Supramolecular stacking in a high Z′ calix[8]arene–porphyrin assembly†
Jimi M.
Alex
,
Patrick
McArdle
and
Peter B.
Crowley
 *
*
School of Chemistry, National University of Ireland Galway, University Road, Galway, H91 TK33, Ireland. E-mail: peter.crowley@nuigalway.ie; Tel: +353 91 49 24 80
First published on 15th November 2019
Abstract
A co-crystal structure of sulfonato-calix[8]arene (sclx8) and trimethylanilinium-porphyrin (tmap) at 1.0 Å resolution is reported. The oppositely charged macrocycles formed an alternating stacked assembly. Crystal packing was completed by peripheral porphyrins that interlinked neighbouring stacks. Numerous cavities, presented by the calixarenes, were occupied by 2-methyl-2,4-pentanediol (the precipitant) resulting in an unusually high Z′.
Calixarenes1,2 and porphyrins3,4 are versatile supramolecular building blocks with wide-ranging applications in sensing,2 catalysis,5,6 and light harvesting.7–9 The upper and lower rims of calixarenes and the porphyrin ring periphery can be substituted with diverse groups to favour (bio)molecular recognition, and/or to facilitate self-assembly.3,4,10,11 Sheet12 or layered13 calixarene assemblies and ring-14 and pillar-like15 structures of porphyrins have been achieved. Porphyrin–calixarene complexation has yielded hybrid assemblies including porous frameworks.16–21 In these cases, the calixarene appears to be a templating agent for porphyrin assembly. Co-crystallization of N-methyl-4-pyridyl-porphyrin with a calix[4]arene derivative resulted in complexes with varying calixarene
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) porphyrin ratios,16,17 that could be modulated either by pH or metal ions.19–21
porphyrin ratios,16,17 that could be modulated either by pH or metal ions.19–21
Porphyrins and calixarenes are useful reagents for protein recognition and assembly.10,11,22–27 Recently, we have shown that the highly anionic sulfonato-calix[8]arene (sclx8) forms relatively large interfaces (∼550 Å) with cationic proteins.25,27 Highly porous crystalline frameworks have been obtained with cytochrome c (cytc) in which the calixarene plays pivotal roles.25,27 The present study began as an attempt to co-crystallize sclx8 with cytc and a porphyrin. The tetra(4-N,N,N-trimethylanilinium)porphyrin (tmap; Fig. 1) was selected considering the high affinity of calix[n]arenes for methylated amines.11 Currently, there are at least ten X-ray structures of sclx8 with different guests.28–36 Only limited X-ray data, including a remarkable cubic cage, is available for tetra(4-aminophenyl)-porphyrin, the precursor to tmap.37,38 And although calix[4]arene-porphyrin complexes have been explored extensively,16–21 no data is available for calix[8]arene. Co-crystallization experiments yielded an interesting result that contained sclx8 and tmap but no protein. Here, we report a 1.0 Å resolution crystal structure of sclx8–tmap. The porphyrin occurs in two distinct conformations depending on its location within the assembly. The molecular recognition features that appear to stabilize the assembly are discussed.
