state structure is achieved at some point in every run. When looking at the average number of basin hopping moves required to reach the ground state, we find for SrTiO3 that the number of moves is almost the same when using BVS directed swapping compared to random swapping. When directing swaps using the site potential, there is a factor of 1.4 improvement in the number of moves required to find the ground state, and a factor of 1.2 improvement when using the BVS+ method. This suggests that the site potential is the better method to use when directing swaps for SrTiO3. The relative success of the site potential compared to the BVS for this system may be down to the fact that cation–cation repulsions are directly accounted for by the site potential, and that these repulsions may play a larger role in SrTiO3 compared to TiO2 since the cation![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) anion ratio is higher.
anion ratio is higher.
| Random swapping | BVS swapping | Site potential swapping | BVS+ swapping | |
|---|---|---|---|---|
| Swapping in optimised geometries | ||||
| No. of runs that achieve SrTiO3 | 30/30 | 30/30 | 30/30 | 30/30 |
| Average no. of BH moves to find SrTiO3 | 219 | 211 | 154 | 176 |
| Variance of the distribution of BH moves | 3.54 × 104 | 2.50 × 104 | 2.67 × 104 | 2.11 × 104 |
| Skewness of the distribution of BH moves | 1.30 | 0.774 | 1.85 | 1.76 |
| Kurtosis of the distribution of BH moves | 1.16 | −0.473 | 3.89 | 4.17 |
| Swapping in unoptimised geometries | ||||
| No. of runs that achieve SrTiO3 | 27/30 | 28/30 | 28/30 | 28/30 |
| Average no. of BH moves to find SrTiO3 | 813 | 610 | 561 | 548 |
| Variance of the distribution of BH moves | 4.52 × 105 | 2.87 × 105 | 2.82 × 105 | 1.92 × 105 |
| Skewness of the distribution of BH moves | 0.519 | 0.926 | 1.27 | 0.884 |
| Kurtosis of the distribution of BH moves | −1.09 | 0.198 | 1.10 | −0.290 |
Choice of geometry for basin hopping
There are two variants of the basin hopping algorithm, which differ by the geometry in which we perform the move from one structure to the next, i.e., swap atoms in ChemDASH. We have focussed so far on swapping atoms in optimised structures. Alternatively, the basin hopping algorithm can be implemented where atoms are swapped in the original, unoptimised geometries. ChemDASH can run in both modes, so we have taken the opportunity to directly compare the performance of both implementations of the method for finding the ground state of SrTiO3. The results for swapping in unoptimised structures are shown in the lower part of Table 3. We can immediately see that the number of basin hopping moves required to find the ground state is considerably more than it was for swapping in optimised geometries. In fact, for the random, BVS, site potential and BVS+ methods of swapping, we find a factor of ∼3 improvement in the number of BH moves required to find the ground state for SrTiO3 when swapping in optimised geometries. Similarly, the variance of the distribution when swapping in unoptimised geometries is an order of magnitude greater than the variance found when swapping in optimised geometries. These results tie in with findings for other methods based on the basin hopping algorithm.42–44Yttrium titanate
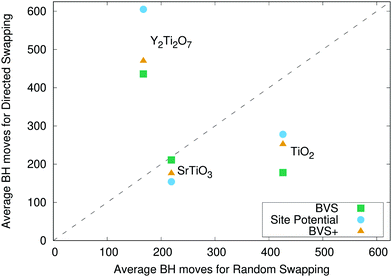
