Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Stability of the chaperonin system GroEL–GroES under extreme environmental conditions†
Michel W.
Jaworek
,
Simone
Möbitz
,
Mimi
Gao
and
Roland
Winter
 *
*
Physical Chemistry I – Biophysical Chemistry, Faculty of Chemistry and Chemical Biology, Technical University Dortmund, Otto-Hahn-Straße 4a, 44227 Dortmund, Germany. E-mail: roland.winter@tu-dortmund.de; Tel: +49 231 755 3900
First published on 21st January 2020
Abstract
The chaperonin system GroEL–GroES is present in all kingdoms of life and rescues proteins from improper folding and aggregation upon internal and external stress conditions, including high temperatures and pressures. Here, we set out to explore the thermo- and piezostability of GroEL, GroES and the GroEL–GroES complex in the presence of cosolvents, nucleotides and salts employing quantitative FTIR spectroscopy and small-angle X-ray scattering. Owing to its high biological relevance and lack of data, our focus was especially on the effect of pressure on the chaperonin system. The experimental results reveal that the GroEL–GroES complex is remarkably temperature stable with an unfolding temperature beyond 70 °C, which can still be slightly increased by compatible cosolutes like TMAO. Conversely, the pressure stability of GroEL and hence the GroEL–GroES complex is rather limited and much less than that of monomeric proteins. Whereas GroES is pressure stable up to ∼5 kbar, GroEl and the GroEl–GroES complex undergo minor structural changes already beyond 1 kbar, which can be attributed to a dissociation-induced conformational drift. Quite unexpectedly, no significant unfolding of GroEL is observed even up to 10 kbar, however, i.e., the subunits themselves are very pressure stable. As for the physiological relevance, the structural integrity of the chaperonin system is retained in a relatively narrow pressure range, from about 1 to 1000 bar, which is just the pressure range encountered by life on Earth.
1. Introduction
Chaperonins represent an important class of proteins in biological organisms (archaea, bacteria and eukarya), which have an essential role in assisting protein folding by transient encapsulation of nascent proteins in an ATP-driven mechanism to prevent misfolding or aggregation. Moreover, stress-induced denaturation of proteins can be counteracted by the chaperonin's support of correct refolding.1–4 Here, we investigated the effect of temperature and pressure stress on the conformational stability of the heat shock protein (Hsp) complex GroEL–GroES of Escherichia coli (Fig. 1a), which belongs to the group I chaperonins. Chaperonins, like the GroEL–GroES complex, are oligomeric proteins, which consist of a large double-ring structure stacked back to back, thereby enclosing a central cavity with its lid-like cofactor. In this case, the lid-like cofactor refers to GroES, which contains seven identical ∼10 kDa subunits assembled as a heptamer ring and caps the ends of the GroEL cylinder. Two heptameric rings of GroEL (each subunit contains ∼57 kDa) form the cavity, exposing hydrophobic amino acid residues for the binding of folding intermediates. Each subunit can be divided into an apical, intermediate and equatorial domain that feature different functions during the reaction cycle.1,2,5 In the first step of the reaction cycle (Fig. 1b), the non-native protein binds to the free end of a GroEL–GroES complex. The ATP-mediated conformational rearrangement of each subunit of GroEL (light green) and the binding of GroES leads to the encapsulation. Within the chaperonin nanocage, the folding takes place for the time required for ATP hydrolysis (∼2–10 s) on each subunit of the heptameric ring (pink). ATP binding to the opposite ring triggers the release of GroES and the folded protein, which completes the protein folding cycle of the chaperonin system. In case of an incomplete folding of the protein, it will rebind after release.1,2,4–7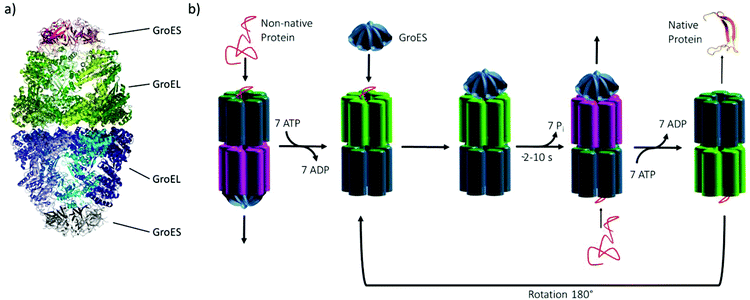 | ||
| Fig. 1 (a) The GroEL–GroES complex in secondary structure representation (PDB: 3WVL), (b) Schematic of the reaction cycle of the GroEL–GroES system. | ||
Besides the “quality control” during the translation process of proteins, chaperonins help to properly refold denatured proteins. But what about the stability of the chaperonin system itself under extreme environmental conditions? A series of biochemical and structural studies have been carried out on the complex and its components looking into the effects of salts, ligands and temperature.7–12 Almost nothing is known about the effect of pressure on the stability of the chaperonin system despite the fact that a huge fraction of organisms in the global biosphere is thriving at high hydrostatic pressure (HHP) conditions in the deep sea and sub-sea floor, where pressures of 1000 bar and more can be reached.13–16
In fact, marine sediments have been shown recently to represent one of the largest microbial habitats on Earth, even down to about 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) km below the ocean floor, where next to high pressures also increasing temperatures are encountered.15 Interestingly, a relatively high concentration of particular osmolytes, such as trimethylamine-N-oxide (TMAO), was found in cells of deep sea organisms, which are thought to help rescuing cells from pressure-induced deterioration, including protein unfolding at high hydrostatic pressures.17–20 Hence, HHP studies on biomolecular systems are prerequisite for understanding current and ancient life in the deep sea, an environment which is also the potential birth place of life on Earth.13,14
km below the ocean floor, where next to high pressures also increasing temperatures are encountered.15 Interestingly, a relatively high concentration of particular osmolytes, such as trimethylamine-N-oxide (TMAO), was found in cells of deep sea organisms, which are thought to help rescuing cells from pressure-induced deterioration, including protein unfolding at high hydrostatic pressures.17–20 Hence, HHP studies on biomolecular systems are prerequisite for understanding current and ancient life in the deep sea, an environment which is also the potential birth place of life on Earth.13,14
Remarkably, very little is known about the temperature and pressure stability of the chaperonin system and the effect of cosolutes on its stability. What is known, however, is that the expression level in E. coli is not only upregulated by temperature stress, but also under high pressure.13,14 Horowitz et al. explored the dissociation of GroEL and GroES in the presence of Mg2+ and nucleotides as a function of pressure using light-scattering data, indicating dissociation around 2 kbar.21–23 Furthermore, different chaotropic cosolvents, like urea or guanidinium hydrochloride, have revealed an adverse influence on the stability of both proteins.24 In this study, we set out to explore the thermo- and piezostability of GroEL, GroES and the GroEL–GroES complex in the presence of cosolvents, nucleotides and salts in more detail employing quantitative Fourier-transform infrared (FTIR) spectroscopy, complemented by small-angle X-ray scattering (SAXS).
