Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Probing self-regeneration of essential protein factors required for in vitro translation activity by serial transfer†
Kai
Libicher
a and
Hannes
Mutschler
 *ab
*ab
aBiomimetic Systems, Max Planck Institute of Biochemistry, Am Klopferspitz 18, 82152 Martinsried, Germany
bFaculty of Chemistry and Chemical Biology, TU Dortmund University, Otto-Hahn-Str. 4a, 44227 Dortmund, Germany. E-mail: hannes.mutschler@tu-dortmund.de
First published on 19th November 2020
Abstract
The bottom-up construction of bio-inspired systems capable of self-maintenance and reproduction is a central goal in systems chemistry and synthetic biology. A particular challenge in such systems is the continuous regeneration of key proteins required for macromolecular synthesis. Here, we probe self-maintenance of a reconstituted in vitro translation system challenged by serial transfer of selected key proteins. We find that the system can simultaneously regenerate multiple essential polypeptides, which then contribute to the maintenance of protein expression after serial transfer. The presented strategy offers a robust methodology for probing and optimizing continuous self-regeneration of proteins in cell-free environments.
The field of bottom-up synthetic biology aims to create lifelike systems from inanimate building blocks.1–4 A major feature of living matter is its ability to undergo self-maintenance and growth by continuous reproduction of their key building blocks. In biomimetic systems based on DNA, RNA, and proteins, this could be achieved with self-maintaining in vitro translation systems in which both polynucleotides and polypeptides of the system are continuously regenerated5,6
We have recently reported the self-replication of large DNA molecules encoding most of the non-ribosomal translation factors (TFs) of the PURE (protein synthesis using recombinant elements) system7 using in vitro transcription-translation (IVTT) coupled DNA replication (TTcDR).8 During TTcDR, a considerable fraction of these genes was expressed in amounts equal or exceeding their respective input levels. However, whether the expressed proteins were functional remained unclear.
A direct method to mimic continuous growth and/or degradation in self-regenerating systems far from thermodynamic equilibrium are serial transfer (ST) experiments.9,10 Here, a fraction of an autocatalytic reaction is recursively transferred into new solutions with fresh substrate molecules, which are consumed during autocatalytic regeneration. ST has been successfully used to study self-replication and evolution of nucleic acid-based replicators11–14 including IVTT-based systems.15–17 In contrast, the corresponding autocatalytic regeneration of essential PURE proteins has remained largely unexplored. Here, we sought to explore if PURE systems can successfully regenerate a subset of its essential protein components during ST to an extent that IVTT activity is maintained throughout the process.
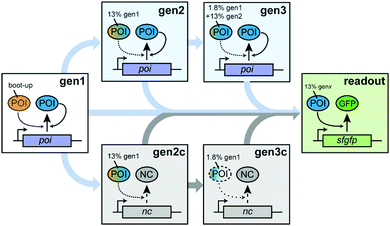
To probe autocatalytic maintenance of specific proteins of interest (POIs) that are essential for IVTT activity, we established a ST protocol based on custom PURE systems (Scheme 1). In the first step, a PURE master mix containing all components required for IVTT initiated synthesis of a specific essential POI from a plasmid template. After incubation for 1 h at 37 °C, a fraction of this seed “generation” (gen1) was transferred to a fresh POI-depleted PURE mastermix (PUREΔPOI) programmed with a POI expression plasmid (gen2). After a further 1 h incubation of gen2, a second transfer of gen2 into a PUREΔPOI mastermix containing the POI-plasmid yielded gen3. We anticipated that both the dilution from gen1 to gen2 (∼13% of remaining input POI) and in particular from gen2 to gen3 (∼1.8% of remaining input POI) would severely reduce IVTT activity in the absence of sufficient POI in situ regeneration and that three generations of ST would be sufficient to address potential POI maintenance in PUREΔPOI systems. To confirm the expected loss in IVTT activity in non-regenerating PURE systems and the maintenance in regenerative systems, we carried out additional control experiments in which the STs were carried out as described above. However, the key difference was that PUREΔPOI samples at gen2 and gen3 were mixed with non-POI plasmids (control generations; gen2c, gen3c) whose expression imposed a similar synthetic burden on the PURE system.
