Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Activity-directed expansion of a series of antibacterial agents†
Abbie
Leggott
ab,
Justin E.
Clarke
 bc,
Shiao
Chow
bc,
Shiao
Chow
 ab,
Stuart L.
Warriner
ab,
Stuart L.
Warriner
 ab,
Alex J.
O’Neill
ab,
Alex J.
O’Neill
 *bc and
Adam
Nelson
*bc and
Adam
Nelson
 *ab
*ab
aSchool of Chemistry, University of Leeds, LS2 9JT, Leeds, UK. E-mail: a.s.nelson@leeds.ac.uk
bAstbury Centre for Structural Molecular Biology, University of Leeds, Leeds, LS2 9JT, UK. E-mail: a.j.oneill@leeds.ac.uk
cSchool of Molecular and Cellular Biology, University of Leeds, Leeds, LS2 9JT, UK
First published on 15th June 2020
Abstract
The feasibility of using activity-directed synthesis to drive antibacterial discovery was investigated. An array of 220 Pd-catalysed microscale reactions was executed, and the crude product mixtures were evaluated for activity against Staphylococcus aureus. Scale-up of the hit reactions, purification and evaluation, enabled expansion of a class of antibacterial quinazolinones. The novel antibacterials had MICs from 0.016 μg mL−1 (i.e. 38 nM) to 2–4 μg mL−1 against S. aureus ATCC29213.
The discovery of bioactive small molecules is generally driven through iterative design-make-purify-test cycles. The process is generally underpinned by a remarkably narrow toolkit1 of reliable reaction classes which limits the diversity of explored chemical space.2 We have recently introduced activity-directed synthesis3 (ADS), a structure-blind and function-driven discovery approach. In ADS, inherently promiscuous chemistry is harnessed in which arrays of reactions are performed, and each reaction has multiple possible outcomes. The crude reaction mixtures are then directly screened for biological activity. We have previously harnessed metal carbenoid chemistry to drive the activity-directed discovery of diverse series of androgen receptor agonists and protein–protein interaction inhibitors.3
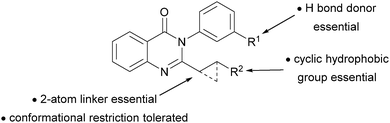
Here, we demonstrate the generality of ADS by showing that other reaction classes (here, Pd-catalysed carbonylation4) and assay types (here, whole-cell antibacterial activity against the ESKAPE5 pathogen S. aureus) may be harnessed. We show that the approach may be used to expand a series of quinazolinones6 that was developed to target penicillin-binding proteins, and displays promising activity (minimum inhibitory concentrations [MICs] as low as 0.003 μg mL−1) against multiple strains of S. aureus.6a
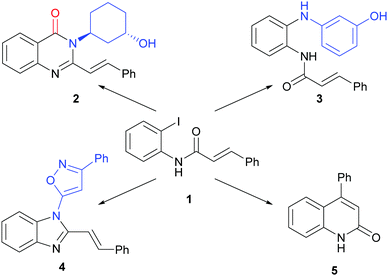
We envisaged that Pd-catalysed reactions may complement the metal carbenoid chemistry that we have previously harnessed in ADS (Fig. 1). Specifically, we noted its potential to yield alternative scaffolds from a common substrate (such as 1). For example, Pd-catalysed carbonylation, followed by reaction with an amine and cyclisation, could yield 2,3-disubstituted quinazolinones such as 2. Alternatively, Pd-catalysed amination, without carbonylation, could yield anilines (e.g.3) or, with subsequent cyclisation, benzimidazoles (e.g.4). Finally, intramolecular Heck reaction could yield quinolinones (e.g.5).
Initially, we adapted a Pd-catalysed carbonylation reaction7 for execution on a 300 μl scale (100 μmol substrate) in microscale vials in 96-well plate format (Scheme 1). Accordingly, the o-iodo anilide 6a (final concentration: 333 mM), the aniline (1.2 eq.), Mo(CO)6 (1 eq.), Pd2(dba)3 (0.1 mol%), P(tBu3)·HBF4 (0.6 mol%) and NEt3 (2.5 eq.) were dissolved in o-xylene; after sealing, the vial was heated at 105 °C for 48 h. It was shown that the yield of 8a, determined by NMR spectroscopy using an internal standard, was broadly similar to that obtained on a preparative scale in a crimp-top vial. At this stage, the quinazolinones 8b and 8c6a were also prepared. Crucially, we had shown that Pd-catalysed chemistry was viable in a format suitable for ADS, and that stirring and an inert atmosphere were not necessary.