2. Materials and methods
2.1 Chemicals and sample preparation
GroEL and GroES genes were produced using the pET-11a vector, cloned in E. coli BL21(DE3)-Codon plus RIPL competent cells from (Agilent Technologies) in LB-medium containing ampicillin and chloramphenicol and incubated overnight. Upon reaching an OD600nm-value of 0.5, protein expression was induced with 1 mM IPTG (isopropyl β-D-1-thiogalactopyranoside). After 3 hours, the cells were centrifuged at 4 °C and 6000 rpm. The cell pellets obtained were resuspended in purification buffer (50 mM Tris/HCl buffer, pH 7.5, 20 mM NaCl, 1 mM EDTA (ethylenediaminetetraacetic acid) and 1 mM DTT (dithiothreitol)). Subsequently, protease inhibitor, DNase und lysozyme was added to the solution, sonicated and then centrifuged at 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm and 10 °C. For purification of GroEL, the supernatant was added to a HiTrapQHP column (GE Healthcare), washed with 12% purification buffer containing 1 M NaCl and finally eluted over 150 mL with a linear gradient from 0% to 100% (purification buffer containing 1 M NaCl). Using SDS-PAGE, the protein was identified, fractions were pooled and added to the GeFi Superdex 75 26/600, which was equilibrated with purification buffer containing 250 mM NaCl. GroEL was re-identified via SDS-PAGE, fractions were collected and frozen at −80 °C. The purification of GroES was carried out by adding the supernatant to the HiTrapQHP column, washed with 5% purification buffer containing 0.5 M NaCl and finally eluted over 150 mL with a linear gradient from 0% to 100% (purification buffer containing 0.5 M NaCl). Protein purity was verified by SDS-PAGE, fractions were pooled and added to the GeFi Superdex 75 26/600, which was equilibrated with purification buffer containing 250 mM NaCl. Then, GroES was re-identified via SDS-PAGE, fractions were collected and also frozen at −80 °C. Under physiological conditions, GroEL (14mer) and GroES (heptamer) only form the asymmetric complex despite the higher concentration of GroES. For H/D-exchange, which is needed for the FTIR spectroscopic studies, the proteins were dialysed against D2O using Amicon Ultra (2 mL) centrifugation units with 10 kDa cutoff and subsequently lyophilized. Considering the effect of temperature or pressure on the pH, 50 mM phosphate buffer or 50 mM Tris buffer was chosen for the FTIR measurements, respectively. Different from phosphate buffer, Tris buffer is suitable for pressure experiments because of its small pKa dependence upon hydrostatic pressurization. Both buffers have a pD-value of 7.5, which was adjusted by adding DCl or NaOD. Here, buffer A is the neat buffer system, while buffer B contains additional 100 mM KCl. All chemicals were purchased from Sigma Aldrich and were used without further purification. For the FTIR measurements, anhydrous trimethylamine N-oxide (TMAO) and isotope labeled 13C-urea were used to avoid overlapping of the amide I’ band with the IR absorption of the deformation (OD)-band of water or the asymmetric ν(CO)-band of urea.
000 rpm and 10 °C. For purification of GroEL, the supernatant was added to a HiTrapQHP column (GE Healthcare), washed with 12% purification buffer containing 1 M NaCl and finally eluted over 150 mL with a linear gradient from 0% to 100% (purification buffer containing 1 M NaCl). Using SDS-PAGE, the protein was identified, fractions were pooled and added to the GeFi Superdex 75 26/600, which was equilibrated with purification buffer containing 250 mM NaCl. GroEL was re-identified via SDS-PAGE, fractions were collected and frozen at −80 °C. The purification of GroES was carried out by adding the supernatant to the HiTrapQHP column, washed with 5% purification buffer containing 0.5 M NaCl and finally eluted over 150 mL with a linear gradient from 0% to 100% (purification buffer containing 0.5 M NaCl). Protein purity was verified by SDS-PAGE, fractions were pooled and added to the GeFi Superdex 75 26/600, which was equilibrated with purification buffer containing 250 mM NaCl. Then, GroES was re-identified via SDS-PAGE, fractions were collected and also frozen at −80 °C. Under physiological conditions, GroEL (14mer) and GroES (heptamer) only form the asymmetric complex despite the higher concentration of GroES. For H/D-exchange, which is needed for the FTIR spectroscopic studies, the proteins were dialysed against D2O using Amicon Ultra (2 mL) centrifugation units with 10 kDa cutoff and subsequently lyophilized. Considering the effect of temperature or pressure on the pH, 50 mM phosphate buffer or 50 mM Tris buffer was chosen for the FTIR measurements, respectively. Different from phosphate buffer, Tris buffer is suitable for pressure experiments because of its small pKa dependence upon hydrostatic pressurization. Both buffers have a pD-value of 7.5, which was adjusted by adding DCl or NaOD. Here, buffer A is the neat buffer system, while buffer B contains additional 100 mM KCl. All chemicals were purchased from Sigma Aldrich and were used without further purification. For the FTIR measurements, anhydrous trimethylamine N-oxide (TMAO) and isotope labeled 13C-urea were used to avoid overlapping of the amide I’ band with the IR absorption of the deformation (OD)-band of water or the asymmetric ν(CO)-band of urea.