First, we tested the ability of the customized PURE systems to individually regenerate the three major PURE enzymes T7-RNA polymerase (T7-RNAP), adenylate kinase (AK) and nucleoside diphosphate kinase (NDK) during ST. While T7 RNAP is required to transcribe the plasmid-encoded genes before translation, NDK is the key enzyme for regeneration of NTP pools from ATP. AK is a component of the PURE energy regeneration system and is vital for the regeneration of AMP produced during tRNA aminoacylation by aminoacyl-tRNA synthetases.18 Direct regeneration of full-length POI IVTT in PUREΔPOI samples programmed with POI-expression plasmids was monitored by in-gel imaging of samples from ST experiments that were carried out in the presence of BODIPY FL-lysyl-tRNAlys (FluoroTec GreenLys in vitro Translation Labeling System). As expected, we observed IVTT of full-length POIs in gen1 in the presence of the encoding plasmids pT7 (expressing T7-RNAP), pAK1 (expressing AK), and pNDK (expressing NDK) (Fig. 1). Intriguingly, we also observed enhanced de novo synthesis of all three POIs after ST into gen2 and gen3. For T7-RNAP, gen2 contained about 70% of the initial gen1 levels followed only by a minor drop to ∼60% in gen3. In contrast, ST of PUREΔT7-RNAP reactions in absence of pT7 led to a severe loss of BODIPY-Lys labelled T7-RNAP at gen2c (∼30%) and gen3c (∼10%), suggesting that the remaining levels of T7-RNAP from gen1 were insufficient for T7-RNAP self-regeneration above background levels. For NDK, apparent self-regeneration was even more pronounced with stable levels of labelled protein throughout gen1 to gen3, whereas serial dilutions of NDK gen1 into PUREΔNDK samples for which pDNK had been replaced with pAK1 led to a drastic gradual decrease of BODIPY-Lys-labelled NDK from ∼50% in gen2c to ∼10% in gen3c. Finally, for AK, ST of gen1 into PUREΔAK containing pAK1 caused stable, though slowly decreasing, levels of labelled AK with ∼80% of gen1 levels at gen2 and 50% at gen3. As for the other POIs, labelled AK levels reduced drastically in the absence of pAK1 (∼30% at gen2c and ∼5% at gen3c). In conclusion, all three tested POIs were expressed in PUREΔPOI samples containing the respective POI expression plasmid throughout two STs (Table S5, ESI†). As expected, POI synthesis rapidly decreased after gen1 upon the respective POI expression plasmid being replaced with a control plasmid (Fig. 1).
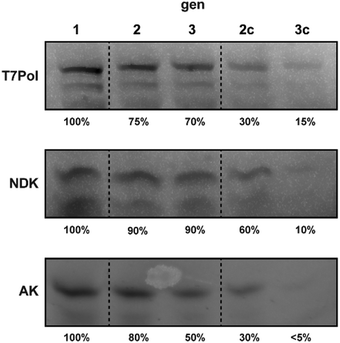 | ||
| Fig. 1 Gel images of PURE reactions after in situ fluorescence labelling of regenerated T7Pol, NDK, and AK using BODIPY FL-lysyl-tRNAlys. Approximate percentages of the main POI band (Fig. S1, ESI†) in relation to gen1 (excluding truncated products) are indicated. | ||
While in-gel expression levels gave an initial estimate of POI regeneration through sustained IVTT, it provided little information regarding to which extent the newly produced POI could contribute to the maintenance of overall IVTT activity in POI-depleted PURE systems. Therefore, we probed the general IVTT activity of each generation by diluting ∼13% of the reaction after 1 h POI regeneration into a PUREΔPOI reaction containing an expression plasmid for superfolder green fluorescent protein (sfGFP)19 (Scheme 1, green box). We anticipated that, in case a generation had produced sufficient amounts of active POI, IVTT should produce similar amounts of sfGFP in gen1, gen2, and gen3. In contrast, the absence of active POI regeneration should largely prevent sfGFP synthesis.