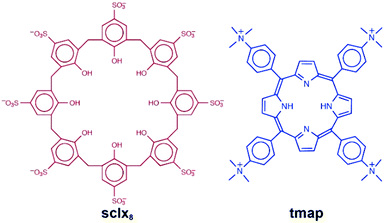 | ||
| Fig. 1 Molecular ions used in this study. Note, at least two of the phenolic groups can be deprotonated. | ||
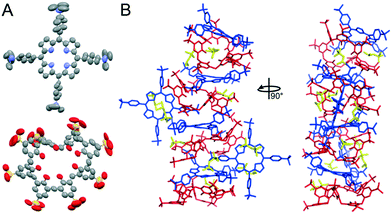
A sparse matrix screen (JCSG++ Biosciences) and an Oryx8 Robot (Douglas Instrument) were used for co-crystallization trials. 1 mM cytc, 10 mM sclx8 and 6.5 mM tmap yielded a single crystal (Fig. S1†) in condition A11 with 50% 2-methyl-2,4-pentanediol (mpd), 0.1 M TRIS-HCl pH 8.5 and 0.2 M ammonium di-hydrogen phosphate (no crystals were obtained in the other 95 conditions). A dataset extending to 1.0 Å resolution was collected at SOLEIL synchrotron. The crystal belonged to the monoclinic space group P21 (Table S1†) and the electron density map was generated ab initio in ACORN (CCP4 suite).39 Unambiguous electron density was evident for the two macrocycles. Interestingly, the asymmetric unit comprised four sclx8, six tmap, ten mpd and two glycerols, resulting in Z′ = 22 (Fig. 2).40,41 The structure was refined using ShelxL42 (version 2018/3) with the crystal data listed in Table S1.† The complex was highly solvated with 145 waters (deposited as CCDC 1956108). However, the R factor was greater than 20% and there were large parameter shifts. Application of the Platon-Squeeze program43 resulted in greatly reduced R factors and parameter shifts (deposited as CCDC 1956128).
Currently, about ten structures with Z′ > 16 are available (http://zprime.co.uk/).40,41 There are 12 structures in the CCDC (Table S2†) with more than 999 atoms, none of which are high Z′ structures. The present structure (without Squeeze – non-H: 1005; H: 1639 and with Squeeze – non-H: 860; H: 1494) provides insight into the conditions/interactions that lead to high Z′. Four tmap alternated with four sclx8 in a stacked assembly (Fig. 2B). The remaining two porphyrins bound the stack peripherally. The stacked porphyrins were puckered4 while the peripheral porphyrins were planar (Fig. 2A and S2†). Using least squares planes defined by the four pyrrole ring atoms plus the bridging carbons the maximum deviation in the stacked and peripheral porphyrins were 0.9 Å and 0.2 Å, respectively (ORTEX module in Oscail44). sclx8 adopted a pleated conformation that consisted of ‘calix[2]arene’ and ‘calix[3]arene’ cavities. These cavities hosted the trimethylanilinium group of the porphyrins, or a mpd or a pyrrole group of the peripheral porphyrin (Fig. 2B). mpd was the precipitant and the 10 mpd molecules in the asymmetric unit are mostly occupying cavities and appear to improve the packing.
Porphyrin–calixarene co-crystals have been characterized previously.16,19–21 These structures comprised porphyrin stacks with peripherally-bound calixarenes. In contrast, in the sclx8–tmap complex both macrocycles participated in the stack and formed multiple noncovalent interactions (Fig. 2). This binding mode appears to have been favoured by the charge and shape complementarity of the trimethylanilinium with the sclx8 conformation. For example, in a sclx8–tmap–sclx8 sandwich the trimethylanilinium groups interacted with the flanking calixarene cavities via charge–charge interactions, cation–π bonds, CH–π and in some cases π–π interactions. Furthermore, the pyrrole rings of each stacking porphyrin formed CH–π bonds with the methylene groups of the flanking calixarenes.
The conformations adopted by the stacking macrocycles appear to have facilitated interactions with the peripheral tmap. A ‘calix[2]arene’ cavity of sclx8 accommodated the pyrrole ring of a peripheral tmap. Interestingly, the trimethylamine group of this porphyrin was inserted close to the (deprotonated) phenolic OH groups, consistent with charge–charge interactions. The peripheral tmap acted as “bridges” between the neighbouring stacks, facilitating crystal packing/assembly (Fig. 3).