2.2 Temperature- and pressure-dependent FTIR spectroscopy
Temperature-dependent Fourier-transform infrared (FTIR) spectra were collected in a Nicolet 5700 (Thermo Fisher Scientific) equipped with a liquid-nitrogen cooled MCT-detector (HgCdTe) in the wavenumber range between 4000 to 1100 cm−1. A sample volume of about 15 μL was placed in the IR cell between two CaF2 windows, which is separated by a mylar spacer of 50 μm thickness. An external, circulating water thermostat controlled the required temperature in the cell with an accuracy of ±0.1 °C. To ensure that the sample was equilibrated, each temperature was maintained for ∼10 min before collecting the spectra. Pressure-dependent FTIR measurements were performed using a MAGNA 550 (Thermo Fisher Scientific) equipped with a liquid-nitrogen cooled MCT-detector (HgCdTe) in the wavenumber range between 4000 to 650 cm−1 and a temperature of 25 °C. To achieve pressures up to 12 kbar (1.2 GPa), a membrane-driven diamond anvil cell (Diacell® VivoDac, Almax easyLab) with type IIa diamonds (Almax easyLab), equipped with an automated pneumatic pressure controller (Diacell® iGM Controller, Almax easyLab), was used. A 50 μm thick gasket of brass with 0.5 mm drilling was placed between two diamonds, holding 2.5 μL of the sample (effective sample volume: 9.8 nL). Using the pressure-dependent stretching vibration of SO42− (983.5 cm−1 at 1 bar), BaSO4 was used as an internal pressure standard.25 A protein concentration of 15 mg mL−1 was used for all FTIR experiments. To avoid CO2 and humidity, the sample chamber was continuously purged with CO2-free and dry air. After placing the sample inside the cell, the waiting time was 30 min to ensure a good signal-to-noise ratio. For each temperature or pressure, spectra were averaged over 128 scans in a row and were processed with Happ–Genzel apodization using the Omnic 7.2 spectral processing software. The resolution was set at 2 cm−1. Spectra were processed and analyzed with Thermo Grams 8.0 software as follows: after buffer subtraction and smoothing, the area of the amide I′ band (1700–1600 cm−1) was normalized. To identify the number and position of the subbands and to detect conformational changes, Fourier self-deconvoluted (FSD) and 2nd derivative spectra were analyzed. The amide I′ band region of GroEL and GroES can be decomposed into seven subbands. To determine the relative changes in the population of secondary structure elements, mixed Gaussian–Lorentzian line shape functions were used in the linear combination fitting procedure.26 Based on the assumption that a two-state unfolding or dissociation process of the protein occurs, a Boltzmann function can be fitted to the pressure- and temperature-dependent sigmoidal curve progression of the secondary structural elements. The pressure- and temperature-induced intensity changes are given by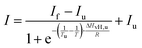 | (1) |
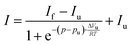 | (2) |
2.3 Small-angle X-ray scattering (SAXS)
SAXS measurements at ambient pressure at 25 °C were carried out on an Anton Paar SAXSess mc2 (Graz, Austria) using a monochromatic X-ray beam (λ = 1.54 Å) and an image plate detector. A protein concentration of 15 mg mL−1 with a volume of 20 μL was loaded into the quartz capillary (μCell of Anton Paar). Scattering patterns were collected over 30 min per image and the data were treated and analysed with 2D-SAXSquant and SAXS Quant. Further, the background of the capillary and buffer was subtracted from the data and the scattering curves were desmeared with the scattering length profile. Pressure-dependent (SAXS) measurements were performed at beamline BL9 of the synchrotron radiation facility DELTA (Dortmund, Germany) using a home-built high-pressure sample cell with flat diamond windows.27–29 The energy of the X-ray was 10 keV, corresponding to a wavelength of λ = 1.2 Å. An image plate detector MAR345 (d = 345 mm, 100 μm pixel size) was exposed 15 min per measurement in the pressure range from 1 to 3500 bar at 50 °C. Silver behenate was used to calibrate the momentum transfer Q of the X-rays, which is given by Q = (4π/λ)sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ, where λ is the wavelength of the X-rays and 2θ is the scattering angle. The GroEL–GroES complex (molar ratio of 1
θ, where λ is the wavelength of the X-rays and 2θ is the scattering angle. The GroEL–GroES complex (molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) was prepared freshly in 50 mM Tris buffer containing 10 mM MgCl2, 100 mM KCl and 3 mM ATP. Here, the data were processed with fit2D, matlab and SAXSutilities.30 The pair distance distribution function, p(r), which depends on the molecular particle shape and on the intra-particle scattering distribution, is given by the indirect Fourier transform of the measured scattered intensity, I(Q). For a particle of uniform electron density, it is given by
2) was prepared freshly in 50 mM Tris buffer containing 10 mM MgCl2, 100 mM KCl and 3 mM ATP. Here, the data were processed with fit2D, matlab and SAXSutilities.30 The pair distance distribution function, p(r), which depends on the molecular particle shape and on the intra-particle scattering distribution, is given by the indirect Fourier transform of the measured scattered intensity, I(Q). For a particle of uniform electron density, it is given by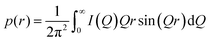 | (3) |
The function p(r) represents the frequency of vector length r connecting small volume elements within the volume of the scattering particle, that is, the protein molecule, with maximum dimension Dmax, and allows to characterize the size and shape of the scattering particle. The p(r) function was calculated by the method of Svergun using the ATSAS software package.31
3. Results and discussion
3.1 Small-angle X-ray scattering (SAXS) measurements
SAXS measurements were employed to verify the correct structure of GroEL, GroES and the whole chaperonin system, GroEL + GroES (at molar ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, the ratio within E. coli cells) (Fig. 2a and b). Fig. 2a shows the SAXS intensity profiles, I(Q), and (c) the corresponding pair-distance distribution functions, p(r), of GroEL, GroES and the GroEL–GroES complex in the presence of 3 mM ATP in aqueous buffer solution at ambient temperature and pressure. The scattering patterns measured agree very well with literature data,32 confirming the correct structures of the proteins and the monodispersity of the samples. The maximal dimension, Dmax, of GroES is about 10 nm, and that of the GroEL–GroES complex about 17.5 to 20 nm (Fig. 2c and d), in good agreement with literature data for the asymmetric chaperonin complex, which is known to be prevalent under these and also under physiological conditions.32
2, the ratio within E. coli cells) (Fig. 2a and b). Fig. 2a shows the SAXS intensity profiles, I(Q), and (c) the corresponding pair-distance distribution functions, p(r), of GroEL, GroES and the GroEL–GroES complex in the presence of 3 mM ATP in aqueous buffer solution at ambient temperature and pressure. The scattering patterns measured agree very well with literature data,32 confirming the correct structures of the proteins and the monodispersity of the samples. The maximal dimension, Dmax, of GroES is about 10 nm, and that of the GroEL–GroES complex about 17.5 to 20 nm (Fig. 2c and d), in good agreement with literature data for the asymmetric chaperonin complex, which is known to be prevalent under these and also under physiological conditions.32
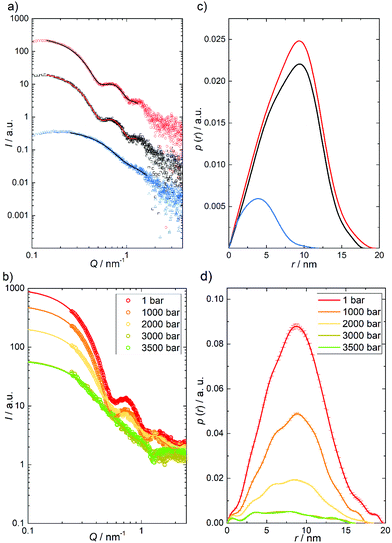 | ||
Fig. 2 (a) SAXS intensity curves, I(Q), and (c) the pair-distance distribution functions, p(r), of GroEL (15 mg mL−1, black), GroES (15 mg mL−1, blue) and the GroEL-GroES complex (molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) in the presence of 3 mM ATP (red) in aqueous solution (50 mM Tris buffer, 100 mM KCl, 10 mM MgCl2, pH 7.5) at room temperature (T ≈ 25 °C). (b) Pressure-dependent SAXS curves and (d) the pair-distance distribution functions, p(r), of the GroEL–GroES complex (25 mg mL−1 GroEL, 4.5 mg mL−1 GroES) in aqueous solution (50 mM Tris buffer, 100 mM KCl, 10 mM MgCl2 and 3 mM ATP, pH 7.5). Symbols show experimental data and solid lines in (a) are fits representing the Fourier-transformations of p(r). Owing to strong background scattering by the high-pressure cell and the smaller Q-range covered, the p(r) data in Fig. 2d appear to be a bit noisier compared to those measured in quartz capillaries at ambient conditions (Fig. 2b). 2) in the presence of 3 mM ATP (red) in aqueous solution (50 mM Tris buffer, 100 mM KCl, 10 mM MgCl2, pH 7.5) at room temperature (T ≈ 25 °C). (b) Pressure-dependent SAXS curves and (d) the pair-distance distribution functions, p(r), of the GroEL–GroES complex (25 mg mL−1 GroEL, 4.5 mg mL−1 GroES) in aqueous solution (50 mM Tris buffer, 100 mM KCl, 10 mM MgCl2 and 3 mM ATP, pH 7.5). Symbols show experimental data and solid lines in (a) are fits representing the Fourier-transformations of p(r). Owing to strong background scattering by the high-pressure cell and the smaller Q-range covered, the p(r) data in Fig. 2d appear to be a bit noisier compared to those measured in quartz capillaries at ambient conditions (Fig. 2b). | ||
Fig. 2b and d show the SAXS profiles of the GroEL–GroES complex in the presence of 3 mM ATP as a function of hydrostatic pressure and the corresponding pair distance distribution functions, p(r). As can be clearly seen, the minima of the I(Q) curve, which are typical for the shape of the chaperonin complex, become less pronounced upon compression beyond 2 kbar, and have finally disappeared at 3.5 kbar. The concomitant decrease of the scattering intensity with increasing pressure is due to a pressure-induced decrease of the electron density contrast between the sample and the solvent, which is due to a drastic increase of the electron density of the water at high pressures. In agreement with the marked changes observed in the p(r) function, these results indicate dissociation of the GroEL–GroES complex at pressures beyond about 2 kbar, which would be consistent with conclusions based on indirect measurements using light scattering methodology.21 Even above 3.5 kbar, no complete dissociation into monomers is observed, as can be deduced from the slope of I(Q) and shape of the p(r) function, which can be attributed to expanded 2D arrays of protein assemblies.
