Open Access Article
Open Access ArticlePhoto-reactive oligodeoxynucleotide-embedded nanovesicles (PROsomes) with switchable stability for efficient cellular uptake and gene knockdown†
Beob Soo
Kim
 a,
Mitsuru
Naito
a,
Mitsuru
Naito
 b,
Rimpei
Kamegawa
a,
Hyun Jin
Kim
b,
Rimpei
Kamegawa
a,
Hyun Jin
Kim
 b,
Ryo
Iizuka
c,
Takashi
Funatsu
c,
Shingo
Ueno
d,
Takanori
Ichiki
ad,
Akihiro
Kishimura
e and
Kanjiro
Miyata
b,
Ryo
Iizuka
c,
Takashi
Funatsu
c,
Shingo
Ueno
d,
Takanori
Ichiki
ad,
Akihiro
Kishimura
e and
Kanjiro
Miyata
 *a
*a
aDepartment of Materials Engineering, Graduate School of Engineering, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8656, Japan. E-mail: miyata@bmw.t.u-tokyo.ac.jp
bCenter for Disease Biology and Integrative Medicine, Graduate School of Medicine, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan
cGraduate School of Pharmaceutical Sciences, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan
dInnovation Center of NanoMedicine, Kawasaki Institute of Inductrial Promotion, 3-25-14 Tonomachi, Kawasaki-ku, Kawasaki 210-0821, Japan
eCenter for Molecular Systems, Kyushu University, 744 Moto-oka, Nishi-ku, Fukuoka 819-0395, Japan
First published on 17th July 2020
Abstract
A photo-responsive nanovesicle is fabricated by polyion complex (PIC) formation between poly(ethylene glycol) (PEG)-block-polypeptides and photo-reactive oligodeoxynucleotides (PROs)/anti-sense oligonucleotides (ASOs). The ultraviolet (UV) light triggers reversible crosslinking between PROs and ASOs in the vesicular membrane, providing the nanovesicle with switchable stability under physiological conditions. The resulting nanovesicle allows efficient cellular internalization, leading to significant UV-triggered gene knockdown in cultured cells.
Polymeric vesicles, termed polymersomes, have been widely investigated as a desirable platform for biochemical and biomedical applications because they can stably encapsulate both hydrophobic and hydrophilic substances in the membrane and hollow aqueous cavity, respectively.1 Indeed, various drugs, such as anticancer drugs, proteins/enzymes, and genetic materials, have been loaded into polymersomes for controlled drug release and targeted drug delivery.2 Notably, such drug payloads can be selectively released from polymersomes by controlling the membrane permeability with various stimuli.2b,3 As an external physical stimulus, light is one of the attractive stimuli because it can trigger drug release through precise spatial and temporal control without additional substances.4
Very recently, we reported nano-sized vesicular PICs that consist of a PIC membrane embedded with small nucleic acids, i.e., small interfering RNAs (siRNAs), and PEG-block-polypeptides.5 In this formulation, siRNAs played two roles; one was a structural component for nanovesicle formation and the other was a pharmaceutical substance. This siRNA-embedded nanovesicle demanded additional stabilization to maintain the vesicle structure under physiological conditions. To this end, glutaraldehyde was employed as a crosslinker between polypeptide segments (or primary amines) but not siRNAs to avoid ineffective release of siRNA payloads. However, glutaraldehyde, which is a homobifunctional crosslinking reagent, likely elicits uncontrollable and irreversible reactions, including self-polymerization and nanoparticle aggregation. Thus, more controllable and reversible crosslinking techniques are required for diverse applications of small nucleic acid-embedded nanovesicles.
Previously, PROs were synthesized by attaching photo-reactive vinyl groups, e.g., cyanovinylcarbazole (CNVK) and CNVK-modified D-threoninol (CNVD), to the nucleobases.6 The PROs installed with CNVK or CNVD can be reversibly reacted with an adjacent pyrimidine base in the complementary strand via [2+2] cycloaddition under UV irradiation at 366 nm (UV366) for crosslinking and 312 nm (UV312) for de-crosslinking. These PROs with high reactivity, selectivity, and reversibility have been utilized for in situ DNA manipulation, cytosine to uracil conversion, gene repair, and genetic engineering.6,7
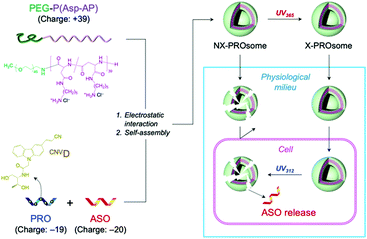
This UV-triggered reversible crosslinking chemistry has inspired us to fabricate a PRO-embedded vesicular PIC, termed PROsome, with switchable stability that enables the stable loading of ASOs in extracellular milieu and the triggered release of ASOs in cells for efficient gene knockdown (Fig. 1). The PROsome is fabricated through PIC formation among three components, PROs as an anionic reversible crosslinker, ASOs as an anionic therapeutic agent, and PEG-block-polypeptides as a cationic membrane component. In this way, UV365-triggered crosslinking should stably hold ASO payloads in the PIC membrane, while UV312-triggered de-crosslinking can compromise the stability of PIC membrane for the facilitated payload release. Herein, we demonstrate the successful fabrication of PROsomes and their photo-responsive functionalities for enhanced gene knockdown in cultured cells.
