Open Access Article
Open Access ArticleTransfecting tissue models with CRISPR/Cas9 plasmid DNA using peptide dendrimers†
Susanna J.
Zamolo
 ,
Tamis
Darbre
,
Tamis
Darbre
 * and
Jean-Louis
Reymond
* and
Jean-Louis
Reymond
 *
*
Department of Chemistry and Biochemistry, University of Bern, Freiestrasse 3, 3012 Bern, Switzerland. E-mail: jean-louis.reymond@dcb.unibe.ch
First published on 31st August 2020
Abstract
There is currently a lack of efficient reagents to transfect cells with large plasmid DNA, which would be enabling tools for gene editing using CRISPR/Cas9 technology. Herein, we report the discovery of peptide dendrimer Z22 as a non-viral vector for transfecting large CRISPR/Cas9 pDNA into 3D-tumor spheroids with exceptionally high efficiency, low cytotoxicity and low immunogenicity.
Gene editing using clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) has revolutionized biology and provides fascinating opportunities as a therapeutic tool.1–3 One of the key challenges in this technology is to deliver CRISPR/Cas9 plasmid DNA (pDNA) into the target cells because the plasmid is relatively large, limiting the use of viral vectors such as adeno-associated virus (AAV) due to their low encapsulation capacity.4,5 Non-viral vectors, typically consisting of cationic lipids or polycationic polymers, should be better suited to deliver CRISPR/Cas9 pDNA because they can accept larger payloads, while having generally a better safety profile in terms of cytotoxicity and immunogenicity and being easier to manufacture than viral vectors.6 Unfortunately non-viral vectors often have limited tissue penetration,7,8 and perform only very poorly with pDNA, for example requiring the use of detached cells by transfection or electroporation when editing organoids.9,10 Here we focused on transfecting 3D-tumor spheroids, which present several unique features found in tissues, such as chemical gradients of oxygen and nutrients at diameters starting from 200 μm and the production of an extracellular matrix as a structural and biochemical support.11 Transfecting such 3D-tumor spheroids is very difficult and has so far only been achieved with very low efficiency using transfection reagents based on poly(β-amino ester) and glycogen nanoparticles.12,13
We aimed for a peptide dendrimer14 as a non-viral vector because dendrimers15–17 and peptide-type oligomers18–20 are both known to be favorable for this task. Specifically, we set out to obtain a peptide dendrimer for pDNA transfection by combining a hydrophobic core similar to the core of peptide dendrimers DMH13 and DMH18, which we recently reported as single component transfection reagents for siRNA,21,22 with the branches of peptide dendrimer G123KL acting as a co-transfection reagent with lipofectin for DNA,23 siRNA24,25 and oligonucleotides.26
We synthesized peptide dendrimers by standard Fmoc solid-phase peptide synthesis (SPPS, Fig. 1 and Table 1). We tested pDNA transfection on HEK293 and HeLa cells using a commercial 9 Kbp “all-in-one” pDNA vector expressing CRISPR/Cas9 and GFP linked by a self-cleavable 2A peptide,27 together with a gRNA sequence targeting β-glucuronidase. In this assay DMH13, DMH18 and G123KL/lipofectin were all less active than the reference reagent lipofectamine L2000.