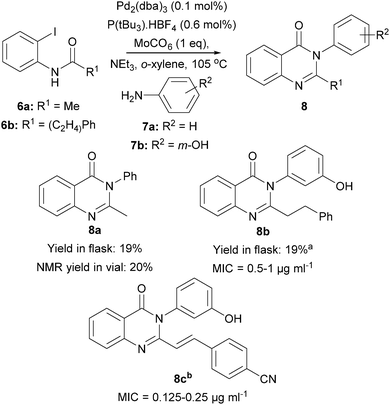 | ||
Scheme 1 Configuration of Pd-catalysed chemistry for use in ADS. a![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LC-MS analysis of the reaction performed in a microscale vial revealed that the product had been formed successfully. b LC-MS analysis of the reaction performed in a microscale vial revealed that the product had been formed successfully. b![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Prepared using a precedented method (ref. 6a). Prepared using a precedented method (ref. 6a). | ||
We adapted standard susceptibility testing methodology8 to assess antibacterial activity against S. aureus ATCC29213. Quinazolinone solubility was improved by using Iso-Sensitest Broth (ISB)9 as the growth medium, and MICs were determined for 8b–c [8b: 0.5–1 μg mL−1 (1.45–2.90 μM); 8c: 0.125–0.25 μg mL−1 (0.34–0.68 μM)]. The reaction of 6b and 7b (which would yield the antibacterial 8b) was then repeated in a microscale vial, the reaction filtered through silica, evaporated, and the crude reaction mixture dissolved in DMSO (total product concentration: 333 mM). The stock solution of the crude product was screened at multiple total product concentrations (100 μM, 50 μM, 25 μM and 10 μM; entries 1 and 9–11, Table 1): visual growth inhibition was only detected at the two highest concentrations. Crucially, the reaction of 6a and 7b did not yield an active crude product at 50 μM total product concentration (entry 2). In addition, the reactions of the individual reactants 6a, 6b and 7b (entries 3–5), as well as the individual components P(tBu3)·HBF4, Mo(CO)6 and Pd2(dba)3 (entries 6–8), also did not result in detectable activity. The assay was deemed suitable for activity-directed antibacterial discovery since activity was only observed for reactions that could have yielded the positive control 8b.
| Entrya | Substrate | Co-Substrateb | Reagentsc | Conc.d/μM | Activitye |
|---|---|---|---|---|---|
| a Reactions performed in microscale vials at 105 °C; crude reaction mixtures were filtered through silica, evaporated and dissolved in DMSO (total product concentration: 333 mM) before evaluation. b 1.2 eq. c Mo(CO)6 (1 eq.), 0.1 mol% Pd2dba3, 0.6 mol% P(tBu3)·HBF4, NEt3 (2.5 eq.), o-xylene. d Total product concentration based on limiting substrate. e Activity against S. aureus ATCC29213 (see Fig. 2 for colour scale); a tick indicates visual growth inhibition (see ESI). f LC-MS revealed that a quinazolinone had been formed. | |||||
| 1 | 6b | 7b | All | 50 |
|
| 2 | 6a | 7b | All | 50 |
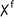
|
| 3 | 6b | — | All | 50 |
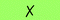
|
| 4 | 6a | — | All | 50 |
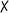
|
| 5 | — | 7b | All | 50 |
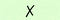
|
| 6 | — | — | P(tBu3)·HBF4 | 50 |
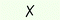
|
| 7 | — | — | Mo(CO)6 | 50 |
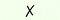
|
| 8 | — | — | Pd2(dba)3 | 50 |
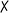
|
| 9 | 6b | 7b | All | 100 |
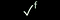
|
| 10 | 6b | 7b | All | 25 |
|
| 11 | 6b | 7b | All | 10 |
|
Having established appropriate synthetic and biological methods for ADS, we designed a reaction array based on ten substrates S1–S10 (and no substrate) and nineteen co-substrates C1–C19 (and no co-substrate). The array of 220 reactions was designed to enable formation of a diverse range of quinazolinones, some related to known analogues,6 as well as other products of Pd-catalysed processes. The substrates were mainly analogues of S1 (e.g. dehomologated, constrained), whilst the aniline and amine co-substrates bore structurally-diverse aryl, hetaryl and other substituents. To expand the possible reaction outcomes, some substrates and co-substrates bore alternative (e.g.S2 and S9) or additional (e.g.C6, C12 and C16) nucleophiles. The substrates and co-substrates were all inactive against S. aureus ATCC29213 at 50 μM. The array was performed in a 96-well OptiBlock and assembled from stock solutions using multi-channel pipettes. After 48 h at 105 °C, the crude reaction mixtures were filtered through silica, evaporated and dissolved in DMSO (total product concentration: 333 μM). Randomly-selected product mixtures (n = 23) were analysed by LC/MS; 16 of these contained carbonylated intermolecular products, 13 of which had been dehydrated. In addition, one product mixture (S3 + C1) contained the product of an intramolecular reaction of S3, later shown to stem from a reductive Heck reaction10 (see ESI†).