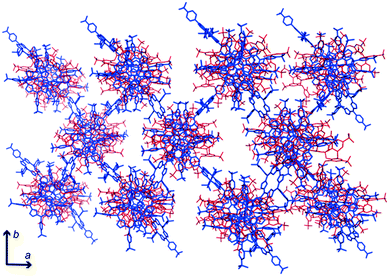 | ||
| Fig. 3 Crystal packing involves sclx8–tmap stacks (top-down view) interacting with each other via bridging peripheral porphyrins. | ||
To determine how well the macrocycles were packed, the number of intermolecular van der Waals contacts were measured in ORTEX (Table 1).44 In the stack, ∼60% of the tmap atoms were in contact with the flanking sclx8. Only ∼35% of the peripheral tmap atoms contacted the calixarenes. Similar results were obtained by measuring the solvent accessible surface areas (Table 1).45 ∼70% of the surface of the stacking porphyrin was buried by the calixarenes, while 20% of the peripheral porphyrin was buried. Protonation of the porphyrin core results in puckering.4 Apparently, the chemical environment afforded by the flanking sclx8 favoured protonation of the stacking porphyrins. In contrast, the more exposed peripheral porphyrins were planar.
| % van der Waal contacts | % buried surface | |||||
|---|---|---|---|---|---|---|
| tmap p | tmap s | sclx 8 | tmap p | tmap s | sclx 8 | |
| pperipheral; sstacking. | ||||||
| tmap p | — | 25 | 35 | — | 15 | 20 |
| tmap s | 25 | — | 60 | 15 | — | 70 |
| sclx 8 | 35 | 55 | — | 20 | 70 | — |
sclx 8 –tmap interactions were investigated in solution by UV/vis spectroscopy (ESI† methods and Fig. S3). In the presence of increasing concentrations of sclx8, the tmap Soret band exhibited a ∼6 nm bathochromic shift (from 412 to 418 nm) as well as gradual hypochromicity followed by gradual hyperchromicity. These spectral changes are consistent with complexation, and suggest the formation of complexes with varying stoichiometry.
In conclusion, we have described an alternating stacked assembly of sclx8 and tmap in the solid state, as well as complexation in solution. Numerous interactions, including charge–charge, CH–π and cation–π, appear to stabilize the complex. The numerous weak intermolecular contacts, the non-planar macrocycle conformations, and the inclusion of crystallization additives (mpd) all contributed to this high Z′ assembly.40,41 The inherent “floppiness” of the calix[8]arene scaffold ensured a suitable conformation that complements the topology and charge of the partner porphyrin. Thus, compared to the rigid calix[4]arenes,11,16,19–21 the conformational flexibility offered by calix[8]arenes may be utilized to yield stacking hybrid architectures. Such complexes, containing porphyrins, may have applications in the development of sensors or light-harvesting systems.2,7–9
This research was funded by NUI Galway (Hardiman Scholarship to JMA) and Science Foundation Ireland (grant 13/CDA/2168 to PBC). We thank SOLEIL synchrotron (France) for beam time allocation and the staff at beamline PROXIMA-2A for their assistance with data collection. We thank A. L. Spek for help with Platon-Squeeze and M. Hoyland for help with IUCr CheckCIF on this large structure.
Conflicts of interest
There are no conflicts to declare.References
- D.-S. Guo and Y. Liu, Acc. Chem. Res., 2014, 47, 1925 CrossRef CAS
.
- R. Kumar, A. Sharma, H. Singh, P. Suating, H. S. Kim, K. Sunwoo, I. Shim, B. C. Gibb and J. S. Kim, Chem. Rev., 2019, 119, 9657 CrossRef CAS PubMed
.
- W. Auwärter, D. Écija, F. Klappenberger and J. V. Barth, Nat. Chem., 2015, 7, 105 CrossRef
.
- M. Kielmann and M. O. Senge, Angew. Chem., Int. Ed., 2019, 58, 418 CrossRef CAS
.
- C. Schöttle, E. Guan, A. Okrut, N. A. Grosso-Giordano, A. Palermo, A. Solovyov, B. C. Gates and A. Katz, J. Am. Chem. Soc., 2019, 141, 4010 CrossRef
.
- P. T. Smith, B. P. Benke, Z. Cao, Y. Kim, E. M. Nichols, K. Kim and C. J. Chang, Angew. Chem., Int. Ed., 2018, 57, 9684 CrossRef CAS
.