| No. | Sequencea | Yieldb mg (%) | MSc calc./obs. | % GFP pos. HEK cellsd | % GFP pos. HeLa cellsd | % free DNAe | % cytotox. HEK cellsg | % viability HEK cellsg |
|---|---|---|---|---|---|---|---|---|
| a One-letter code amino acids are used, K is the branching lysine residue, C-termini are carboxamide CONH2, and all N-termini are free. Alkyl chains in the structure are represented by “C” followed by their number of carbon atoms. b Isolated yields as trifluoroacetate salt after preparative HPLC purification. c ESI-MS, see also the ESI. d Transfection efficiency in HEK and HeLa cells measured as the percentage of GFP positive cells after 48 h of transfection. Experiments were carried out in triplicate in three independent experiments (N/P 5, 175 pmol of peptide dendrimers in 100 μL Optimem per well, 1.7 μM, 250 ng pDNA). e Fluorescence from intercalation in the pDNA/peptide dendrimer complex at N/P 5 (200 μL, final concentration of 0.085 nM pDNA and 0.35 μM or 1 μg mL−1 L2000) using the Quant-It PicoGreen assay and normalized to the value of pDNA alone. f Previously published peptide dendrimer as a co-transfection reagent with lipofectin displaying good pDNA transfection efficiency. g Viability and cytotoxicity assays were performed after 4 h of incubation of HEK cells with peptide dendrimers/pDNA complexes at N/P 5. Cytotoxicity was then determined using the Pierce LDH Cytotoxicity Assay Kit according to the manufacturer's instructions, and cell viability was assessed using the PrestoBlue reagent. | ||||||||
| L2000 | — | — | — | 44.5 ± 6.2 | 34.2 ± 10.0 | 21.7 ± 1.9 | 6.6 ± 8.5 | 85 ± 6.4 |
| DMH13 | (kl)8(kkl)4(kll)2k(C16)k(C16) | 128.4 (7) | 4992.83/4992.84 | 28.9 ± 2.1 | 2.6 ± 2.4 | 13.1 ± 2.2 | 0.9 ± 1.3 | 96 ± 5.6 |
| DMH18 | (kl)8(kkl)4(kll)2kllll | 73.6 (11) | 4712.51/4712.52 | 5.8 ± 2.5 | 0.6 ± 0.3 | 11.7 ± 2.3 | 1.8 ± 1.1 | 91 ± 13 |
| G123KL | (KL)8(KKL)4(KKL)2KGSCf | 7.23 (9) | 4539.0/4540.1 | 8.0 ± 2.7 | 4.0 ± 1.9 | 34.8 ± 6.2 | 2.3 ± 1.3 | 52 ± 8.6 |
| (1) Optimization of the lipid core: | ||||||||
| Z1 | (KL)8(KKL)4(KKL)2KK(C18) | 8.8 (2) | 4684.55/4684.57 | 10.7 ± 3.2 | 2.4 ± 2.2 | 7.3 ± 1.7 | 11.3 ± 1.1 | 64 ± 7.0 |
| Z2 | (KL)8(KKL)4(KKL)2KK(C18)K(C18) | 1.1 (1) | 5078.91/5078.92 | 11.7 ± 3.6 | 2.0 ± 1.4 | 12.4 ± 2.8 | 2.3 ± 0.6 | 79 ± 11 |
| (2) Cysteine insertion | ||||||||
| Z12 | (KL)8(KKL)4(KKL)2KK(C18)C | 1.5 (2) | 4787.56/4787.57 | 12.0 ± 3.0 | 3.8 ± 2.5 | 5.4 ± 1.5 | 3.8 ± 0.7 | 80 ± 5.3 |
| Z13 | (KL)8(KKL)4(KKL)2KK(C18)K(C18)C | 4.4 (3) | 5181.92/5181.93 | 12.8 ± 2.1 | 1.4 ± 0.5 | 7.5 ± 6.9 | 2.9 ± 0.9 | 60 ± 10 |
| Z14 | (KL)8(KKL)4(KKL)2KK(C18)S | 3.6 (3) | 4771.59/4771.60 | 7.4 ± 1.6 | 2.1 ± 1.9 | 6.7 ± 3.5 | 4.9 ± 0.9 | 62 ± 6.5 |
| Z15 | (KL)8(KKL)4(KKL)2KK(C18)A | 8.3 (5) | 4755.59/4755.60 | 10.1 ± 2.4 | 1.0 ± 0.9 | 5.8 ± 5.0 | 3.0 ± 0.2 | 71 ± 6.9 |
| Z16 | (KL)8(KKL)4(KKL)2KK(C18)CC | 5.1 (3) | 4890.57/4890.58 | 9.5 ± 2.6 | 1.7 ± 1.0 | 7.1 ± 1.3 | 1.1 ± 0.7 | 62 ± 11 |
| Z17 | ((KL)8(KKL)4(KKL)2KK(C18)C)2 | 1 (1) | 9573.11/9573.10 | 15.0 ± 5.7 | 3.5 ± 1.0 | 11.7 ± 8.6 | 3.7 ± 0.8 | 106 ± 12 |
| (3) Stereochemistry and cationic residues | ||||||||