When swapping at random for Y2Ti2O7, we find the pyrochlore ground state in each run of ChemDASH, in an average of 167 basin hopping moves as shown in Table 4. However, when we consider directed swapping, the situation is very different. For all methods of directed swapping, there are runs of the code where the ground state is not achieved, and the average number of moves required to reach the ground state is 2.6–3.6 times as many as the moves required when swapping atoms at random. Looking at the other moments of the distributions, we can see that the variance is an order of magnitude larger for directed swapping compared to random swapping, and the distribution of BH moves for random swapping is more strongly skewed and has a considerably larger kurtosis compared to the distributions for directed swapping. Indeed, the kurtosis for random swapping implies that this distribution features more outliers than a normal distribution, whereas the distributions for directed swapping all imply the presence of fewer outliers than a normal distribution. The performance of each method of directed swapping compared to random swapping for TiO2, SrTiO3, and Y2Ti2O7 is shown in Fig. 6. All of the points for TiO2 and SrTiO3 lie below the diagonal, demonstrating that each method of directed swapping performs better than random swapping for these systems, although the ordering of the directed swapping methods is different for the two systems. For Y2Ti2O7, all three points lie considerably above the diagonal, showing a significant drop in performance for all methods of directed swapping compared to random swapping. From both Table 4 and Fig. 6, we can clearly see that for Y2Ti2O7, directed swapping – especially using the site potential – makes the performance of ChemDASH considerably worse.| Random swapping | BVS swapping | Site potential swapping | BVS+ swapping | |
|---|---|---|---|---|
| No. of runs that achieve Y2Ti2O7 | 30/30 | 27/30 | 29/30 | 28/30 |
| Average no. of BH moves to find Y2Ti2O7 | 167 | 436 | 605 | 470 |
| Variance of the distribution of BH moves | 1.23 × 104 | 1.58 × 105 | 1.31 × 105 | 1.43 × 105 |
| Skewness of the distribution of BH moves | 2.62 | 1.09 | 1.08 | 1.24 |
| Kurtosis of the distribution of BH moves | 9.27 | −0.0663 | 0.421 | 1.09 |
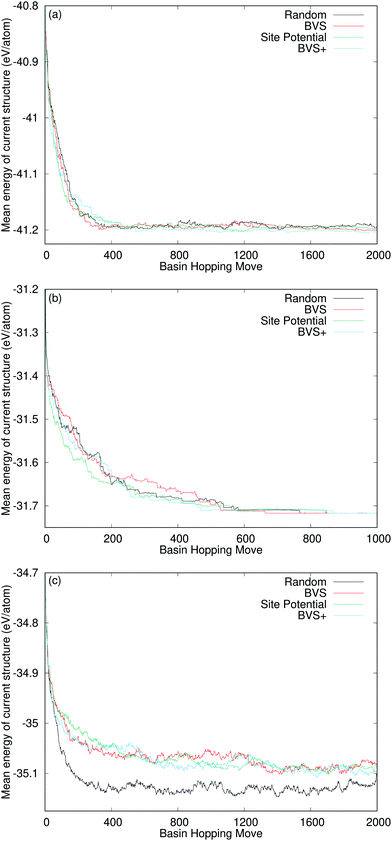
Fig. 7 shows the maximum, minimum and mean energies across each of the thirty runs for each method of swapping for Y2Ti2O7 at each point in the run. For all of the methods of swapping, one of the runs achieves the ground state energy fairly quickly, so the minimum energy rapidly reaches the ground state in all cases. When comparing random swapping to all of the methods of directed swapping, two observations are apparent. Firstly, the maximum energy is generally higher for directed swapping than it is for random swapping, particularly early in the run. Secondly, when swapping at random the mean energy can be seen to track more closely towards the minimum energy than the maximum energy, but for all methods of directed swapping, the average energy follows a trajectory closer to halfway between the maximum and minimum energies. This suggests that the maximum energy is a relative outlier for random swapping, with most energies much closer to the minimum energy, but for directed swapping there is a more even spread of energies between the thirty runs.