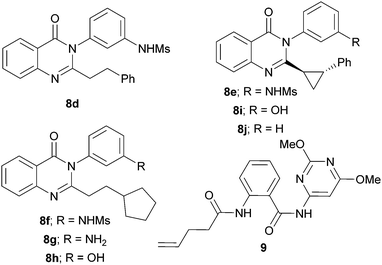
The crude reaction mixtures were screened in duplicate against S. aureus ATCC29213 (final total product concentration of 50 μM in 1% DMSO in ISB) (Fig. 2). Six of the 220 reactions displayed significant antibacterial activity in both replicate cultures: substrate S1 (with C1 or C18); substrate S7 (with C18); and substrate S10 (with C1, C6 or C18). Reassuringly, the combination of S1 and C1 had been previously validated in our method development work (Table 1). The remaining five hit reactions were scaled fifty-fold, and the products purified by mass-directed HPLC (Table 2). In each case, quinazolinones with an extended hydrophobic substituent in the 2-position, and a meta-substituted phenyl group in the 3-position, were obtained. The products had MIC values against S. aureus ATCC29213 ranging from 0.016 μg mL−1 (i.e. 38 nM) for 8d to 2–4 μg mL−1 (i.e. 6–12 μM) for 8g. Notably, 8d was ∼10-fold more active than the positive control6a8c. The products 8d–h had comparable activity against the methicillin-resistant USA300 JE2 strain, but were significantly less active against laboratory strain SH1000. In addition, we purified the products of three reactions that were not identified as hits: S7 + C1; S7 + aniline; and S4 + C9; two of these reactions also yielded quinazolinones, and the third an uncyclised amide 9, and, crucially, all of these products displayed significantly lower (or no detectable) antibacterial activity.
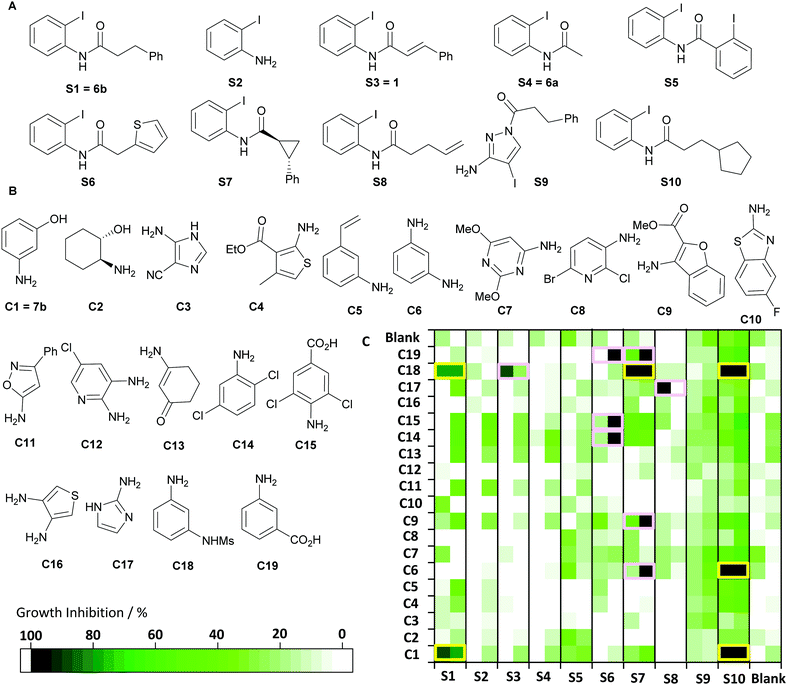 | ||
| Fig. 2 Activity-directed antibacterial discovery (see Table 1 for reaction conditions). Panels A and B: Structures of substrates (A) and co-substrates (B). Panel C: Activity against S. aureus ATCC29213 (total product concentration: 50 μM), with active combinations that were scaled up shown (yellow). Product mixtures that displayed activity against only one colony (pink) were re-assayed, but none of these combinations were validated as hits (see ESI†). | ||
| Hit? | Substrates | Producta (yieldb) | MIC (μg mL−1) | |||
|---|---|---|---|---|---|---|
| ATCC29213 | USA300 JE2 | SH1000 | ||||
| a Co-Substrate (1.2 eq.), Mo(CO)6 (1 eq.), 0.1 mol% Pd2bda3, 0.6 mol% P(tBu3)·HBF4, NEt3, o-xylene. b After mass-directed HPLC. All compounds were also screened against yeast (Candida albicans, Ca6) and were found to be inactive at 16 μg mL−1. | ||||||