- P. Kubát, J. Šebera, S. Záliš, J. Langmaier, M. Fuciman, T. Polívka and K. Lang, Phys. Chem. Chem. Phys., 2011, 13, 6947 RSC
.
- J. Otsuki, J. Mater. Chem. A, 2018, 6, 6710 RSC
.
- P. Pathak, W. Yao, K. D. Hook, R. Vik, F. R. Winnerdy, J. Q. Brown, B. C. Gibb, Z. F. Pursell, A. T. Phan and J. Jayawickramarajah, J. Am. Chem. Soc., 2019, 141, 12582 CrossRef CAS PubMed
.
- H. Zhou, L. Baldini, J. Hong, A. J. Wilson and A. D. Hamilton, J. Am. Chem. Soc., 2006, 128, 2421 CrossRef CAS
.
-
(a) M. Giuliani, I. Morbioli, F. Sansone and A. Casnati, Chem. Commun., 2015, 51, 14140 RSC
; (b) C. S. Beshara, C. E. Jones, K. D. Daze, B. J. Lilgert and F. Hof, ChemBioChem, 2010, 11, 63 CrossRef CAS
.
- D. Shetty, T. Skorjanc, J. Raya, S. K. Sharma, I. Jahovic, K. Polychronopoulou, Z. Asfari, D. S. Han, S. Dewage, J.-C. Olsen, R. Jagannathan, S. Kirmizialtin and A. Trabolsi, ACS Appl. Mater. Interfaces, 2018, 23, 17359 CrossRef
.
- M. Moradi, N. L. Opara, L. G. Tulli, C. Wäckerlin, S. J. Dalgarno, S. J. Teat, M. Baljozovic, O. Popova, E. van Genderen, A. Kleibert, H. Stahlberg, J. P. Abrahams, C. Padeste, P. F.-X. Corvini, T. A. Jung and P. Shahgaldian, Sci. Adv., 2019, 5, e4489 CrossRef
.
- R. Haver, L. Tejerina, H.-W. Jiang, M. Rickhaus, M. Jirasek, I. Grübner, H. J. Eggimann, L. M. Herz and H. L. Anderson, J. Am. Chem. Soc., 2019, 141, 7965 CrossRef CAS
.
- P. Martinez-Bulit, C. A. O'Keefe, K. Zhu, R. W. Schurko and S. J. Loeb, Cryst. Growth Des., 2019, 19, 5679 CrossRef CAS
.
- L. Di Costanzo, S. Geremia, L. Randaccio, R. Purrello, R. Lauceri, D. Sciotto, F. G. Gulino and V. Pavone, Angew. Chem., Int. Ed., 2001, 40, 4245 CrossRef CAS
.
- G. Moschetto, R. Lauceri, F. G. Gulino, D. Sciotto and R. Purrello, J. Am. Chem. Soc., 2002, 124, 14536 CrossRef CAS PubMed
.
- F. G. Gulino, R. Lauceri, L. Frish, T. Evan-Salem, Y. Cohen, R. De Zorzi, S. Geremia, L. Di Costanzo, L. Randaccio, D. Sciotto and R. Purrello, Chem. – Eur. J., 2006, 12, 2722 CrossRef CAS
.
- R. De Zorzi, N. Guidolin, L. Randaccio, R. Purrello and S. Geremia, J. Am. Chem. Soc., 2009, 131, 2487 CrossRef CAS PubMed
.
- R. De Zorzi, N. Guidolin, L. Randaccio and S. Geremia, CrystEngComm, 2010, 12, 4056 RSC
.
- G. Brancatelli, R. De Zorzi, N. Hickey, P. Siega, G. Zingone and S. Geremia, Cryst. Growth Des., 2012, 12, 5111 CrossRef CAS
.
- M. Goel, R. S. Damai, D. K. Sethi, K. J. Kaur, B. G. Maiya, M. J. Swamy and D. M. Salunke, Biochemistry, 2005, 44, 5588 CrossRef CAS
.
- P. B. Crowley, P. Ganji and H. Ibrahim, ChemBioChem, 2008, 9, 1029 CrossRef CAS
.
- O. Kokhan, N. Ponomarenko, P. R. Pokkuluri, M. Schiffer and D. M. Tiede, Biochemistry, 2014, 53, 5070 CrossRef CAS
.
- M. L. Rennie, G. C. Fox, J. Pérez and P. B. Crowley, Angew. Chem., Int. Ed., 2018, 57, 13764 CrossRef CAS PubMed
.
- J. M. Alex, M. L. Rennie, S. Engilberge, G. Lehoczki, H. Dorottya, Á. Fizil, G. Batta and P. B. Crowley, IUCrJ, 2019, 6, 238–247 CrossRef CAS PubMed
.
- S. Engilberge, M. L. Rennie, E. Dumont and P. B. Crowley, ACS Nano, 2019, 13, 10343 CrossRef CAS
.
- S. J. Dalgarno, M. J. Hardie, J. L. Atwood, J. E. Warren and C. L. Raston, New J. Chem., 2005, 29, 649 RSC
.
- F. Perret, V. Bonnard, O. Danylyuk, K. Suwinska and A. W. Coleman, New J. Chem., 2006, 30, 987 RSC
.
- C. B. Smith, L. J. Barbour, M. Makha, C. L. Raston and A. N. Sobolev, New J. Chem., 2006, 30, 991 RSC
.
- O. Danylyuk, F. Perret, A. W. Coleman and K. Suwinska, Open Crystallogr. J., 2008, 1, 18 CrossRef CAS
.
- W. He, Y. Bi, W. Liao and D. Li, J. Mol. Struct., 2009, 937, 95–99 CrossRef CAS
.
- Y. Liu, W. Liao, Y. Bi, M. Wang, Z. Wu, X. Wang, Z. Sub and H. Zhang, CrystEngComm, 2009, 11, 1803 RSC
.
- B. Leśniewska, F. Perret, K. Suwińska and A. W. Coleman, CrystEngComm, 2014, 16, 4399 RSC
.
- B. Leśniewska, A. W. Coleman, Y. Tauran, F. Perret and K. Suwińska, CrystEngComm, 2016, 18, 8858 RSC
.
- O. Danylyuka, H. Butkiewicza, A. W. Coleman and K. Suwińska, J. Mol. Struct., 2017, 1150, 28 CrossRef
.
- W. Meng, B. Breiner, K. Rissanen, J. D. Thoburn, J. K. Clegg and J. R. Nitschke, Angew. Chem., Int. Ed., 2011, 50, 3479 CrossRef CAS PubMed
.
- Y. Park, J. Y. Koo and H. C. Choi, Cryst. Growth Des., 2018, 18, 7239 CrossRef CAS
.
- E. J. Dodson and M. M. Woolfson, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2009, 65, 881 CrossRef CAS PubMed
.
- K. M. Steed and J. W. Steed, Chem. Rev., 2015, 115, 2895 CrossRef CAS PubMed
.
- W. Clegg, Acta Crystallogr., Sect. C: Struct. Chem., 2019, 75, 833 CrossRef CAS PubMed
.
- G. M. Sheldrick, Acta Crystallogr., Sect. C: Struct. Chem., 2015, 71, 13 Search PubMed
.
- A. L. Spek, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2009, 65, 148 CrossRef CAS PubMed
.
- P. McArdle, J. Appl. Crystallogr., 2017, 50, 320 CrossRef CAS
.
- J. Ribeiro, C. Ríos-Vera, F. Melo and A. Schüller, Bioinformatics, 2019, 35, 3499 CrossRef PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available: Methods, figures and X-ray statistics table. CCDC 1956108 and 1956128. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c9ce01646e |
| This journal is © The Royal Society of Chemistry 2020 |