| Z18 | (kl)8(KKL)4(KKL)2KK(C18)C | 5.3 (5) | 4787.56/4787.57 | 16.7 ± 3.9 | 2.9 ± 1.1 | 4.2 ± 2.7 | 1.7 ± 0.7 | 70 ± 7.0 |
| Z19 | ((kl)8(KKL)4(KKL)2KK(C18)C)2 | 1.5 (5) | 9573.11/9574.13 | 15.4 ± 5.6 | 4.2 ± 2.0 | 13.7 ± 2.1 | 3.3 ± 0.7 | 62 ± 5.6 |
| Z20 | (kl)8(kkl)4(kkl)2kk(C18)c | 3.1 (5) | 4787.56/4787.57 | 51.6 ± 12.5 | 34.2 ± 13.1 | 10.6 ± 2.1 | 3.7 ± 1.4 | 85 ± 8.7 |
| Z21 | ((kl)8(kkl)4(kkl)2kk(C18)c)2 | 3.2 (5) | 9573.11/9573.13 | 55.0 ± 10.6 | 37.2 ± 12.6 | 10.1 ± 4.0 | 5.0 ± 2.8 | 63 ± 9.6 |
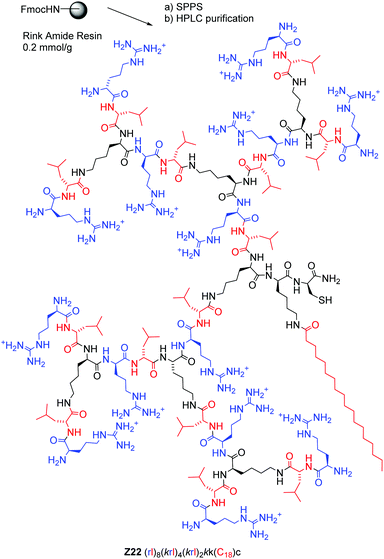
| Z22 | (rl)8(krl)4(krl)2kk(C18)c | 7.8 (4) | 5179.65/5179.66 | 62.5 ± 9.5 | 47.3 ± 9.3 | 6.5 ± 1.8 | 4.7 ± 2.5 | 83 ± 8.5 |
An initial series of dendrimers showed a significant transfection level when using one (Z1) or two (Z2) side-chain stearoylated (C18) lysines as the dendrimer core, while dendrimers with other lipidations or buffering histidines at the core or in the branches were less active or totally inactive (Z3–Z11, Table S1, ESI†). Transfection improved slightly by adding a core cysteine residue to Z1 (→Z12) or Z2 (→Z13), but not when adding a serine (→Z14) or alanine (→Z15). Furthermore, while inserting a second cysteine reduced transfection (Z12 → Z16), the disulfide bridged dimer Z17 was slightly more active than Z12. Following up on both of these compounds, we obtained a further increase in pDNA transfection efficiency by using D-enantiomeric residues in the third generation dipeptide branches (→Z18, Z19). The increase was particularly large with the full D-enantiomers (→Z20, Z21), which both surpassed L2000 in transfection efficiency. Finally, the exhaustive replacement of lysine residues with arginines in Z20 provided Z22, which transfected up to 62.5% of HEK293 cells and 47.3% of HeLa cells with the CRISP/Cas9 pDNA vector.
All dendrimers bound pDNA slightly better than L2000 or G123KL/lipofectin as measured using the picoGreen assay (Table 1).28 However, there was no correlation between pDNA binding and transfection efficiency, which is not surprising considering that the percentage of GFP-positive cells reflects overall cellular uptake, intracellular pDNA release and protein expression.
The pDNA/dendrimer complexes did not reduce cell viability and were not cytotoxic in both cell lines (Table 1 and Table S2, ESI†). The best transfection dendrimer Z22 was among the least problematic in the series, and only Z1 showed significant toxicity but no reduction in cell viability. Additional testing showed that Z20, Z21 and Z22 preserved their excellent transfection efficiencies and low cytotoxicity at different N/P ratios, low concentration and in the presence of serum proteins, an important parameter for potential therapeutic applications (Fig. S1, ESI†). Furthermore, exposing THP-1 cells, which are monocytes involved in innate immunity,29 to the dendrimers alone or with pDNA did not induce any significant release of the pro-inflammatory cytokines Tumor necrosis factor α (TNF-α) or interleukin-1β (IL-1β) in comparison to the positive controls LPS30 and PMA.31 We observed a moderate toxicity of the dendrimers on THP-1 cells; however, the toxic effect was entirely absent in the presence of 10% serum (Fig. S2, ESI†).