| ✓ | S1, C1 | 8b (19%) | 0.5–1 | 0.5 | 4 |
|
| ✓ | S1, C18 | 8d (4%) | 0.016 | 0.016 | 0.5–1 | |
| ✓ | S7, C18 | 8e (2%) | 1–2 | 2 | 32–64 | |
| ✓ | S10, C1 | 8f (8%) | 1 | 0.5–1 | 4–8 | |
| ✓ | S10, C6 | 8g (8%) | 2–4 | 2–4 | 16 | |
| ✓ | S10, C18 | 8h (14%) | 0.5 | 0.5 | 1–2 | |
| ✗ | S7, C1 | 8i (21%) | 4–8 | 4–8 | 16 | |
| ✗ | S4, C9 | 9 (5%) | >128 | 64 | 32–64 | |
| ✗ | S7, aniline | 8j (18%) | >128 | >128 | >128 | |
Our experiments enabled the SAR of the quinazolinones to be significantly expanded (Fig. 3).6 We found that a –CH2CH2– linker (as in 8d and 8f–h) could be replaced by a trans-disubstituted cyclopropane (as in 8e); however, reactions involving dehomologated (e.g.S6) or truncated (e.g.S4) substrates, or those offering alternative reaction pathways (e.g.S2, S3, S5, S8 and S9), did not yield antibacterial products. In some cases, the meta OH or NHMs group (in 8f and 8h) could be replaced by a NH2 group (in 8g); however, none of the co-substrates that lacked such a m-substituted phenyl group ever yielded antibacterial products. Crucially, the products of reactions that had not been identified as hits were substantially less (or not) active. Although the quinazolinone 8i is matched pair of 8a, 8e and 8h (MICs: 0.5–2 μg mL−1), it is significantly less active (MIC: 4–8 μg mL−1). The quinazolinones 8j and the amide 9 were not active, even at 128 μg mL−1.
In conclusion, we have shown that ADS is feasible with a novel reaction class (Pd-catalysed chemistry) and with a phenotypic assay (rather than, as previously,3 a biophysical assays). An array of 220 reactions enabled efficient exploration of the function of products based on several alternative product scaffolds. Based on our analysis of reaction outcomes by LC-MS, and the range of substrates used, we estimate that most of these reactions productively yielded previously unexplored products. Scale-up of the identified hit reactions, and purification, yielded a range of antibacterial quinazolinones and, thereby, expansion of their SAR. We envisage that, by harnessing alternative reaction classes to explore diverse chemical space, ADS may be exploited in the discovery of novel series of antimicrobials not subject to existing antibiotic resistance mechanisms.11
We thank EPSRC (EP/N025652/1) for funding and Luiza Galarion, Merianne Mohamad and Adam Green for useful discussions.