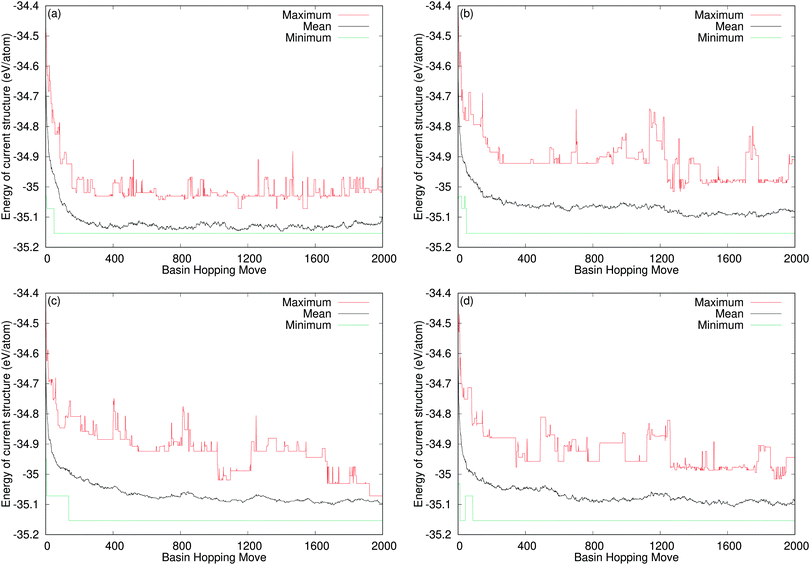
Fig. 8 shows the mean energy values across the thirty runs for all of our systems. This allows us to compare the performance of the directed swapping methods for each system throughout the runs. The plots for TiO2 and SrTiO3 show all of the curves following the same trajectory of decreasing average energy, approaching the ground state. For TiO2, the average energy values at the end of each set of runs oscillate between the energies of the stable polymorphs. For SrTiO3, we find that average energy reaches the ground state energy in all cases, i.e., all of the runs for each method of swapping achieve the ground state and finish there. The plot for Y2Ti2O7 shows a different picture. We can immediately see that this time the four curves do not follow the same trajectory. From 100 basin hopping moves onwards, the average energy for random swapping (black line in Fig. 8(c)), always tracks a lower energy than the average energy for all of the methods of directed swapping. Clearly then, our methods of chemically directed swapping have an unexpected effect on the search for the ground state of Y2Ti2O7, such that it is more difficult to achieve the correct ground state structure when using directed swapping. To understand how this is the case, we must examine more closely how well the bond valence sum and site potential apply to the ground state structure of Y2Ti2O7.
Understanding directed swapping for yttrium titanate
To understand why directed swapping performs badly for Y2Ti2O7, we have looked into the values of the bond valence sum and site potential for the atoms in the experimental ground state structure,45 and the ground state structures that result from optimising this structure using our force-field and DFT (with VASP). For the DFT optimisation, we used the PBE functional46 with the projector augmented wave approach to treat core electrons.47 A 3 × 3 × 3 k-point grid was used with a plane wave cutoff of 600 eV. The cell vectors and atomic positions were optimized until forces fell below 0.01 eV Å−1. We note that the temperature of the experimental structure is 293 K,45 whereas the computed structures are evaluated at 0 K. The ground state of Y2Ti2O7 takes on the pyrochlore structure with the space group Fd![[3 with combining macron]](https://www.rsc.org/images/entities/char_0033_0304.gif) m. This means that Y atoms lie on the 16d sites, Ti atoms lie on the 16c sites, whilst O atoms are split between the 8b and 48f sites. The lattice parameters, bond valence sum and site potential values for the experimental, force-field, and DFT ground states are shown in Table 5. For the experimental structure, the bond valence sum values for the Y, Ti, and O(48f) atoms are very close to the expected values. However, the O(8b) atoms are overbonded, with a BVS value much larger than expected. This is often found to be the case for pyrochlore structures.28,48,49 The effect of the force-field on this structure is to isotropically expand the unit cell, i.e., the lattice parameter is larger. As a result, the bond valence sum values for Y, O(8b) and O(48f) atoms are all quite far away from the expected values. For the DFT ground state, we can see in Table 5 that the lattice parameter is smaller than that for the force-field ground state, and consequently the BVS and site potential values are much closer to the values for the experimental ground state compared to the force-field ground state. Given that there is not an ideal value to compare to for the site potential, it is harder to determine how well the site potential for each atom is modelled, however, we can compare the experimental structure with the force-field structure. When we apply our force-field to Y2Ti2O7, Table 5 shows that the Ti and O(8b) atoms lie in deeper potentials compared to the experimental structure, whereas the Y and O(48f) atoms lie in shallower potentials after the force-field is applied. This follows the same trend as for the bond valence sum. In particular, we can see that when considering the O atoms, in the experimental structure the O(48f) atoms lie in a deeper potential than the O(8b) atoms, but this is reversed in the force-field structure.
