Ribosomal incorporation of cyclic β-amino acids into peptides using in vitro translation†
Abstract
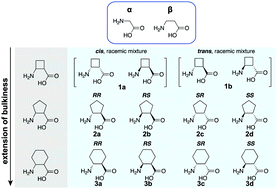
We demonstrate in vitro incorporation of cyclic β-amino acids into peptides by the ribosome through genetic code reprogramming. Further, we show that incorporation efficiency can be increased through the addition of elongation factor P.


 Please wait while we load your content...
Please wait while we load your content...