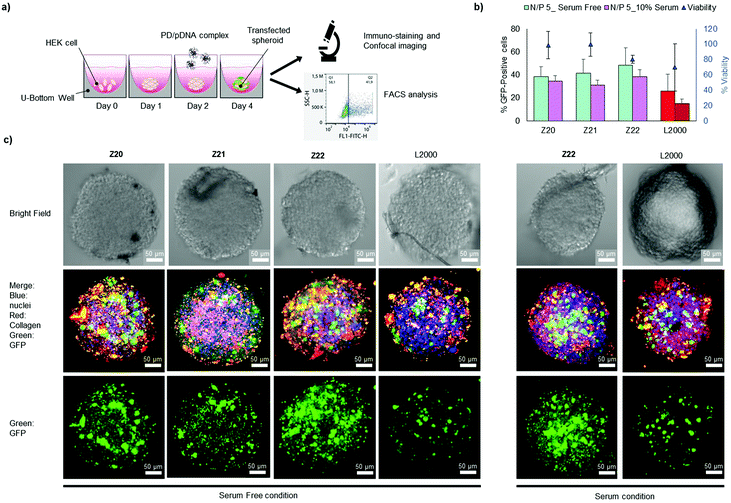
In view of their favourable transfection activities combined with low toxicity and low immunogenicity, we tested Z20, Z21 and Z22 for transfecting the CRISPR/Cas9 pDNA into 3D tumor spheroids from HEK cells and quantified transfection by FACS analysis upon spheroid destruction (Fig. 2a). All three peptide dendrimers had higher transfection efficiency than L2000, both in the presence and in the absence of serum proteins, while also preserving spheroid viability better than L2000 (Fig. 2b). Transfection was further evidenced by confocal imaging, which showed that cells throughout the spheroids were efficiently transfected (Fig. 2c and Fig. S3, ESI†).
To better understand pDNA transfection by our dendrimers, we further characterized Z20–Z22, as well as Z1 and Z12 as two less potent analogues. Dynamic light scattering showed that dendrimer/pDNA complexes formed large nanoparticles (Rh = 300 nm) at pH 7.4 similar to those formed between pDNA and the commercial reagents jetPEI or PEI (Fig. S4a, ESI†).32,33 These nanoparticles were only moderately stable as indicated by zeta potential values close to zero (Fig. S4b, ESI†), and by the fact that pDNA was readily displaced by heparin (Fig. S5, ESI†). The dendrimers formed aggregates even in the absence of pDNA at pH 7.4 at a critical micellar concentration (CMC ∼ 100 μg mL−1) corresponding to pDNA complexation as evidenced by the Nile Red assay.34 However, the dendrimers were not aggregated at pH 5.0 corresponding to the acidified endosome (Fig. S6, ESI†), and had a different conformation at pH 7.4 and pH 5.0 as evidenced by circular dichroism spectra (Fig. S7, ESI†). These effects are probably triggered by the protonation of the eight amino termini, which occurs around pH 6.5 with such polycationic peptide dendrimers,21,35 leading to increased positive charge density and electrostatic repulsion between dendrimers.
These experiments suggest that Z22 and its analogues mediate pDNA transfection through the same mechanism as dendrimers DMH13 and DMH18 for siRNA transfection,21,22 despite the very different sizes of their oligonucleotide cargo. This mechanism involves the aggregation and complexation of the oligonucleotide at pH 7.4 to form nanoparticles, which enter cells by endocytosis. Endosome acidification then induces protonation of the eight amino termini on the peptide dendrimer, resulting in partial disaggregation and endosome escape. Indeed, dendrimer mediated pDNA transfection was entirely blocked by inhibiting endosome acidification with bafilomycin A1 (Fig. S8, ESI†).
In summary, we identified peptide dendrimer Z22 as an exceptionally highly efficient reagent for transfecting large pDNA such as the CRISPR/Cas9 plasmid into cells and 3D-tumor spheroids, with low cytotoxicity and low immunogenicity. Similar to related peptide dendrimers,35–38Z22 is accessible in pure form by standard SPPS, which greatly facilitates its use.