3.2 Temperature- and pressure-dependent FTIR spectroscopic measurements
To reveal also the secondary structural changes of the chaperonin system over a wide range of temperatures and pressures, FTIR spectroscopy was employed. Fig. 3 shows the effect of temperature on the amide I′ band of GroEL and GroES in neat buffer and their corresponding changes in secondary structure elements. Details of the assignment and band fitting procedure to determine the various secondary structure elements are illustrated in Fig. S1, ESI.† The normalized amide I′ band of GroEL as a function of temperature (Fig. 3a) shows a broad band at ∼1650 cm−1, which decreases with increasing temperature, while two peaks appear at ∼1618 cm−1 and ∼1683 cm−1, respectively. These two bands are characteristic for aggregation upon unfolding of proteins at high temperature and can be assigned to intermolecular β-sheets. Based on the fitting procedure, the main secondary structural components at 25 °C and ambient pressure are α-helices (∼39%), in agreement with literature values,33 and intramolecular β-sheets (∼23%). These results are in rather good agreement with crystallographic data obtained by Braig et al.,34 who found ∼45% α-helices and ∼17% intramolecular β-sheets. The small differences observed might be due to different absorption coefficients of the secondary structure elements. Here, we consider and discuss essentially relative changes, only. The amide I′ band shows no significant changes between 25 and 65 °C, where thermal unfolding sets in, which is accompanied by decreasing amounts of α-helices (∼−10%) and random coils (∼−7%) and, concomitantly, an increasing amount of intermolecular β-sheets (∼+13%). Boltzmann fits to the experimental data (eqn (1)) allow to determine the unfolding temperature, Tu, and the van’t Hoff enthalpy change, ΔHvH,u, upon (partial) unfolding of the protein: Tu = 69.1 ± 0.2 °C and ΔHvH,u = 1000 ± 60 kJ mol−1 (Table SI 1). The unfolding temperature is in accord with the literature, which reports denaturation of GroEL to occur between 65 and 70 °C.35 The ΔHvH,u-value must be handled with caution, however, due to the irreversibility of the heat-induced protein unfolding and subsequent aggregation process, i.e., it reflects no true thermodynamic data and serves only for the comparison with ΔHvH,u values determined in the presence of different salts, nucleotides and cosolvents (see below).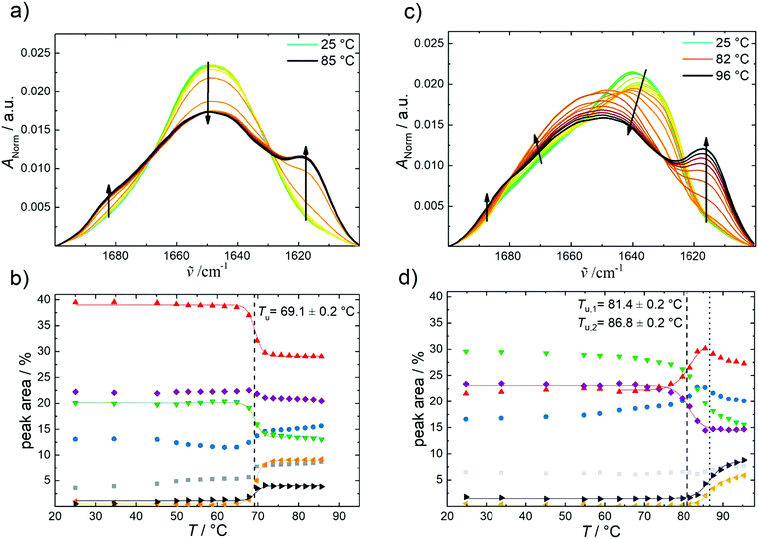 | ||
| Fig. 3 Temperature-dependent FTIR absorption data of GroEL and GroES in neat buffer at ambient pressure. (a) Temperature-dependent changes of the normalized amide I′ band region of GroEL and (b) corresponding changes in secondary structure elements by temperature: intermolecular β-sheets (1683 cm−1, grey); turns and loops (1672 cm−1, blue); α-helices (1654 cm−1, red); random coils (1642 cm−1, green); intramolecular β-sheets (1636 cm−1, purple); intermolecular β-sheets (1618 cm−1, orange) and side chains (1612 cm−1, black). (c) Temperature-dependent changes of the normalized amide I′ band region of GroES and (d) corresponding changes in secondary structure elements by temperature: intermolecular β-sheets (1685 cm−1, grey); turns and loops (1672 cm−1, blue); α-helices (1654 cm−1, red); random coils (1641 cm−1, green); intramolecular β-sheets (1630 cm−1, purple); intermolecular β-sheets (1618 cm−1, orange) and side chains (1613 cm−1, black). Lines display the Boltzmann fits to the experimental data using eqn (1). | ||
In contrast to GroEL, the normalized amide I′ band of GroES shows a broad band at ∼1640 cm−1 (Fig. 3c), which indicates a different distribution of secondary structure elements. Upon increasing temperature, we observe, on the one hand, a decrease of the main peak and on the other hand, a distinct structural transition (turquoise to orange line) before aggregation commences, which is indicated by the intermolecular β-sheet peaks appearing at ∼1617 cm−1 and ∼1684 cm−1. GroES shows high contents of random coils (∼29%) and intramolecular β-sheets (∼23%) at 25 °C and ambient pressure, and the amount of α-helices is much lower than in GroEL. Valencia et al. reported similar values for the secondary structure elements.36 The secondary structure of GroES does not exhibit significant changes between 25 and 70 °C. Beyond 70 °C, a structural transition involving intramolecular β-sheets transforming to more α-helices takes place. The aggregation of the protein starts at ∼82 °C, where the percentage of intermolecular β-sheets increases to 7%, while the percentage of α-helices decreases slightly (∼−4%). During the overall phase transition, a continuous decrease of the percentage of random coils (∼−12%) is observed. The temperatures of the two structural transitions are located at Tu,1 = 81.4 ± 0.2 °C and Tu,2 = 86.8 ± 0.2 °C, respectively. Hence, GroES displays an about 12 °C higher temperature stability than GroEL. As shown by Boudker et al., the stability of GroES seems to be concentration-dependent, because the Tu-value was found to increase with increasing protein concentration.37 DSC- and CD-experiments revealed a Tu-value of about 76.4 °C for a GroES concentration of 175 μM. Here, we used a much higher GroES concentration, ∼1.44 mM.