Conflicts of interest
There are no conflicts to declare.Notes and references
- (a) J. Boström, D. G. Brown, R. J. Young and G. M. Kererü, Nat. Rev. Drug Discovery, 2018, 17, 709–727 CrossRef PubMed; (b) D. G. Brown and J. Boström, J. Med. Chem., 2016, 59, 4443–4458 CrossRef CAS PubMed; (c) T. W. Cooper, I. B. Campbell and S. J. Macdonald, Angew. Chem., Int. Ed., 2010, 49, 8082–8091 CrossRef CAS PubMed.
- (a) N. Schneider, D. M. Lowe, R. A. Sayle, M. A. Tarselli and G. A. Landrum, J. Med. Chem., 2016, 59, 4385–4402 CrossRef CAS PubMed; (b) W. P. Walters, J. Green, J. R. Weiss and M. A. Murcko, J. Med. Chem., 2011, 54, 6405–6416 CrossRef CAS PubMed.
- (a) G. Karageorgis, S. Warriner and A. Nelson, Nat. Chem., 2014, 6, 872–876 CrossRef CAS PubMed; (b) G. Karageorgis, M. Dow, A. Aimon, S. Warriner and A. Nelson, Angew. Chem., 2015, 127, 13742–13748 CrossRef; (c) A. Green, F. Hobor, C. Tinworth, S. Warriner, A. Wilson and A. Nelson, Chem. – Eur. J., 2020 DOI:10.1002/chem.202002153.
- L. Åkerbladh, L. R. Odell and M. Larhed, Synlett, 2019, 141–155 Search PubMed.
- J. N. Pendleton, S. P. Gorman and B. F. Gilmore, Expert Rev. Anti-Infect. Ther., 2013, 11, 297–308 CrossRef CAS PubMed.
- (a) R. Bouley, D. Ding, Z. Peng, M. Bastian, E. Lastochkin, W. Song, M. A. Suckow, V. A. Schroeder, W. R. Wolter, S. Mobashery and M. Chang, J. Med. Chem., 2016, 59, 5011–5021 CrossRef CAS PubMed; (b) S. Gatadi, T. V. Lakshmi and S. Nanduri, Eur. J. Med. Chem., 2019, 170, 157–172 CrossRef CAS PubMed; (c) Y. Qian, et al. , J. Med. Chem., 2020, 63, 5287–5296 CrossRef PubMed.
- J.-B. Peng, H.-Q. Geng, W. Wang, X. Qi, J. Ying and X.-F. Wu, J. Catal., 2018, 365, 10–13 CrossRef CAS.
- F. R. Cockerill III, M. A. Wikler, J. Alder, M. N. Dudley, G. M. Eliopoulos, M. J. Ferraro, D. J. Hardy, D. W. Hecht, J. A. Hindler, J. B. Patel, M. Powell, J. M. Swenson, R. B. Thomson Jr., M. Traczewski, J. D. Turnidge, M. P. Weinstein and B. L. Zimmer, CLSI, M07-A9: Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 9th edn, Clinical and Laboratory Standards Institute, 2012 Search PubMed.
- D. J. Farrell, M. Robbins, W. Rhys-Williams and W. G. Love, Antimicrob. Agents Chemother., 2010, 55, 1177–1181 CrossRef PubMed.
- (a) S. Mannathan, S. Raoufmoghaddam, J. N. H. Reek, J. G. de Vries and A. J. Minnaard, ChemCatChem, 2017, 9, 551–554 CrossRef CAS; (b) A. Minatti, X. Zheng and S. L. Buchwald, J. Org. Chem., 2007, 72, 9253–9258 CrossRef CAS PubMed.
- (a) P. Vikesland, E. Garner, S. Gupta, S. Kang, A. Maile-Moskowitz and N. Zhu, Acc. Chem. Res., 2019, 52, 916–924 CrossRef CAS PubMed; (b) N. A. Turner, B. K. Sharma-Kuinkel, S. A. Maskarinec, E. M. Eichenberger, P. P. Shah, M. Carugati, T. L. Holland and V. G. Fowler, Nat. Rev. Microbiol., 2019, 17, 203–218 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures and NMR spectra. CCDC 1991665. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/d0cc02361b |
| This journal is © The Royal Society of Chemistry 2020 |