This work was supported financially by the Marie Curie Training Network MMBIO (grant no. 721613) and the Swiss National Science Foundation (grants no. 200020_178998). We thank Dr Sacha Javor for his help in data interpretation and text editing.
Conflicts of interest
There are no conflicts to declare.Notes and references
- J. A. Doudna and E. Charpentier, Science, 2014, 346, 1258096 CrossRef.
- L. Cong, F. A. Ran, D. Cox, S. Lin, R. Barretto, N. Habib, P. D. Hsu, X. Wu, W. Jiang, L. A. Marraffini and F. Zhang, Science, 2013, 339, 819–823 CrossRef CAS.
- L. Naldini, Nature, 2015, 526, 351–360 CrossRef CAS.
- Z. Wu, H. Yang and P. Colosi, Mol. Ther., 2010, 18, 80–86 CrossRef CAS.
- G. Gernoux, A. M. Gruntman, M. Blackwood, M. Zieger, T. R. Flotte and C. Mueller, Mol. Ther., 2020, 28, 747–757 CrossRef CAS.
- H.-X. Wang, M. Li, C. M. Lee, S. Chakraborty, H.-W. Kim, G. Bao and K. W. Leong, Chem. Rev., 2017, 117, 9874–9906 CrossRef CAS.
- R. K. Jain and T. Stylianopoulos, Nat. Rev. Clin. Oncol., 2010, 7, 653–664 CrossRef CAS.
- L. Zhang, P. Hao, D. Yang, S. Feng, B. Peng, D. Appelhans, T. Zhang and X. Zan, J. Mater. Chem. B, 2019, 7, 953–964 RSC.
- G. Schwank, B.-K. Koo, V. Sasselli, J. F. Dekkers, I. Heo, T. Demircan, N. Sasaki, S. Boymans, E. Cuppen, C. K. van der Ent, E. E. S. Nieuwenhuis, J. M. Beekman and H. Clevers, Cell Stem Cell, 2013, 13, 653–658 CrossRef CAS.
- M. Matano, S. Date, M. Shimokawa, A. Takano, M. Fujii, Y. Ohta, T. Watanabe, T. Kanai and T. Sato, Nat. Med., 2015, 21, 256–262 CrossRef CAS.
- M. Zanoni, F. Piccinini, C. Arienti, A. Zamagni, S. Santi, R. Polico, A. Bevilacqua and A. Tesei, Sci. Rep., 2016, 6, 19103 CrossRef CAS.
- M. Wojnilowicz, Q. A. Besford, Y.-L. Wu, X. J. Loh, J. A. Braunger, A. Glab, C. Cortez-Jugo, F. Caruso and F. Cavalieri, Biomaterials, 2018, 176, 34–49 CrossRef CAS.
- N. S. Bhise, R. S. Gray, J. C. Sunshine, S. Htet, A. J. Ewald and J. J. Green, Biomaterials, 2010, 31, 8088–8096 CrossRef CAS.
- R. Sapra, R. P. Verma, G. P. Maurya, S. Dhawan, J. Babu and V. Haridas, Chem. Rev., 2019, 119, 11391–11441 CrossRef.
- C. C. Lee, J. A. MacKay, J. M. J. Frechet and F. C. Szoka, Nat. Biotechnol., 2005, 23, 1517–1526 CrossRef CAS.
- A.-M. Caminade, D. Yan and D. K. Smith, Chem. Soc. Rev., 2015, 44, 3870–3873 RSC.
- Y. Dong, T. Yu, L. Ding, E. Laurini, Y. Huang, M. Zhang, Y. Weng, S. Lin, P. Chen, D. Marson, Y. Jiang, S. Giorgio, S. Pricl, X. Liu, P. Rocchi and L. Peng, J. Am. Chem. Soc., 2018, 140, 16264–16274 CrossRef CAS.
- K. Ezzat, S. E. Andaloussi, E. M. Zaghloul, T. Lehto, S. Lindberg, P. M. Moreno, J. R. Viola, T. Magdy, R. Abdo, P. Guterstam, R. Sillard, S. M. Hammond, M. J. Wood, A. A. Arzumanov, M. J. Gait, C. I. Smith, M. Hallbrink and U. Langel, Nucleic Acids Res., 2011, 39, 5284–5298 CrossRef CAS.