Fig. 4 highlights the effect of pressure on the amide I′ band and the fraction of secondary structures of GroEL and GroES derived from the analysis of the underlying subbands (Fig. SI2, ESI†). For both proteins, a pressure range between 1 bar and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 bar (1 GPa) has been covered. The amide I′ band of GroEL is shifting towards lower wavenumbers upon application of pressure (Fig. 4a). This linear pressure-induced elastic shift can be explained by a pressure-induced compression of the chemical bonds, equivalent to changes of the force constant of the C
000 bar (1 GPa) has been covered. The amide I′ band of GroEL is shifting towards lower wavenumbers upon application of pressure (Fig. 4a). This linear pressure-induced elastic shift can be explained by a pressure-induced compression of the chemical bonds, equivalent to changes of the force constant of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration.
O stretching vibration.
 | ||
| Fig. 4 Pressure-dependent FTIR absorption data of GroEL and GroES in neat buffer at 25 °C. (a) Pressure-dependent changes of the normalized amide I′ band region of GroEL and (b) corresponding changes in secondary structure elements by pressure: intermolecular β-sheets (1683 cm−1, grey); turns and loops (1672 cm−1, blue); α-helices (1654 cm−1, red); random coils (1641 cm−1, green); intramolecular β-sheets (1635 cm−1, purple) and side chains (1611 cm−1, black). (c) Pressure-dependent changes of the normalized amide I′ band region of GroES and (d) corresponding changes in secondary structure elements by pressure: intermolecular β-sheets (1683 cm−1, grey); turns and loops (1671 cm−1, blue); α-helices (1654 cm−1, red); random coils (1641 cm−1, green); intramolecular β-sheets (1630 cm−1, purple) and side chains (1612 cm−1, black). Lines display Boltzmann fits to the experimental data using eqn (2). | ||
Regarding the percentage of secondary structure elements, a slightly reduction of the α-helix content (∼−5%) is observed with increasing pressure, whereas the content of intramolecular β-sheets increases concomitantly (∼ +5%) beyond about 1 kbar. Up to 10 kbar, only this small conformational change takes place, which is accompanied by a volume change as small as ΔVu = −8.8 ± 3 mL mol−1 as determined using eqn (2). The pressure dependence of the amide I′ band and relative changes of secondary structure elements of the GroEL–GroES complex are similar to those of GroEL (Fig. SI 3, ESI†).
The pressure dependence of the secondary structure of GroES differ significantly from that of GroEL. GroES undergoes partial unfolding at pu of 5.4 ± 0.3 kbar (Fig. 4c and d), which is accompanied by an increase of α-helices (∼+4%), whereas the content of intramolecular β-sheets decreases concomitantly (∼−5%) upon compression. Using eqn (2), the volume change of partial unfolding of GroES is determined to be ΔVu = −53 ± 12 mL mol−1. This value is typical for ΔVu-values found for unfolding of monomeric proteins.38–41
By favoring states with a smaller partial molar volume, pressure generally shifts an equilibrium toward a state with smaller overall volume, in accord with Le Châtelier's principle.16,41 Hence, owing to the fact that the pressure stability of proteins in general, and of oligomeric proteins in particular, is largely due to their internal void volume and packing defects, which get filled with water upon unfolding,38 we can conclude that the packing of the two proteins at ambient temperature is different. But the difference is very small, comparable to the volume of just 2–3 water molecules, only. GroEL exhibits a small ΔVu-value of about −8.8 mL mol−1 and experiences small conformational changes above 1 kbar only, where the amount of intramolecular β-sheets increases slightly at the expense of α-helices, whereas GroES tolerates pressures up to about 5.4 kbar, where partial unfolding of the protein commences, which is accompanied by a volume change of about −53 mL mol−1.