m. This means that Y atoms lie on the 16d sites, Ti atoms lie on the 16c sites, whilst O atoms are split between the 8b and 48f sites. The lattice parameters, bond valence sum and site potential values for the experimental, force-field, and DFT ground states are shown in Table 5. For the experimental structure, the bond valence sum values for the Y, Ti, and O(48f) atoms are very close to the expected values. However, the O(8b) atoms are overbonded, with a BVS value much larger than expected. This is often found to be the case for pyrochlore structures.28,48,49 The effect of the force-field on this structure is to isotropically expand the unit cell, i.e., the lattice parameter is larger. As a result, the bond valence sum values for Y, O(8b) and O(48f) atoms are all quite far away from the expected values. For the DFT ground state, we can see in Table 5 that the lattice parameter is smaller than that for the force-field ground state, and consequently the BVS and site potential values are much closer to the values for the experimental ground state compared to the force-field ground state. Given that there is not an ideal value to compare to for the site potential, it is harder to determine how well the site potential for each atom is modelled, however, we can compare the experimental structure with the force-field structure. When we apply our force-field to Y2Ti2O7, Table 5 shows that the Ti and O(8b) atoms lie in deeper potentials compared to the experimental structure, whereas the Y and O(48f) atoms lie in shallower potentials after the force-field is applied. This follows the same trend as for the bond valence sum. In particular, we can see that when considering the O atoms, in the experimental structure the O(48f) atoms lie in a deeper potential than the O(8b) atoms, but this is reversed in the force-field structure.
| Experimental structure | Force-field structure | DFT structure | |
|---|---|---|---|
| Lattice parameter | 10.09 Å | 10.32 Å | 10.17 Å |
| Y BVS | 3.05 | 2.42 | 2.95 |
| Ti BVS | 4.19 | 4.03 | 3.93 |
| O(8b) BVS | 2.58 | 2.25 | 2.46 |
| O(48f) BVS | 1.98 | 1.78 | 1.89 |
| Y site potential (V) | −31.1 | −29.2 | −31.1 |
| Ti site potential (V) | −45.6 | −46.1 | −44.9 |
| O(8b) site potential (V) | 23.4 | 23.9 | 22.9 |
| O(48f) site potential (V) | 24.1 | 23.4 | 23.9 |
This then helps to explain why we find that directed swapping is so ineffective for Y2Ti2O7. The directed swapping method leads the search of the PES towards structures that have the ideal BVS or site potential values for each species. However, the ground state of the force-field has atoms with values that differ significantly from the ideal values. Hence, the effect of directed swapping is in fact to push the exploration of the PES for Y2Ti2O7 away from the ground state. For the DFT ground state, the lattice parameter, BVS and site potential values are much closer to the values for the experimental ground state compared to the force-field ground state. This suggests that the directed swapping method would be more effective if DFT were used for energy calculations with Y2Ti2O7. However, the BVS still suggests that the O(8b) atoms are still overbonded in the DFT ground state structure, which will limit the overall effectiveness of directed swapping for Y2Ti2O7. It is important to note however, that this does not necessarily mean that the directed swapping method cannot work for Y2Ti2O7, merely that the use of the BVS and site potential to rank atoms is not appropriate for Y2Ti2O7. Indeed, we have already seen for SrTiO3 that site potential directed swapping proved more effective than BVS directed swapping. If another model was used to calculate the chemical environment of the atoms that did fit well with the ground state of our force-field for Y2Ti2O7, then the directed swapping method may prove effective once more.