Indeed, while sfGFP expression during active T7-RNAP regeneration remained close to constant from gen1 to gen3, the control transfer gen2c and gen3c showed a massive decrease in sfGFP yields (Fig. 2a). Thus, the in situ produced T7-RNAP was active and the yield was sufficient to maintain IVTT activity throughout both STs. The residual sfGFP expression observed in gen3c suggested that the remaining T7-RNAP from the initial gen1 PURE mix, the de novo synthesised T7-RNAP in the same generation, the remaining pT7 plasmid, as well as T7-RNAP mRNA might allow very low basal levels of background transcription for at least two STs into samples containing PUREΔT7-RNA.
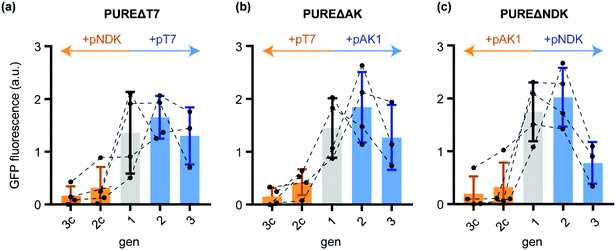 | ||
| Fig. 2 Normalised sfGFP fluorescence endpoints (100 min IVTT, 37 °C) after diluting different generations of the ST protocol (Scheme 1) into PUREΔPOI programmed with a sfGFP expression plasmid. (a) sfGFP IVTT performance of different PUREΔT7 generations starting from gen1 (grey) and further dilutions in presence of either pT7 (blue) or the control plasmid pNDK (orange). (b) sfGFP IVTT performance of PUREΔAK generations. After gen1 (grey), reactions were subsequently diluted either in the presence of pAK1 (blue) or the control plasmid pT7 (orange). (c) For PUREΔAK, dilutions were carried out either in the presence of pNDK (blue) or pAK1 (orange). Error bars show standard deviations. | ||
Similar evidence for sustained regeneration of active POI were obtained for gen1 to gen3 for AK and NDK (Fig. 2b and c). For the latter, sfGFP expression in gen3 was decreased compared to gen1 indicating that not all of the regenerated protein was functionally active. In all cases, sfGFP expression in the control reactions expressing non-depleted proteins (Table S5, ESI†) collapsed to minimal sfGFP expression levels after gen1. We could confirm these results by in-gel imaging of sfGFP expression (Fig. S2, ESI†). In conclusion, in situ regeneration of individually depleted PURE proteins enables stable maintenance during ST experiments.
Having shown that custom PURE systems can regenerate missing POIs upon depletion by ST, we sought to probe whether regeneration and maintenance of IVTT activity is also possible if multiple TFs require regeneration during gen2 and gen3. To this end, we explored the parallel regeneration of twelve essential tRNA synthetases and release factor 1 (RF1). All of these proteins are encoded on the large plasmid pLD1, which was designed to facilitate in-house production of PURE systems.20 We were recently able to show that expression of all thirteen pLD1-encoded TFs can be achieved in non-depleted PURE systems compatible with DNA replication.8 To probe if the in situ synthesised TFs can feed back into IVTT activity and, thus, contribute to a partial self-regeneration in the semi-autonomous PURE system, we challenged a PURE system depleted from all thirteen pLD1-encoded TFs (PUREΔLD1) through ST. Using BODIPY-Lys-tRNA labelling, we found evidence for stable expression of the TFs during gen1, gen2, and gen3 (Fig. 3a and Table S6, ESI†). In contrast, TF expression rapidly ceased in gen2c and gen3c when the control plasmid pLD2 was used during ST instead of pLD1. pLD2 encodes a subset of 9 PURE TFs, which are, however, present in PUREΔLD1.20
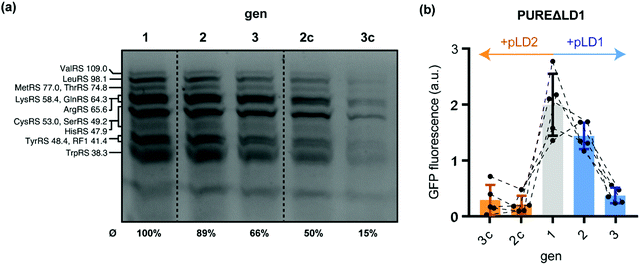 | ||
| Fig. 3 Regeneration of pLD1-encoded translation factors during serial dilution of PUREΔLD1. (a) Fluorescent gel image of PURE reactions after in situ fluorescence labelling of 13 regenerated pLD1 proteins using BODIPY FL-lysyl-tRNAlys. Annotated bands are based on previous annotations, which were confirmed by mass spectrometry.8,20 Average expression yields of pLD1 proteins relative to gen1 were estimated by fluorescence densitometry of the seven annotated bands. The relative yields of each assigned band are shown in Table S6 (ESI†). (b) sfGFP IVTT performance of PUREΔLD1 generations. Following gen1 (grey), reactions were subsequently diluted either in the presence of pLD1 (blue) or the control plasmid pLD2 (orange) that encode different subsets of PURE translation factors.20 | ||