3.3 Effects of cosolvents, salts and nucleotides on the folding stability of the chaperonin system
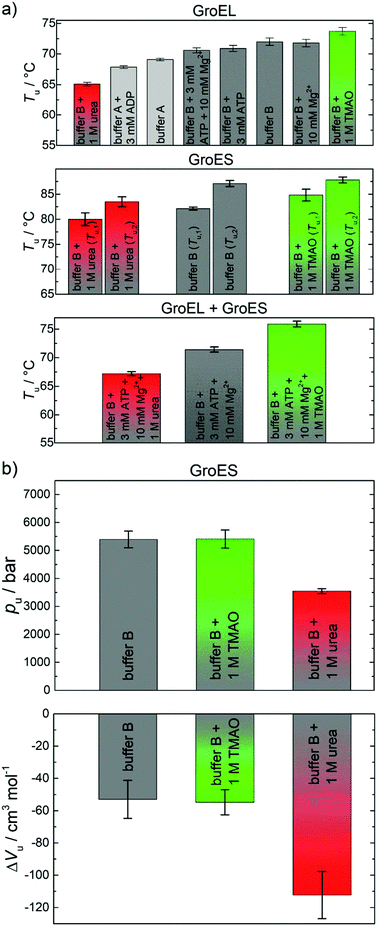
Cations, nucleotides and osmolytes have been shown to be important for the maintenance of the reaction cycle of the chaperonin system and for osmotic pressure regulation.42,43 Hence, studies of the effects of salts, ATP, ADP, and of TMAO on the thermo- and piezostability of GroEL, GroES and the GroEL–GroES complex have been included in our studies using FTIR spectroscopy. All results obtained are depicted in Fig. 5a and are listed in Table SI 1 (ESI†). In the presence of 100 mM K+ (buffer B) and additionally 1 M TMAO, the unfolding temperature of GroEL is significantly shifting towards higher temperatures (ΔTu ≈ 5 °C), i.e. these cosolutes increase the stability of the proteins. Addition of 3 mM of the nucleotide ADP and addition of 1 M urea show the opposite effect. Differently, GroES experiences no significant changes of the Tu-value in the presence of salts and nucleotides. However, the unfolding temperature of GroES is found to be highly dependent on the cosolvents TMAO and urea, which also have a positive (1 M TMAO) or negative (1 M urea) effect on the temperature stability of the protein. Interestingly, the stability of GroEL is also enhanced by addition of GroES (molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), in good agreement with the literature,11 both in the absence and presence of cosolvents (Fig. 5a). The spectroscopically determined van’t Hoff enthalpy changes (Table SI 1, ESI†) are not significantly affected by the addition of salts, nucleotides and TMAO. The effect of salts and nucleotides on the thermostability of neat GroEL are in good agreement with data derived from differential scanning calorimetry by Surin et al.11 The destabilizing effect of the nucleotide can be explained by conformational changes in the open state of the GroEL subunit structure, which occur probably near the nucleotide binding site.44,45 Also urea exhibits a negative influence on the stability, which results most likely from direct interactions of the urea molecules with the protein backbone and its side chains. Hence, the unfolded state is energetically favored and stabilized relative to the native conformation.46,47 In contrast, TMAO is able to enhance the protein stability without direct interaction with the protein. It prevents unfolding by favorable interaction with water, leading to an increase in hydrogen bonding and structuring of the solvent.40,48 Such preferential exclusion effect of TMAO from the protein interface leads to an increase of the stability of folded and compact structures.48,49 The presence of K+ ions as well as GroES also affect the temperature stability in a positive way. Both factors regulate and stabilize the quaternary structure of GroEL. GroES interacts with the apical domain of GroEL197–376 and triggers a conformational change.24,50,51 K+ and Mg2+ ions play also an important role for ATP binding and hydrolysis and have an impact on the stability of GroEL.52–54
2), in good agreement with the literature,11 both in the absence and presence of cosolvents (Fig. 5a). The spectroscopically determined van’t Hoff enthalpy changes (Table SI 1, ESI†) are not significantly affected by the addition of salts, nucleotides and TMAO. The effect of salts and nucleotides on the thermostability of neat GroEL are in good agreement with data derived from differential scanning calorimetry by Surin et al.11 The destabilizing effect of the nucleotide can be explained by conformational changes in the open state of the GroEL subunit structure, which occur probably near the nucleotide binding site.44,45 Also urea exhibits a negative influence on the stability, which results most likely from direct interactions of the urea molecules with the protein backbone and its side chains. Hence, the unfolded state is energetically favored and stabilized relative to the native conformation.46,47 In contrast, TMAO is able to enhance the protein stability without direct interaction with the protein. It prevents unfolding by favorable interaction with water, leading to an increase in hydrogen bonding and structuring of the solvent.40,48 Such preferential exclusion effect of TMAO from the protein interface leads to an increase of the stability of folded and compact structures.48,49 The presence of K+ ions as well as GroES also affect the temperature stability in a positive way. Both factors regulate and stabilize the quaternary structure of GroEL. GroES interacts with the apical domain of GroEL197–376 and triggers a conformational change.24,50,51 K+ and Mg2+ ions play also an important role for ATP binding and hydrolysis and have an impact on the stability of GroEL.52–54
Temperature-induced unfolding of proteins is mainly driven by the conformational entropy gain of the peptide chain, whereas the overall volume change plays the decisive role in the pressure-induced unfolding scenario.16,41 The system tends to occupy a smaller volume state at high pressure, which includes the filling and elimination of internal void volume and cavities as possible mechanism for pressure-induced unfolding.38,41,55 The thermodynamic data for GroES in the presence of the two cosolvents studied compared to the neat buffer data are displayed in Fig. 5b and Table SI 2 (ESI†). Within the accuracy of the experiment, 1 M TMAO has no significant effect on pu and ΔVu. A similar behavior has been observed for GroEL (data not shown).
Differently, marked stabilizing effects of some monomeric proteins (e.g., SNase, RNase A) by TMAO against pressure-induced unfolding and favorable effects of TMAO on enzyme function, protein polymerization and channel activity under high-pressure stress have been reported.41,56–63 The minor (possibly stabilizing) effect observed here is probably due to a smaller degree of unfolding and exposure of solvent accessible surface area (SASA), rendering the excluded volume effect imposed by TMAO less effective.
The marine-osmolyte TMAO has been discussed as a potential piezolyte, i.e. an osmolyte-type solute that is able to counteract deteriorating pressure effects, in many deep sea vertebrate animals (e.g., chondrichthyans (such as sharks and rays), teleost fishes, crustaceans), which in addition has the ability to counteract destabilizing effects of urea.17,60 In deep sea invertebrates, cells contain high levels of neutral free amino acids and some other methylamines,64 which show different cytoprotective properties against environmental stressors. For example, hypotaurine seems to protect cells as an antioxidant from sulfide toxicity.18 Glycine, in comparison to TMAO, appears to stabilize proteins essentially against temperature stress.60
Conversely, pu markedly decreases by ∼1800 bar in the presence of 1 M urea and, consequently, ΔVu is much more negative compared to the cosolvent-free buffer (buffer B). The pressure-induced destabilization of urea is expected to be due to favorable enthalpic interactions of urea with the protein, leading to marked conformational changes. They may lead to an increase of void volume and/or an increase of electrostrictive hydration of a larger exposed SASA upon (partial) unfolding, and hence to the more negative ΔVu-value observed.41,59
4. Conclusions