Conclusion
We have shown how the evolution of structures found in crystal structure prediction can be chemically directed, and practically applied this approach with the method of chemically directed swapping. This method has been implemented in ChemDASH, which explores the PES using a basin-hopping algorithm with new structures generated by swapping atoms and vacancies. Directed swapping is implemented in ChemDASH by using the bond valence sum or electrostatic site potential to evaluate the environment of each atom, ranking the atoms of each species according to how well the environments compare to the ideal. We find that ChemDASH reliably succeeds in finding ground states which have a range of different structure types. For titanium dioxide we find that directed swapping finds both the ground state and one of the stable polymorphs faster than swapping at random. Similarly, site potential-based directed swapping finds the ground state of strontium titanate faster than random swapping. However, for yttrium titanate, we find that directed swapping is less effective than random swapping, because the models we used to rank the environment of each atom are not well suited to the ground state pyrochlore structure of Y2Ti2O7. Nevertheless, ChemDASH succeeds in finding the ground state structure of Y2Ti2O7 in the vast majority of cases. Given these findings, a clear direction for future development of the directed swapping method is to investigate chemical models that can be used as alternatives to the bond valence sum and electrostatic site potential for measuring the chemical environment of atoms and vacancies in crystal structures. This should enable the approach to work effectively for a wider variety of structures. In addition, the atom rankings do not currently inform the choice of how many atoms are swapped. One possibility is to swap all atoms that are beyond a cut-off value of how their environment compares to the ideal environment. The value of such a cut-off would need to be carefully considered, particularly given that swapping atoms will also affect nearby unswapped atoms – although this impact may be mitigated through structural optimisation. Chemically directed structure evolution can be applied to any method used to explore potential energy surfaces, for example as a fitness function in a genetic algorithm, with the potential to enhance their abilities at discovering new materials.Conflicts of interest
There are no conflicts to declare.Appendix: algorithm for swapping atoms
The aim of our algorithm for swapping atoms is to efficiently construct and perform a swap of the atoms where we ensure that all atoms and vacancies involved in the swap are replaced by atoms (or vacancies) of a different species. This approach succeeds in this aim because we can guarantee a non-trivial swap of all of the atoms, and the performance of the algorithm is such that the time taken to swap atoms is negligible compared to the time taken to optimise structures. Within this algorithm, vacancies are simply treated as a special type of atom. This algorithm covers how atoms are selected once the swap group and number of atoms to swap have been established.Generating a shortlist
The first step to ensuring a non-trivial swap of the desired number of atoms, m, from the chosen swap group is to choose the atomic species of each atom that we will swap. In order to do this we first need to generate a shortlist of atomic species to choose from, such that any choice of m atoms from the shortlist will enable a non-trivial swap to be performed.It is impossible to swap atoms non-trivially if there are more than m/2 atoms of a single species. Therefore, the shortlist consists of m/2 entries for each species of atom (rounded down) included in the chosen swap group. If there are fewer than m/2 atoms of a particular species in the structure, then the number of entries in the shortlist for that species is instead equal to the number of atoms available in the structure.
Having constructed the shortlist we then check that it contains at least m entries. If it does not, which can occur if, for example, we wish to swap an odd number of atoms from a swap group that contains only two different species, then we reduce the number of atoms we will swap by one and construct a new shortlist. This ensures that we maintain the principle of being more likely to swap a smaller number of atoms than a larger number, whilst insisting that we perform a non-trivial swap. If the shortlist does contain at least m entries then it is guaranteed that a choice of any m entries results in a list of species that can be swapped non-trivially. Having generated a valid shortlist we choose m entries from it at random, which are the species of the atoms that we will swap. We then need to choose specific atoms of those species in the structure, recording their positions. This is done either at random or according to the appropriate ranked lists in the directed swapping method.
Performing the swap
The method used to efficiently perform a swap involving many atoms is shown in Fig. 9. We first take the list of the species of each atom and rearrange it with the aim that each atom in the original list is changed for one of a different species in the new list. In order to achieve this outcome efficiently, we perform this swap on a species-by-species basis. We begin by distributing the atoms of the most abundant species in the swap list amongst the sites that they did not occupy in the old list. Then, for each subsequent species, we distribute the atoms amongst the sites that they did not occupy in the old list and are unoccupied in the new list. We work from the most abundant species in the list down to the least abundant species. If all of the atoms lie in different sites compared to the original list, then we have completed a non-trivial swap of the species under consideration.However, as in Fig. 9(c), a situation may arise where the number of unoccupied sites that did not contain a particular species is fewer than the number of atoms of that species. In that case, we simply distribute the atoms amongst the remaining unoccupied sites (Fig. 9(d)). We then need to deal with unswapped atoms. We do this by first identifying all atoms that occupy the same position in the new list as they did in the old list. For each of these atoms in turn, we swap it with an atom in a site that is not occupied by an atom of the same species in either the old or new lists, as shown in Fig. 9(e) and (f).
We have now generated a new ordering of the list of the species of the atoms we are swapping. In order to swap the atoms we have selected we reorder the list of their positions in the same way as their species, and apply the new positions to the selected atoms.
Worked example
To illustrate the procedure for swapping atoms, we will consider a real example. For a structure containing two formula units of Strontium Titanate and fifteen vacancies, we have a total of 25 atoms and vacancies. We first need to choose the group of atoms to include in the swap, we will choose the “all” group, and hence consider a swap involving all atoms and vacancies. We now need to determine the maximum number of atoms we can swap non-trivially. We have two Sr atoms, two Ti atoms, six O atoms and fifteen X atoms (vacancies). Neglecting the vacancies (as they are most abundant), we have ten atoms. If we were to swap every atom with a vacancy, this gives a maximum of 20 atoms to swap, which is less than the maximum number of atoms and vacancies in the system. We next need to choose how many atoms to swap, we will consider a swap involving seven atoms.The shortlist for a seven-atom swap involving Sr, Ti, O and X atoms is then:
| O, O, O, Sr, Sr, Ti, Ti, X, X, X. | (6) |
The shortlist is valid as it contains more than seven entries. The atoms that we choose to swap may then be:
| O, O, O, Sr, Ti, Ti, X | (7) |
Our next task is to reorder the swap list (7) according to the procedure in Section A.2 and Fig. 9. This may proceed as follows:
| ′′, ′′, ′′, ′′, ′′, ′′, ′′, ′′, |
| ′′, ′′, ′′, O, ′′, O, O, |
| Ti, ′′, Ti, O, ′′, O, O, |
| Ti, Sr, Ti, O, ′′, O, O, |
| Ti, Sr, Ti, O, X, O, O. |
All atoms have been swapped after the initial repopulation, so we now assign the positions to the atoms according to this new order, and our swap is completed.
Acknowledgements
We thank EPSRC for funding under EP/N004884. We thank the University of Liverpool for computational resources.Notes and references
- K. Momma and F. Izumi, J. Appl. Crystallogr., 2011, 44, 1272–1276 CrossRef CAS.
- F. H. Stillinger, Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top., 1999, 59, 48–51 CrossRef CAS.
- J. C. Schön, K. Doll and M. Jansen, Phys. Status Solidi B, 2010, 247, 23–39 CrossRef.
- M. R. Hoare and J. McInnes, Faraday Discuss. Chem. Soc., 1976, 61, 12–24 RSC.
- C. J. Tsai and K. D. Jordan, J. Phys. Chem., 1993, 97, 11227–11237 CrossRef CAS.
- S. M. Woodley and R. Catlow, Nat. Mater., 2008, 7, 937–946 CrossRef CAS PubMed.
- C. J. Pickard and R. J. Needs, J. Phys.: Condens. Matter, 2011, 23, 053201 CrossRef PubMed.
- S. Kirkpatrick, C. D. Gelatt and M. P. Vecchi, Science, 1983, 220, 671–680 CrossRef CAS PubMed.
- A. Laio and M. Parrinello, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 12562–12566 CrossRef CAS PubMed.
- A. Barducci, G. Bussi and M. Parrinello, Phys. Rev. Lett., 2008, 100, 020603 CrossRef PubMed.
- D. M. Deaven and K. M. Ho, Phys. Rev. Lett., 1995, 75, 288–291 CrossRef CAS PubMed.
- T. S. Bush, C. R. A. Catlow and P. D. Battle, J. Mater. Chem., 1995, 5, 1269–1272 RSC.
- S. M. Woodley, P. D. Battle, J. D. Gale and C. R. A. Catlow, Phys. Chem. Chem. Phys., 1999, 1, 2535–2542 RSC.
- N. L. Abraham and M. I. J. Probert, Phys. Rev. B: Condens. Matter Mater. Phys., 2006, 73, 224104 CrossRef.
- D. C. Lonie and E. Zurek, Comput. Phys. Commun., 2011, 182, 372–387 CrossRef CAS.
- D. J. Wales and J. P. K. Doye, J. Phys. Chem. A, 1997, 101, 5111–5116 CrossRef CAS.
- D. Bochicchio and R. Ferrando, Nano Lett., 2010, 10, 4211–4216 CrossRef CAS PubMed.
- D. Bochicchio and R. Ferrando, Phys. Rev. B: Condens. Matter Mater. Phys., 2013, 87, 165435 CrossRef.
- D. Schebarchov and D. J. Wales, J. Chem. Phys., 2013, 139, 221101 CrossRef CAS PubMed.
- D. Schebarchov and D. J. Wales, Phys. Rev. Lett., 2014, 113, 156102 CrossRef CAS PubMed.
- D. Schebarchov and D. J. Wales, Phys. Chem. Chem. Phys., 2015, 17, 28331–28338 RSC.
- K. Röder and D. J. Wales, J. Phys. Chem. Lett., 2018, 9, 6169–6173 CrossRef PubMed.
- J. D. Gale and A. L. Rohl, Mol. Simul., 2003, 29, 291–341 CrossRef CAS.
- G. Kresse and J. Furthmüller, Phys. Rev. B: Condens. Matter Mater. Phys., 1996, 54, 11169–11186 CrossRef CAS PubMed.
- C. Collins, M. S. Dyer, M. J. Pitcher, G. F. S. Whitehead, M. Zanella, P. Mandal, J. B. Claridge, G. R. Darling and M. J. Rosseinsky, Nature, 2017, 546, 280–284 CrossRef CAS PubMed.
- J. Gamon, B. B. Duff, M. S. Dyer, C. Collins, L. M. Daniels, T. W. Surta, P. M. Sharp, M. W. Gaultois, F. Blanc, J. B. Claridge and M. J. Rosseinsky, Chem. Mater., 2019, 31, 9699–9714 CrossRef CAS PubMed.
- I. D. Brown, The Chemical Bond in Inorganic Chemistry: The Bond Valence Model, Oxford University Press, 2006 Search PubMed.
- I. D. Brown and D. Altermatt, Acta Crystallogr., Sect. B: Struct. Sci., 1985, 41, 244–247 CrossRef.
- N. E. Brese and M. O'Keeffe, Acta Crystallogr., Sect. B: Struct. Sci., 1991, 47, 192–197 CrossRef.
- A. H. Larsen, J. J. Mortensen, J. Blomqvist, I. E. Castelli, R. Christensen, M. Dułak, J. Friis, M. N. Groves, B. Hammer, C. Hargus, E. D. Hermes, P. C. Jennings, P. B. Jensen, J. Kermode, J. R. Kitchin, E. L. Kolsbjerg, J. Kubal, K. Kaasbjerg, S. Lysgaard, J. B. Maronsson, T. Maxson, T. Olsen, L. Pastewka, A. Peterson, C. Rostgaard, J. Schiøtz, O. Schütt, M. Strange, K. S. Thygesen, T. Vegge, L. Vilhelmsen, M. Walter, Z. Zeng and K. W. Jacobsen, J. Phys.: Condens. Matter, 2017, 29, 273002 CrossRef PubMed.
- A. Togo and I. Tanaka, Spglib: a software library for crystal symmetry search, version 1.12, 2018, https://atztogo.github.io/spglib/ Search PubMed.
- M. C. Payne, M. P. Teter, D. C. Allan, T. A. Arias and J. D. Joannopoulos, Rev. Mod. Phys., 1992, 64, 1045–1097 CrossRef CAS.
- V. Eyert, J. Comput. Phys., 1996, 124, 271–285 CrossRef.
- B. G. Pfrommer, M. Côté, S. G. Louie and M. L. Cohen, J. Comput. Phys., 1997, 131, 233–240 CrossRef CAS.
- S. Goedecker, J. Chem. Phys., 2004, 120, 9911–9917 CrossRef CAS PubMed.
- J. D. Farrell and D. J. Wales, J. Phys. Chem. A, 2014, 118, 7338–7348 CrossRef CAS PubMed.
- N. Metropolis, A. W. Rosenbluth, M. N. Rosenbluth, A. H. Teller and E. Teller, J. Chem. Phys., 1953, 21, 1087–1092 CrossRef CAS.
- C. Collins, G. R. Darling and M. J. Rosseinsky, Faraday Discuss., 2018, 211, 117–131 RSC.
- B. W. H. van Beest, G. J. Kramer and R. A. van Santen, Phys. Rev. Lett., 1990, 64, 1955–1958 CrossRef CAS PubMed.
- W. H. Press, S. A. Teukolsky, W. T. Vetterling and B. P. Flannery, Numerical Recipes: The Art of Scientific Computing, Cambridge University Press, 2007 Search PubMed.
- M. R. Hestenes and E. Stiefel, J. Res. Natl. Bur. Stand., 1952, 49, 409–436 CrossRef.
- R. P. White and H. R. Mayne, Chem. Phys. Lett., 1998, 289, 463–468 CrossRef CAS.
- M. A. Zwijnenburg and S. T. Bromley, Phys. Rev. B: Condens. Matter Mater. Phys., 2011, 83, 024104 CrossRef.
- M. A. Zwijnenburg and S. T. Bromley, J. Mater. Chem., 2011, 21, 15255–15261 RSC.
- W.-J. Becker and G. Will, Z. Kristallogr. – Cryst. Mater., 1970, 131, 278–288 CrossRef CAS.
- J. P. Perdew, K. Burke and M. Ernzerhof, Phys. Rev. Lett., 1996, 77, 3865–3868 CrossRef CAS PubMed.
- G. Kresse and D. Joubert, Phys. Rev. B: Condens. Matter Mater. Phys., 1999, 59, 1758–1775 CrossRef CAS.
- I. D. Brown and K. R. Poeppelmeier, Bond Valences, Springer, 2014 Search PubMed.
- B. A. Trump, S. M. Koohpayeh, K. J. T. Livi, J.-J. Wen, K. E. Arpino, Q. M. Ramasse, R. Brydson, M. Feygenson, H. Takeda, M. Takigawa, K. Kimura, S. Nakatsuji, C. L. Broholm and T. M. McQueen, Nat. Commun., 2018, 9, 2619 CrossRef CAS PubMed.
| This journal is © the Owner Societies 2020 |