- J. Hoyer and I. Neundorf, Acc. Chem. Res., 2012, 45, 1048–1056 CrossRef CAS.
- C. Douat, M. Bornerie, S. Antunes, G. Guichard and A. Kichler, Bioconjugate Chem., 2019, 30, 1133–1139 CrossRef CAS.
- M. Heitz, S. Javor, T. Darbre and J.-L. Reymond, Bioconjugate Chem., 2019, 30, 2165–2182 CrossRef CAS.
- M. Heitz, S. Zamolo, S. Javor and J.-L. Reymond, Bioconjugate Chem., 2020, 31, 1671–1684 CrossRef CAS.
- A. Kwok, G. A. Eggimann, J.-L. Reymond, T. Darbre and F. Hollfelder, ACS Nano, 2013, 7, 4668–4682 CrossRef CAS.
- A. Kwok, G. A. Eggimann, M. Heitz, J. L. Reymond, F. Hollfelder and T. Darbre, ChemBioChem, 2016, 17, 2223–2229 CrossRef CAS.
- M. Heitz, A. Kwok, G. A. Eggimann, F. Hollfelder, T. Darbre and J. L. Reymond, Chimia, 2017, 71, 220–225 CrossRef CAS.
- O. Saher, C. S. J. Rocha, E. M. Zaghloul, O. P. B. Wiklander, S. Zamolo, M. Heitz, K. Ezzat, D. Gupta, J. L. Reymond, R. Zain, F. Hollfelder, T. Darbre, K. E. Lundin, S. El Andaloussi and C. I. E. Smith, Eur. J. Pharm. Biopharm., 2018, 132, 29–40 CrossRef CAS.
- A. L. Szymczak, C. J. Workman, Y. Wang, K. M. Vignali, S. Dilioglou, E. F. Vanin and D. A. A. Vignali, Nat. Biotechnol., 2004, 22, 589–594 CrossRef CAS.
- S. J. Ahn, J. Costa and J. R. Emanuel, Nucleic Acids Res., 1996, 24, 2623–2625 CrossRef CAS.
- O. Takeuchi and S. Akira, Cell, 2010, 140, 805–820 CrossRef CAS.
- S. Mariathasan, D. S. Weiss, K. Newton, J. McBride, K. O’Rourke, M. Roose-Girma, W. P. Lee, Y. Weinrauch, D. M. Monack and V. M. Dixit, Nature, 2006, 440, 228–232 CrossRef CAS.
- E. Kontny, M. ZiÓŁKowska, A. Ryżewska and W. Maśliński, Cytokine, 1999, 11, 839–848 CrossRef CAS.
- X. Chen, Z. Yuan, X. Yi, R. Zhuo and F. Li, Nanotechnology, 2012, 23, 415602 CrossRef.
- Z. Tan, Y. Jiang, W. Zhang, L. Karls, T. P. Lodge and T. M. Reineke, J. Am. Chem. Soc., 2019, 141, 15804–15817 CrossRef CAS.
- M. C. A. Stuart, J. C. van de Pas and J. B. F. N. Engberts, J. Phys. Org. Chem., 2005, 18, 929–934 CrossRef CAS.
- B.-H. Gan, T. N. Siriwardena, S. Javor, T. Darbre and J.-L. Reymond, ACS Infect. Dis., 2019, 5, 2164–2173 CrossRef CAS.
- J.-L. Reymond and T. Darbre, Org. Biomol. Chem., 2012, 10, 1483–1492 RSC.
- T. N. Siriwardena, M. Stach, R. He, B.-H. Gan, S. Javor, M. Heitz, L. Ma, X. Cai, P. Chen, D. Wei, H. Li, J. Ma, T. Köhler, C. van Delden, T. Darbre and J.-L. Reymond, J. Am. Chem. Soc., 2018, 140, 423–432 CrossRef CAS.
- T. N. Siriwardena, A. Capecchi, B. H. Gan, X. Jin, R. He, D. Wei, L. Ma, T. Kohler, C. van Delden, S. Javor and J. L. Reymond, Angew. Chem., Int. Ed., 2018, 57, 8483–8487 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0cc04750c |
| This journal is © The Royal Society of Chemistry 2020 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: