Open Access Article
Open Access ArticleMultifunctional, histidine-tagged polymers: antibody conjugation and signal amplification†
Yichuan
Zhang‡
 ab,
Alessia
Gambardella‡
a,
Muhammed
Üçüncü
ab,
Alessia
Gambardella‡
a,
Muhammed
Üçüncü
 ac,
Jin
Geng
ac,
Jin
Geng
 ab,
Jessica
Clavadetscher
a,
Mark
Bradley
ab,
Jessica
Clavadetscher
a,
Mark
Bradley
 *a and
Annamaria
Lilienkampf
*a and
Annamaria
Lilienkampf
 *a
*a
aEaStCHEM School of Chemistry, University of Edinburgh, EH9 3FJ, Edinburgh, UK. E-mail: mark.bradley@ed.ac.uk; annamaria.lilienkampf@ed.ac.uk
bShenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China
cDepartment of Analytical Chemistry, Faculty of Pharmacy, Izmir Katip Celebi University, Izmir, Turkey
First published on 9th October 2020
Abstract
A polymer scaffold, with multiple reactive centres, was synthesised by RAFT polymerisation and conjugated to the antibody herceptin. A hexahistidine RAFT agent enabled the rapid and simple purification of polymer–protein conjugates, while the tetrazine conjugation strategy allows myriad cargos to be attached and amplified.
The ability to amplify reporter signals and cargos is important in a number of areas ranging from fluorescent imaging to radiotherapy and is of particular importance with low abundance targets. Several signal enhancement methods have been developed with numerous techniques associated with the polymerase chain reaction and variants.1 Non-nucleic acid based amplification methods include horseradish peroxidase-driven signal amplification of tyramide-conjugates2 or luminol,3 and alkaline phosphatase driven amplification.4 Although these enzyme-based amplification methods work well, they typically require the use of secondary antibodies and bespoke substrates, with colorimetric or fluorescent outputs often able to diffuse away from the site of generation. Non-enzymatic fluorescence amplification techniques include e.g. the recruitment of antibody-GFP fusion proteins to a target protein tagged with multiple copies of the antibody epitope,5 Avidin-conjugated viral capsids loaded with dyes for the detection of biotinylated targets,6 and biological self-assembly of biotin and streptavidin surface engineered quantum dots.7
Synthetic polymers are an attractive approach to biomolecule labelling and fluorescence or mass-tag signal amplification.8 Conjugation of polymers to proteins can be categorised as “grafting-from” and “grafting-to” approaches,9 the former starting with the formation of a “macro-activator” (i.e., linkage of an initiator or a chain transfer agent to a protein), followed by polymerisation; however, this approach is limited by the need to avoid protein denaturation and the restricted choice of suitable functional monomers.10 The “grafting-to” approach uses pre-synthesised polymers for bioconjugation but suffers from complications, such as how to separate conjugated proteins from non-conjugated materials.11
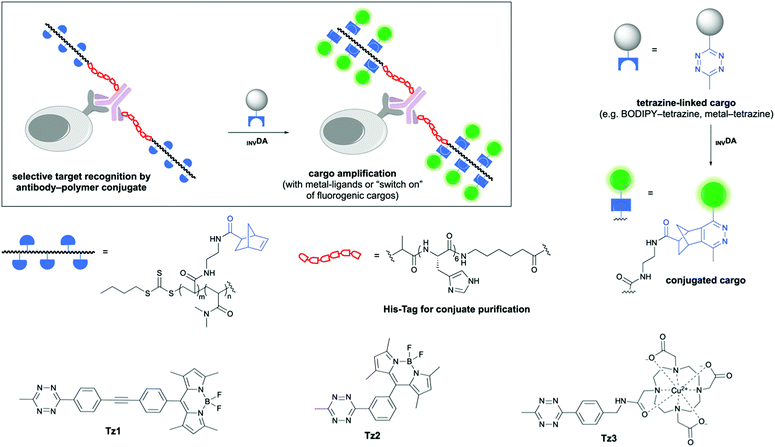
Here, we report a rapid amplification approach, based on poly(acrylamide)–antibody conjugates bearing norbornene reactive centres for both “switch-on” of quenched tetrazine-linked fluorophores and cargo amplification in the form of metal ions (Fig. 1). The polymers were synthesised through reversible addition–fragmentation chain transfer (RAFT) polymerisation, using the synthesised chain transfer agent 2 that contained the ubiquitous hexahistidine tag (His-Tag), designed to allow rapid polymer–antibody conjugate purification by metal affinity chromatography. Upon treatment with a quenched BODIPY–tetrazine conjugate, rapid “switch-on” and amplification in fluorescence were observed.
Five polymers with different molecular weights and densities of the norbornene reactive centres were synthesised (P1–P5) to explore how composition affected conjugation, antibody binding affinity, and the level of cargo amplification (Fig. 2a). N,N-Dimethylacrylamide (DMA) was used due to its low cytotoxicity and the high water solubility of its polymers,12 with the norbornene-based acrylamide 1 (Scheme S1, ESI†) providing the site for tetrazine ligation. The polymers were efficiently synthesised by RAFT polymerisation with the desired molecular weights (7–30 kDa) with polydispersities between 1.2–1.8 (Table S1 and Fig. S1, ESI†) using a His-Tag-RAFT agent 2 (synthesised on solid-phase13 in high yield/purity, Scheme S2, ESI†), which allowed the widely used method of metal ion affinity chromatography to be applied to polymer-conjugate purification.
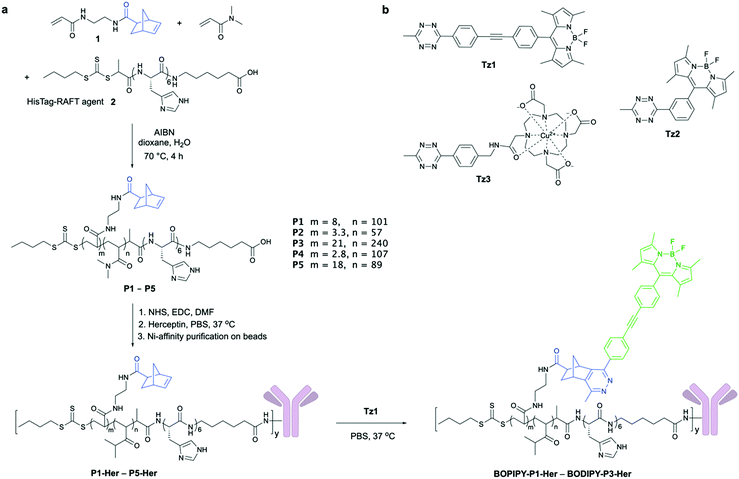 | ||
| Fig. 2 (a) Synthesis of the multifunctional polymers P1–P5 and the ratios of 1, 2 and DMA found in the synthesised polymers (based on 1H NMR analysis). All polymers were synthesised as random copolymers (see ESI,† Table S1 for full characterisation). Polymer synthesis was followed by their antibody conjugation (10 eq. polymer per antibody added) and metal-affinity chromatography purification of the polymer–antibody conjugates (P1-Her–P5-Her) and subsequent INVDA mediated dequenching to give the fluorescent conjugates. The NHS-conjugation strategy produces heterogenous polymer–antibody population, with multiple conjugation sites possible; however, only one modification site on the antibody is shown for clarity. (b) The tetrazines explored for signal amplification. | ||
To achieve simultaneous “switch-on” and amplification of fluorescence by the polymer–protein conjugates, two fluorogenic BODIPY–tetrazines were synthesised (Fig. 2b). Tetrazine-quenched fluorophores, that are activated via an inverse electron-demand Diels–Alder reaction (INVDA), have found use in fluorescent imaging,14–18 allowing direct target visualisation without extensive washing steps. Tz1 was synthesised as reported previously,14 whereas the published route to Tz215 could not be reproduced and instead was synthesised from 3-formylbenzonitrile, via the tetrazine aldehyde S9,16 which was treated with 2,4-dimethylpyrrole, followed by oxidation with DDQ and complexation with BF3·OEt2 to give the BODIPY–tetrazine Tz2 in high yield (Scheme S3, ESI†).17 The metal chelating tetrazine Tz3 was synthesised by coupling the DOTA–NHS ester to an amino-functionalised tetrazine,19 followed by Cu(II) loading (Scheme S4, ESI†).
The reactivity of the norbornene-decorated polymer P1 (12 μM) with the tetrazines Tz1 and Tz2 (1 μM) was investigated by monitoring the time-dependent increase in fluorescence, triggering fluorescence increases of 21 and 9-fold respectively, in PBS after 30 min (Fig. S3 and S4, ESI†). However, stability studies showed that although Tz1 and Tz2 were robust in PBS and serum-free media, Tz2 underwent undesired tetrazine hydrolysis in serum, while in contrast, Tz1 showed good stability and was thus used in preference for all investigations (Fig. S5, ESI†).
The antibody herceptin facilitates tyrosine kinase receptor HER2 degradation and is used clinically to treat aggressive breast cancer,20 and was used here to demonstrate both cell labelling21 and cargo amplification. Polymers conjugated to herceptin (P1-Her–P5-Her) were purified in 45–88% yields utilising the His-Tag (Fig. 2a and Table S2, ESI†), with SDS PAGE showing successful conjugation (Fig. S9, ESI†). When P1-Her was treated with tetrazine Tz1 (Fig. 2a), intense fluorescent labelling of the antibody resulted, with selectivity between the polymer-tagged antibody and the unmodified variant observed (Fig. 3a, lane 2 vs. 4). In addition, SDS-PAGE analysis indicated that the majority of polymer conjugation took place on the heavy chains of the antibody. The influence of the polymers on the antibody's binding affinity was investigated by flow cytometry analysis of the labelling efficiency of the polymer–antibody conjugates on HER2 receptor positive breast cancer cells (SK-BR-3) with BODIPY-P1-Her, BODIPY-P2-HER and BODIPY-P3-Her and BODIPY-Her. Incubation of SK-BR-3 cells with small molecule labelled BODIPY-Her (Scheme S5, ESI†) resulted in rapid labelling of the cells with fluorescence saturation of cells observed after 2 h. Similar labelling and fluorescence saturation of cells was also observed by polymer conjugated antibodies BODIPY-P2-Her and BODIPY-P3-Her, whereas BODIPY-P1-Her showed slower labelling with fluorescence saturation reached after 4 h incubation (Fig. 3b). MCF-7 cells, which do not express the HER2 receptor, showed no significant changes in fluorescence intensity when incubated with the polymer–herceptin conjugates (Fig. S10, ESI†). None of the polymer–antibody conjugates (P1-Her to P5-Her), BODIPY-Her or Tz1 showed cellular toxicity at 10 nM, 10 nM and 1 μM, respectively (Fig. S11, ESI†).
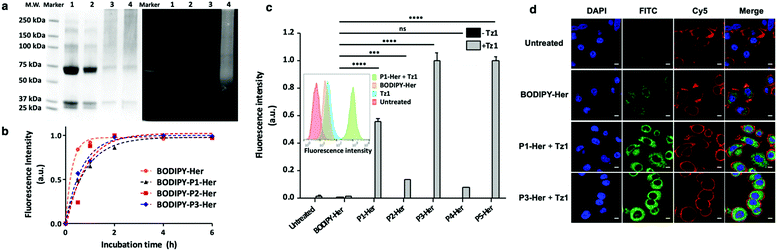 | ||
| Fig. 3 (a) SDS-PAGE showing successful conjugation and fluorescence “switch-on”. Lane 1 herceptin; lane 2 herceptin (2.5 μM) incubated with Tz1 (25 μM); lane 3 P1-Her and lane 4 P1-Her (2.5 μM) incubated with Tz1 (25 μM) (reaction mixtures were purified using centrifugal size exclusion filters prior to analysis). (b) Polymer–herceptin conjugate labelling of SK-BR-3 cells (quantified by flow cytometry with fluorescence intensity normalised to fluorescence saturated cells, n = 3) compared to the traditional small molecule labelled herceptin BODIPY-Her (Scheme S5, ESI†). (c) Fluorescence intensity of HER2 receptor positive SK-BR-3 cells treated first with the polymer–herceptin conjugates (10 nM) followed by Tz1 (1 μM) (normalised to cells treated with P3-Her followed by Tz1). The increase in fluorescence intensity between BODIPY-Her and BODIPY-labelled P1–P5-Her was 45, 10, 83, 6 and 83-fold, respectively. Inset: The flow cytometry histograms of SK-BR-3 cells treated with BODIPY labelled herceptin BODIPY-Her or Tz1, or the polymer–antibody conjugate P1-Her followed by Tz1 (green). Data were analysed using one-way ANOVA with Dunnett post-test (ns, not significant, ***P < 0.001, ****P < 0.0001). (d) Confocal fluorescence microscopy images of SK-BR-3 cells treated with BODIPY-labelled herceptin BODIPY-Her (10 nM), and polymer–antibody conjugates P1-Her and P3-Her (10 nM) followed by Tz1 (1 μM). The cell nucleus was stained with Hoechst 33342 (blue, λex/em = 353/483 nm) and the plasma membrane stained with CellMask™ Deep Red (red, λex/em = 649/666 nm). “Switched-on“ BODIPY from Tz1 is shown in green (λex/em = 488/512 nm). Scale bar = 10 μm. The norbornene content and monomer ratios for P1–P5 (based on 1H NMR analysis) are given in Fig. 2. | ||
The fluorescence “turn-on” and signal amplification with the polymer–antibody conjugates and tetrazine Tz1 was demonstrated in live SK-BR-3 cells by flow cytometry and microscopy (Fig. 3c and d). Cells treated with BODIPY-Her (4 h, 10 nM) showed a moderate increase in fluorescence intensity; however, treatment with the conjugates P1-Her to P5-Her (4 h, 10 nM), followed by the quenched fluorophore Tz1 (30 min, 1 μM) resulted in remarkable increases in cellular fluorescence (83-fold increase for P3-Her and P5-Her, compared to BODIPY-Her). The fluorescence intensity could be tuned by either changing the molecular weight of the polymers (45, 10 and 83-fold increases for P1-Her, P2-Her and P3-Her, respectively) or the density of the reactive centres (6 and 83-fold increase for P4-Her and P5-Her, bearing 2.8 and 18 norbornes per polymer chain, respectively). Confocal microscopy images of the cells treated with P1-Her and P3-Her, which had different numbers of reactive centres (8 vs. 21), followed by Tz1 showed much brighter fluorescence (λex/em = 488/512 nm) than BODIPY-Her treated cells, although with similar cellular localisations, i.e., cytoplasma and membrane (Fig. 3d and Fig. S12, ESI†).
This amplification method could also be translated to amplifying the loading density of radioisotopes.22 As a proof of concept, P5-Her was pre-labelled with Tz3 (Fig. 2a, Fig. S11 and Scheme S6, ESI†) bearing Cu(II) on a DOTA ligand (copper was used as a model as 64Cu is widely used in nuclear medicine23). The amplified metal loading on SK-BR-3 cells was confirmed by inductively coupled plasma-mass spectrometry (ICP-MS) where the Cu(II) loaded polymer–antibody conjugate Cu-P5-Her showed a 6-fold higher copper loading (1.6 pg per cell) in comparison to the conventionally radiolabelled antibody Cu-Her (Fig. S14 and Scheme S7, ESI†).
In summary, a linear poly(acrylamide) scaffold bearing norbornenes and a His-Tag was synthesised, conjugated to a clinically used antibody herceptin, and used for fluorescence and metal amplification. Compared to the standard small molecule fluorophore labelled antibody, the polymer–antibody conjugates gave up to an 83-fold increase in fluorescence without affecting antibody binding. This approach of receptor targeted polymer conjugates enables simultaneous “switch-on” and amplification of fluorescent signal with possible contributions to fluorescence-guided surgery and other fluorescence-based applications, especially those where signals are inherently weak due to low numbers of receptors. The approach was applied to the amplification of metal loading with possible uses in nuclear medicine, but the designed polymer scaffold would also enable the “multiplication” of other molecules (e.g. drugs). The new His-Tag-RAFT agent provides a powerful new approach to access various functional polymers with an endogenous purification handle to enable the rapid and simple purification of both “graft-to” or “graft-from” polymer-conjugates.
We thank European Research Council (ADREEM ERC-2013-340469) for funding and Dr L. Eades for assistance with ICP-MS.
Conflicts of interest
There are no conflicts to declare.Notes and references
- L. Lan, Q. Guo, H. Nie, C. Zhou, Q. Cai, J. Huang and X. Meng, Chem. Sci., 2019, 10, 2034 RSC; Y. Wang, C. Liu, X. Zhang, W. Yang, F. Wu, G. Zou, X. Weng and X. Zhou, Chem. Sci., 2018, 9, 3723 RSC; P. V. Robinson, C. Tsai, A. E. de Groot, J. L. McKechnie and C. R. Bertozzi, J. Am. Chem. Soc., 2016, 138, 10722 CrossRef CAS; T. Sano, C. L. Smith and C. R. Cantor, Science, 1992, 258, 120 CrossRef.
- M. N. Bobrow, T. D. Harris, K. J. Shaughnessy and G. J. Litt, J. Immunol. Methods, 1989, 125, 279 CrossRef CAS; R. P. v. Gijlswijk, H. J. Zijlmans, J. Wiegant, M. N. Bobrow, T. J. Erickson, K. E. Adler, H. J. Tanke and A. K. Raap, J. Histochem. Cytochem., 1997, 45, 375 CrossRef; K. Akama, K. Shirai and S. Suzuki, Anal. Chem., 2016, 88, 7123 CrossRef.
- D. Li, B. Shlyahovsky, J. Elbaz and I. Willner, J. Am. Chem. Soc., 2007, 129, 5804 CrossRef CAS; Y. He, J. Sun, X. Wang and L. Wang, Sens. Actuators, B, 2015, 221, 792 CrossRef.
- T. Nobori, K. Tosaka, A. Kawamura, T. Joichi, K. Kamino, A. Kishimura, E. Baba, T. Mori and Y. Katayama, Anal. Chem., 2017, 90, 1059 CrossRef CAS; F. Patolsky, A. Lichtenstein and I. Willner, Nat. Biotechnol., 2001, 19, 253 CrossRef.
- M. E. Tanenbaum, L. A. Gilbert, L. S. Qi, J. S. Weissman and R. D. Vale, Cell, 2014, 159, 635 CrossRef CAS.
- C. M. Soto, A. S. Blum, G. J. Vora, N. Lebedev, C. E. Meador, A. P. Won, A. Chatterji, J. E. Johnson and B. R. Ratna, J. Am. Chem. Soc., 2006, 128, 5184 CrossRef CAS.
- J. Park, Y. Park and S. Kim, ACS Nano, 2013, 7, 9416 CrossRef CAS.
- D. Duret, Z. Haftek-Terreau, M. Carretier, T. Berki, C. Ladavière, K. Monier, P. Bouvet, J. Marvel, Y. Leverrier, M. T. Charreyre and A. Favier, Polym. Chem., 2018, 9, 1857 RSC; L. Zhang, W. Zhao, X. Liu, G. Wang, Y. Wang, D. Li, L. Xie, Y. Gao, H. Deng and W. Gao, Biomaterials, 2015, 64, 2 CrossRef CAS; H. N. Kim, Z. Guo, W. Zhu, J. Yoon and H. Tian, Chem. Soc. Rev., 2011, 40, 79 RSC; H. J. Avens and C. N. Bowman, Acta Biomater., 2010, 6, 83 CrossRef; B. Allo, X. Lou, A. Bouzekri and O. Ornatsky, Bioconjugate Chem., 2018, 29, 2028 CrossRef; T. Berki, A. Bakunts, D. Duret, L. Fabre, C. Ladavieŕe, A. Orsi, M.-T. Charreyre, A. Raimondi, E. van Anken and A. Favier, ACS Omega, 2019, 4, 12841 CrossRef.
- Y. Wang and C. Wu, Biomacromolecules, 2018, 19, 1804 CrossRef CAS.
- J. Xu, K. Jung, N. A. Corrigan and C. Boyer, Chem. Sci., 2014, 5, 3568 RSC; I. Cobo, M. Li, B. S. Sumerlin and S. Perrier, Nat. Mater., 2015, 14, 143 CrossRef CAS; J. Liu, V. Bulmus, D. L. Herlambang, C. Barner-Kowollik, M. H. Stenzel and T. P. Davis, Angew. Chem., Int. Ed., 2007, 46, 3099 CrossRef.
- J. M. Paloni, E. A. Miller, H. D. Sikes and B. D. Olsen, Biomacromolecules, 2018, 19, 3814 CrossRef CAS; J. Morgenstern, G. Gil Alvaradejo, N. Bluthardt, A. Beloqui, G. Delaittre and J. R. Hubbuch, Biomacromolecules, 2018, 19, 4250 CrossRef.
- M. Schuster, C. Turecek, B. Kaiser, J. Stampfl, R. Liska and F. Varga, J. Macromol. Sci., Part A: Pure Appl. Chem., 2007, 44, 547 CrossRef CAS.
- J. Hentschel, K. Bleek, O. Ernst, J. F. Lutz and H. G. Börner, Macromolecules, 2008, 41, 1073 CrossRef CAS.
- A. Wieczorek, T. Buckup and R. Wombacher, Org. Biomol. Chem., 2014, 12, 4177 RSC.
- J. C. Carlson, L. G. Meimetis, S. A. Hilderbrand and R. Weissleder, Angew. Chem., Int. Ed., 2013, 52, 6917 CrossRef CAS.
- A. Wieczorek, P. Werther, J. Euchner and R. Wombacher, Chem. Sci., 2017, 8, 1506 RSC.
- G. Linden, L. Zhang, F. Pieck, U. Linne, D. Kosenkov, R. Tonner and O. Vazquez, Angew. Chem., Int. Ed., 2019, 58, 12868 CrossRef CAS.
- H. Wu and N. K. Devaraj, Acc. Chem. Res., 2018, 51, 1249 CrossRef CAS; L. G. Meimetis, J. C. Carlson, R. J. Giedt, R. H. Kohler and R. Weissleder, Angew. Chem., 2014, 126, 7661–7664 CrossRef; Y. Lee, W. Cho, J. Sung, E. Kim and S. B. Park, J. Am. Chem. Soc., 2017, 140, 974 CrossRef; E. Kozma, O. Demeter and P. Kele, ChemBioChem, 2017, 18, 486 CrossRef.
- R. Sen, D. Gahtory, J. Escorihuela, J. Firet, S. P. Pujari and H. Zuilhof, Chem. – Eur. J., 2017, 23, 13015 CrossRef CAS.
- C. A. Hudis, N. Engl. J. Med., 2007, 357, 39 CrossRef CAS.
- Y. Urano, D. Asanuma, Y. Hama, Y. Koyama, T. Barrett, M. Kamiya, T. Nagano, T. Watanabe, A. Hasegawa and P. L. Choyke, Nat. Med., 2009, 15, 104 CrossRef CAS; J. B. Haun, N. K. Devaraj, S. A. Hilderbrand, H. Lee and R. Weissleder, Nat. Nanotechnol., 2010, 5, 660 Search PubMed.
- N. A. Thiele, V. Brown, J. M. Kelly, A. Amor-Coarasa, U. Jermilova, S. N. MacMillan, A. Nikolopoulou, S. Ponnala, C. F. Ramogida, A. K. Robertson, C. Rodríguez-Rodríguez, P. Schaffer, C. Williams, J. W. Babich, V. Radchenko and J. J. Wilson, Angew. Chem., Int. Ed., 2017, 56, 14712 CAS; B. Yu, H. Wei, Q. He, C. A. Ferreira, C. J. Kutyreff, D. Ni, Z. T. Rosenkrans, L. Cheng, F. Yu, J. W. Engle, X. Lan and W. Cai, Angew. Chem., Int. Ed., 2018, 57, 218 Search PubMed.
- A. Boschi, P. Martini, E. Janevik-Ivanovska and A. Duatti, Drug Discovery Today, 2018, 23, 1489 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0cc04591h |
| ‡ These authors contribute equally. |
| This journal is © The Royal Society of Chemistry 2020 |