Next, we tested the performance of the regenerated pLD1-encoded TFs using the sfGFP IVTT assay. Intriguingly, we observed gen2 sfGFP expression levels in PUREΔLD1 samples similar to gen1 (Fig. 3b). This implied that sufficient amounts of TFs were regenerated to maintain overall IVTT activity. In contrast, sfGFP expression in gen2c was barely detectable, suggesting that the transfer from gen1 was not sufficient to maintain IVTT activity and that further POI re-synthesis was required. While enough TFs were produced for sustained IVTT activity in gen2 and the additional dilution into the sfGFP reporter mixture, the same sfGFP experiments proved to be overly challenging for the PUREΔLD1 system at gen3 under the current conditions. Here, sfGFP expression levels dropped sharply by about 75% compared to gen2. This loss in IVTT activity suggests that the production of all or some of the thirteen essential pLD1 proteins was not sufficient to provide the concentrations of active enzymes required to enable IVTT after the further ∼1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ST of gen3 into the sfGFP reporter reaction (Scheme 1). One possible interpretation for this loss in activity is that the levels of some pLD1 proteins may be inefficiently regenerated throughout repeated passages of ST. This would be in agreement with the previous observation that pLD1-encoded Gln-tRNA synthetase or release factor 1 (RF1) are expressed at levels below their initial input amounts.8 Furthermore, some of the regenerated pLD1 proteins may have folding defects e.g. due to the absence of chaperones, which would lower the effective regeneration efficiency of the active protein fraction.21,22
10 ST of gen3 into the sfGFP reporter reaction (Scheme 1). One possible interpretation for this loss in activity is that the levels of some pLD1 proteins may be inefficiently regenerated throughout repeated passages of ST. This would be in agreement with the previous observation that pLD1-encoded Gln-tRNA synthetase or release factor 1 (RF1) are expressed at levels below their initial input amounts.8 Furthermore, some of the regenerated pLD1 proteins may have folding defects e.g. due to the absence of chaperones, which would lower the effective regeneration efficiency of the active protein fraction.21,22
In summary, we have shown that a straightforward and easily implemented ST protocol can be used to study the self-coded regeneration of essential PURE proteins. This is an important step towards the creation, prototyping, and evolution of self-regenerating in vitro translation systems. Furthermore, the in situ production of individual proteins essential for IVTT activity could enable the development of new cell-free strategies for protein evolution or library screening of interesting target genes such as orthogonal aminoacyl tRNA synthetases, metabolic enzymes or nucleic acid polymerases.
The parallel regeneration of several proteins over a longer period is still in need of optimization; for example, by adjusting the expression strength of the individual co-factors. Future work will reveal whether the regenerative potential of PURE can be maintained continuously over a larger number of generations. Indeed, during the preparation of this manuscript, the continuous simultaneous regeneration of up to seven tRNA synthetases in a microchemostatic setup was reported.23 The same work also showed that fine-tuning the amounts of added DNA template is a critical factor to achieve robust protein regeneration of multiple proteins in a continuous flow system. Similar optimizations as well as the addition of chaperone systems21 might also improve protein regeneration in discontinuous ST-based experiments that aim to increase the autonomy of PURE systems.
This work was supported by the MaxSynBio consortium, which is jointly funded by the Federal Ministry of Education and Research of Germany (FKZ 031A359K) and the Max Planck Society. We thank S. Şahsuvar for her contributions to the purification of pLD1 protein fraction and E.Y. Song for feedback on the manuscript.
Conflicts of interest
There are no conflicts to declare.References
- V. Noireaux, Y. T. Maeda and A. Libchaber, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 3473–3480 CrossRef CAS.
- J. W. Szostak, D. P. Bartel and P. L. Luisi, Nature, 2001, 409, 387–390 CrossRef CAS.
- P. L. Luisi, Anat. Rec., 2002, 268, 208–214 CrossRef CAS.
- P. Schwille, J. Spatz, K. Landfester, E. Bodenschatz, S. Herminghaus, V. Sourjik, T. J. Erb, P. Bastiaens, R. Lipowsky, A. Hyman, P. Dabrock, J.-C. Baret, T. Vidakovic-Koch, P. Bieling, R. Dimova, H. Mutschler, T. Robinson, T.-Y. D. Tang, S. Wegner and K. Sundmacher, Angew. Chem., Int. Ed., 2018, 57, 13382–13392 CrossRef CAS.
- M. C. Jewett and A. C. Forster, Curr. Opin. Biotechnol., 2010, 21, 697–703 CrossRef CAS.
- K. Le Vay, L. I. Weise, K. Libicher, J. Mascarenhas and H. Mutschler, Adv. Biosyst., 2019, 3, 1800313 CrossRef.
- Y. Shimizu, A. Inoue, Y. Tomari, T. Suzuki, T. Yokogawa, K. Nishikawa and T. Ueda, Nat. Biotechnol., 2001, 19, 751–755 CrossRef CAS.
- K. Libicher, R. Hornberger, M. Heymann and H. Mutschler, Nat. Commun., 2020, 11, 904 CrossRef CAS.
- P. Adamski, M. Eleveld, A. Sood, Á. Kun, A. Szilágyi, T. Czárán, E. Szathmáry and S. Otto, Nat. Rev. Chem., 2020, 4, 386–403 CrossRef.
- W. Hordijk, N. Vaidya and N. Lehman, J. Syst. Chem., 2014, 5, 4 CrossRef.
- D. R. Mills, R. L. Peterson and S. Spiegelman, Proc. Natl. Acad. Sci. U. S. A., 1967, 58, 217–224 CrossRef CAS.
- N. Vaidya, M. L. Manapat, I. A. Chen, R. Xulvi-Brunet, E. J. Hayden and N. Lehman, Nature, 2012, 491, 72–77 CrossRef CAS.
- T. A. Lincoln and G. F. Joyce, Science, 2009, 323, 1229–1232 CrossRef CAS.
- E. Edeleva, A. Salditt, J. Stamp, P. Schwintek, J. Boekhoven and D. Braun, Chem. Sci., 2019, 10, 5807–5814 RSC.
- N. Ichihashi, K. Usui, Y. Kazuta, T. Sunami, T. Matsuura and T. Yomo, Nat. Commun., 2013, 4, 2494 CrossRef.
- R. Mizuuchi and N. Ichihashi, Nat. Ecol. Evol., 2018, 2, 1654–1660 CrossRef.
- Y. Sakatani, R. Mizuuchi and N. Ichihashi, Protein Eng., Des. Sel., 2019, 32, 481–487 CrossRef CAS.
- J. Swartz, Nat. Biotechnol., 2001, 19, 732–733 CrossRef CAS.
- J. D. Pédelacq, S. Cabantous, T. Tran, T. C. Terwilliger and G. S. Waldo, Nat. Biotechnol., 2006, 24, 79–88 CrossRef.
- T. R. Shepherd, L. Du, J. Liljeruhm, Samudyata, J. Wang, M. O. D. Sjödin, M. Wetterhall, T. Yomo and A. C. Forster, Nucleic Acids Res., 2017, 45, 10895–10905 CrossRef CAS.
- T. Niwa, T. Kanamori, T. Ueda and H. Taguchi, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 8937–8942 CrossRef CAS.
- T. Awai, N. Ichihashi and T. Yomo, Biochem. Biophys. Rep., 2015, 3, 140–143 Search PubMed.
- B. Lavickova, N. Laohakunakorn and S. J. Maerkl, Biorxiv DOI:10.1101/2020.07.03.185900.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0cc06515c |
| This journal is © The Royal Society of Chemistry 2020 |