The chaperonin system GroEL–GroES is present in all kingdoms of life and rescues proteins from improper folding and aggregation. As a usual answer to a stress condition, including high temperatures and pressures, production of heat shock proteins such as the GroEL–GroES system, is boosted.13,65,66 Here, we explored the thermo- and piezostability of GroEL, GroES and the GroEL–GroES complex itself, covering a large region of the temperature–pressure plane, and included the effects of particular cosolvents, nucleotides and salts, employing quantitative FTIR spectroscopy, complemented by SAXS data. Owing to its high biological relevance and lack of data, our focus was especially on the effect of pressure on the chaperonin system. The experimental results reveal that the GroEL–GroES complex is remarkably temperature stable with an unfolding temperature beyond ∼70 °C, which can still be slightly increased by compatible cosolutes like TMAO, facilitated by the preferential exclusion of TMAO from the protein surface. Conversely, the pressure stability of GroEL and hence the GroEL–GroES complex is rather limited and much less than that of monomeric proteins, which typically unfold beyond pressures of 4–8 kbar, only.13,17 Whereas GroES is stable up to about 5.5 kbar before it partially unfolds, GroEL and hence the GroEL–GroES complex undergoes minor conformational changes already beyond 1 kbar, which can be attributed to a dissociation-induced conformational drift, leading to a slight decrease of the α-helix content and a concomitant increase of intramolecular β-sheets. The FTIR data are corroborated by the high-pressure SAXS data, which reveal dissociation of the complex at about 2 kbar. Such conclusions would be in line with early light scattering experiments by Horowitz et al., suggesting dissociation of the system at pressures around 2 kbar, and the re-association of the complex being very slow with half-times of several days, which might be due to the conformational drift of the structure upon dissociation of the complex.67 Further, the FTIR data show, quite unexpectedly, that no significant unfolding of GroEL is observed even up to 10 kbar, i.e., the subunits are very pressure stable themselves. The SAXS data measured up to 3.5 kbar also indicate that no complete dissociation into the small monomeric subunits and unfolding of the monomers takes place. Accumulation of particular cosolutes, such as methylamines, polyols and amino acids, are often found in cells to equilibrate cellular osmotic pressure and to maintain stability and functionality of proteins and their assemblies.17–20,40 Therefore, such small organic molecules are termed osmolytes or chemical chaperones. A prominent and effective osmolyte, trimethylamine-N-oxide (TMAO), is even found in tissue of various deep sea animals and its concentration correlates with the ocean depth, i.e. hydrostatic pressure, where those animals were encountered.17,18,60 Here we show that 1 M TMAO is able to increase the temperature stability of the proteins slightly, the pressure stability is hardly affected by the cosolute, only, which might be due to the absence of significant unfolding of the subunits.Hence, overall, the GroEL–GroES complex is rather temperature stable and stable against high hydrostatic pressure in the range of 1 to about 1000 bar, where dissociation of the GroEL–GroES complex sets in. Thus, the chaperonin system is just stable in the pressure range encountered to life on Earth, including the deepest trenches of the ocean. It remains to be learned, how cells of bacteria and archaea are able to withstand more elevated temperature and pressure conditions as encountered in km depths of the sub-seafloor crust.15
Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
Diese Arbeit wurde durch RESOLV unterstützt durch die Deutsche Forschungsgemeinschaft (DFG) im Rahmen der Exzellenzstrategie des Bundes und der Länder – EXC2033 – Projektnummer 390677874. Financial support from the DFG Research Unit FOR 1979 is also gratefully acknowledged. We thank Dr Manajit Hayer-Hartl and her group (MPI Biochemistry, Germany) for providing GroEL and GroES plasmids. The authors thank DELTA for experimental time at beamline BL9. Special thanks go to Dr M. Paulus and Dr C. Sternemann for their support at the beamline and G. Surmeier with the high-pressure setup. We are grateful for experimental help by M. Manisegaran, J.-M. Knop and Dr I. Kiesel.References
- D. Balchin, M. Hayer-Hartl and F. U. Hartl, Science, 2016, 353, aac4354 CrossRef PubMed.
- F. Hartl and M. Hayer-Hartl, Sci. Compass, 2002, 295, 1852–1858 CrossRef CAS PubMed.
- A. Cario, M. Jebbar, A. Thiel, N. Kervarec and P. M. Oger, Sci. Rep., 2016, 6, 1–8 CrossRef PubMed.
- F. U. Hartl, A. Bracher and M. Hayer-Hartl, Nature, 2011, 475, 324–332 CrossRef CAS PubMed.
- A. J. Gupta, S. Haldar, G. Miličić, F. U. Hartl and M. Hayer-Hartl, J. Mol. Biol., 2014, 426, 2739–2754 CrossRef CAS PubMed.
- S. Sharma, K. Chakraborty, B. K. Müller, N. Astola, Y. C. Tang, D. C. Lamb, M. Hayer-Hartl and F. U. Hartl, Cell, 2008, 133, 142–153 CrossRef CAS PubMed.
- H. Taguchi, J. Biochem., 2005, 137, 543–549 CrossRef CAS PubMed.
- W. A. Fenton and A. L. Horwich, Q. Rev. Biophys., 2003, 36, 229–256 CrossRef CAS PubMed.
- D. Thirumalai and G. H. Lorimer, Annu. Rev. Biophys. Biomol. Struct., 2001, 30, 245–269 CrossRef CAS PubMed.
- A. Marx and E. Mandelkow, Eur. Biophys. J., 1994, 22, 405–421 CrossRef CAS PubMed.
- A. K. Surin, N. V. Kotova, I. A. Kashparov, V. V. Marchenkov, S. Y. Marchenkova and G. V. Semisotnov, FEBS Lett., 1997, 405, 260–262 CrossRef CAS PubMed.
- B. Sot, S. Bañuelos, J. M. Valpuesta and A. Muga, J. Biol. Chem., 2003, 278, 32083–32090 CrossRef CAS PubMed.
- F. Meersman, I. Daniel, D. H. Bartlett, R. Winter, R. Hazael and P. F. McMillan, Rev. Mineral. Geochem., 2013, 75, 607–648 CrossRef CAS.
- I. Daniel, P. Oger and R. Winter, Chem. Soc. Rev., 2006, 35, 858–875 RSC.
- B. Orcutt and I. Daniel, in DeepCarbon: Past to Present, ed. R. Dasgupta, Cambridge University Press, 2019 Search PubMed.
- K. Akasaka and H. Matsuki, High Pressure Bioscience, Springer, Heidelberg, 2015 Search PubMed.
- R. H. Kelly and P. H. Yancey, Biol. Bull., 1999, 196, 18–25 CrossRef CAS PubMed.
- P. H. Yancey, W. R. Blake and J. Conley, Comp. Biochem. Physiol., Part A: Mol. Integr. Physiol., 2002, 133, 667–676 CrossRef.
- M. A. Schroer, Y. Zhai, D. C. F. Wieland, C. J. Sahle, J. Nase, M. Paulus, M. Tolan and R. Winter, Angew. Chem., Int. Ed., 2011, 50, 11413–11416 CrossRef CAS PubMed.
- K. Julius, J. Weine, M. Berghaus, N. König, M. Gao, J. Latarius, M. Paulus, M. A. Schroer, M. Tolan and R. Winter, Phys. Rev. Lett., 2018, 121, 038101 CrossRef CAS PubMed.
- M. Panda, J. Ybarra and P. M. Horowitz, J. Biol. Chem., 2001, 276, 6253–6259 CrossRef CAS PubMed.
- M. Panda and P. M. Horowitz, Biochemistry, 2002, 41, 1869–1876 CrossRef CAS PubMed.
- M. Panda, J. Ybarra and P. M. Horowitz, Biochemistry, 2002, 41, 12843–12849 CrossRef CAS PubMed.
- N. A. Ryabova, V. V. Marchenkov, S. Y. Marchenkova, N. V. Kotova and G. V. Semisotnov, Biochemistry, 2013, 78, 1405–1414 CAS.
- P. Wong and D. Moffat, Appl. Spectrosc., 1989, 43, 1279–1281 CrossRef CAS.
- G. Panick and R. Winter, Biochemistry, 2000, 39, 1862–1869 CrossRef CAS PubMed.
- C. Krywka, M. Paulus, C. Sternemann, M. Volmer, A. Remhof, G. Nowak, A. Nefedov, B. Pöter, M. Spiegel and M. Tolan, J. Synchrotron Radiat., 2006, 13, 8–13 CrossRef CAS PubMed.
- C. Krywka, C. Sternemann, M. Paulus, M. Tolan, C. Royer and R. Winter, ChemPhysChem, 2008, 9, 2809–2815 CrossRef CAS PubMed.
- R. Winter, Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol., 2002, 1595, 160–184 CrossRef CAS.
- M. Sztucki and T. Narayanan, J. Appl. Crystallogr., 2007, 40, 459–462 CrossRef.
- D. Franke, M. V. Petoukhov, P. V. Konarev, A. Panjkovich, A. Tuukkanen, H. D. T. Mertens, A. G. Kikhney, N. R. Hajizadeh, J. M. Franklin, C. M. Jeffries and D. I. Svergun, J. Appl. Crystallogr., 2017, 50, 1212–1225 CrossRef CAS PubMed.
- T. Inobe, K. Takahashi, K. Maki, S. Enoki, K. Kamagata, A. Kadooka, M. Arai and K. Kuwajima, Biophys. J., 2008, 94, 1392–1402 CrossRef CAS PubMed.
- J. Chen, H. Yagi, Y. Furutani, T. Nakamura, A. Inaguma, H. Guo, Y. Kong and Y. Goto, Sci. Rep., 2014, 4, 1–7 Search PubMed.
- K. Braig, Z. Otwinowski, R. Hedge, D. C. Boisvert, A. Joachimiak, A. L. Horwich and P. B. Sigler, Nature, 1994, 371, 578–586 CrossRef CAS PubMed.
- J. Martin, A. Horwich and F. Hartl, Science, 1992, 258, 995–998 CrossRef CAS PubMed.
- A. Valencia, T. J. Hubbard, A. Muga, S. Banuelos, O. Llorca, J. L. Carrascosa and J. M. Valpuesta, Proteins: Struct., Funct., Genet., 1995, 22, 199–209 CrossRef CAS PubMed.
- O. Boudker, M. J. Todd and E. Freire, J. Mol. Biol., 1997, 272, 770–779 CrossRef CAS PubMed.
- J. Roche, J. A. Caro, D. N. Norberto, P. B. Barthe, C. R. Roumestand, J. L. Schlessmand, A. E. Garcia, B. García-Moreno and C. A. Royer, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 6945–6950 CrossRef CAS PubMed.
- J. L. Silva, A. C. Oliveira, T. C. R. G. Vieira, G. A. P. de Oliveira, M. C. Suarez and D. Foguel, Chem. Rev., 2014, 114, 7239–7267 CrossRef CAS PubMed.
- C. A. Royer, Biochim. Biophys. Acta, 2002, 1595, 201–2009 CrossRef CAS.
- R. Winter, Annu. Rev. Biophys., 2019, 48, 441–463 CrossRef CAS PubMed.
- G. S. Jackson, R. A. Staniforth, D. J. Haisall, T. Atkinson, J. J. Holbrook, A. R. Clarke and S. G. Burston, Biochemistry, 1993, 32, 2554–2563 CrossRef CAS PubMed.
- P. V. Viitanen, T. H. Lubben, J. Reed, P. Goloubinoff, D. P. O'Keefe and G. H. Lorimer, Biochemistry, 1990, 29, 5665–5671 CrossRef CAS PubMed.
- A. Galán, O. Llorca, J. M. Valpuesta, J. Pérez-Pérez, J. L. Carrascosa, M. Menéndez, S. Bañuelos and A. Muga, Eur. J. Biochem., 1999, 259, 347–355 CrossRef PubMed.
- Q. Zhang, J. Chen, K. Kuwajima, H. M. Zhang, F. Xian, N. L. Young and A. G. Marshall, Sci. Rep., 2013, 3, 1247–1253 CrossRef PubMed.
- C. Pace, D. Laurents and J. Thomson, Biochemistry, 1990, 29, 2564–2572 CrossRef CAS PubMed.
- Q. Zou, S. Habermann-Rottinghaus and K. Murphy, Proteins: Struct., Funct., Genet., 1998, 31, 107–115 CrossRef CAS.
- D. Canchi and A. García, Annu. Rev. Phys. Chem., 2013, 64, 273–294 CrossRef CAS PubMed.
- D. Canchi, P. Jayasimha, D. Rao, G. Makhatadze and A. Garcia, Biophys. J., 2013, 104, 12095–12104 CrossRef.
- B. L. De Groot, G. Vriend and H. J. C. Berendsen, J. Mol. Biol., 1999, 286, 1241–1249 CrossRef CAS PubMed.
- A. M. Roseman, S. Chen, H. White, K. Braig and H. R. Saibil, Cell Press, 1996, 87, 241–251 CAS.
- J. P. Grason, J. S. Gresham, L. Widjaja, S. C. Wehri and G. H. Lorimer, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 17334–17338 CrossRef CAS PubMed.
- J. Wang and D. C. Boisvert, J. Mol. Biol., 2003, 327, 843–855 CrossRef CAS PubMed.
- S. Meena and A. K. Saxena, J. Phys. Chem. Biophys., 2016, 6, 1–6 Search PubMed.
- L. Smeller, Biochim. Biophys. Acta, 2002, 1595, 11–29 CrossRef CAS.
- L. Arns, V. Schuabb, S. Meichsner, M. Berghaus and R. Winter, Z. Phys. Chem., 2018, 232, 615–634 CAS.
- S. R. Al-Ayoubi, P. H. Schummel, M. Golub, J. Peters and R. Winter, Phys. Chem. Chem. Phys., 2017, 19, 14230–14237 RSC.
- P. H. Schummel, M. W. Jaworek, C. Rosin, J. Högg and R. Winter, Phys. Chem. Chem. Phys., 2018, 20, 28400–28411 RSC.
- T. Q. Luong, S. Kapoor and R. Winter, ChemPhysChem, 2015, 16, 3555–3571 CrossRef CAS PubMed.
- P. H. Yancey and J. F. Siebenaller, J. Exp. Biol., 2015, 218, 1880–1896 CrossRef PubMed.
- A. A. Brindley, R. W. Pickersgill, J. C. Partridge, D. J. Dunstan, D. M. Hunt and M. J. Warren, PLoS One, 2008, 3, e2042 CrossRef PubMed.
- R. R. Petrov, B. Rohde, B. Cornell and B. Martinac, Channels, 2012, 6, 262–271 CrossRef PubMed.
- M. Gao, C. Held, S. Patra, L. Arns, G. Sadowski and R. Winter, ChemPhysChem, 2017, 18, 2951–2972 CrossRef CAS PubMed.
- A. Downing, G. Wallace and P. Yancey, Deep Sea Res., Part I, 2018, 138, 1–10 CrossRef CAS.
- T. J. Welch, A. Farewell, F. C. Neidhardt and D. H. Bartlett, J. Bacteriol., 1993, 175, 7170–7177 CrossRef CAS PubMed.
- D. H. Bartlett, Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol., 2002, 1595, 367–381 CrossRef CAS.
- B. Gorovits, C. S. Raman and P. M. Horowitz, J. Biol. Chem., 1995, 270, 2061–2066 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9cp06468k |
| This journal is © the Owner Societies 2020 |