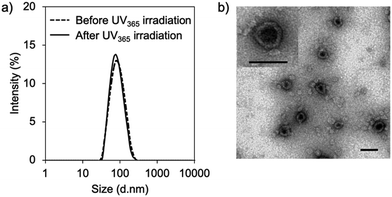
First, vesicular PIC formation was verified for the mixture of PRO, ASO, and PEG-block-polypeptide in an aqueous solution. The structural information of oligodeoxynucleotides is summarized in Table S1 (ESI†). The PRO was designed to have 5 CNVD-installed adenine (A) units in 20-mer polyA for random crosslinking with pyrimidine, i.e., thymine (T) and cytosine, in ASO. The nuclear-enriched abundant transcript 2 (NEAT2)-targeting ASO (asNEAT2) was selected to evaluate the gene knockdown potential because NEAT2 long non-coding RNA (lncRNA) plays as an oncogene transcript and is highly expressed in many types of human cancers, including human lung cancers.8 As a PEG-block-polypeptide, PEG-block-poly[N-(5-aminopentyl)-α,β-aspartamide] (PEG–P(Asp-AP)) was synthesized to have a Mn of PEG: 2000 and a degree of polymerization of P(Asp-AP): 39, as confirmed by size exclusion chromatography (Fig. S1, ESI†) and 1H NMR (Fig. S2, ESI†). Of note, this polymer composition can satisfy the criteria of a fPEG (defined by 100 × [total weight of PEG in a PIC]/[total weight of components in a PIC]) of less than 10% for vesicular PIC formation under a charge-stoichiometric condition (defined by [number of amino groups in PEG–P(Asp-AP)]/[number of phosphate groups in ASO/PRO] = 1).9 Indeed, PEG–P(Asp-AP) has formed vesicular PICs with various polyanions, including siRNA, in previous studies.5,10 Here, PIC samples were prepared by vortex mixing of PEG–P(Asp-AP)/PRO/asNEAT2 at a molar charge ratio of 2/1/1 (fPEG: 8%), followed by irradiation with UV365 (1920 mJ cm−2).6 The formation of vesicular PICs was characterized by dynamic light scattering (DLS) and transmission electron microscopy (TEM). Regardless of UV365 irradiation, PIC samples had a hydrodynamic diameter of approximately 80 nm with a narrow size distribution (polydispersity index (PDI): ∼0.1) (Fig. 2a). The TEM image of UV365-irradiated PICs clearly displays vesicular nanostructures with darker pool and brighter rim (Fig. 2b), similar to those of previously reported vesicular PICs.10 Of note, the vesicular nanostructures were maintained after UV312 irradiation (Fig. S3, ESI†), indicating that the UV312 irradiation did not induce structural changes in vesicular PICs.
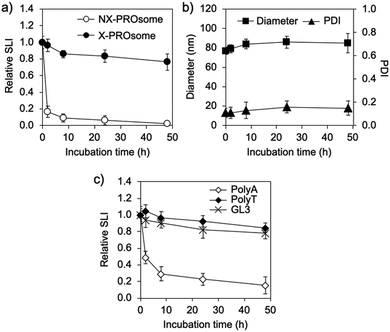
Next, the stability of PROsomes was investigated to confirm whether the CNVD moieties in PRO were effectively photo-crosslinked with asNEAT2 in the PIC membrane. The stability was examined by static light scattering (SLS) and DLS in terms of changes in scattered light intensity (SLI), size, and PDI of PROsomes under a physiological salt condition. UV365-untreated PROsomes, i.e., non-crosslinked (NX-)PROsomes, instantly reduced relative SLI (Fig. 3a). In contrast, the PROsomes treated with 1 min UV365 irradiation, i.e., crosslinked (X-)PROsomes, exhibited apparently constant SLI even after 48 h incubation. Also, the DLS results revealed that X-PROsomes maintained their initial size and PDI without significant changes in physiological milieu (Fig. 3b). These results indicate that the UV365 irradiation to PROsomes dramatically inhibited their structural changes in physiological milieu. Of note, considerable changes in relative SLI, size, and PDI of PROsomes were observed after treatment with less than 1 min UV365 irradiation (Fig. S4, ESI†), suggesting that 1 min UV365 irradiation was required for the stabilization of nanovesicle structures under the physiological milieu.
To validate the effect of photo-crosslinking of PRO (or CNVD) with T in ASO, control PROsomes were further prepared from a series of oligodeoxynucleotide mixtures, i.e., PRO/polyA, PRO/polyT, and PRO/GL3 luciferase-targeted ASO (asGL3) (Table S1, ESI†), and then, were treated with UV365 in the same manner as the PROsomes prepared from PRO/asNEAT2. PolyT and asGL3 including T units clearly suppressed the decrease in relative SLI, whereas such a suppression effect was not observed for polyA (Fig. 3c). This result clearly indicates that the photo-crosslinking between PROs (or CNVD moieties) and T-containing oligodeoxynucleotides should be essential for the significant stability of PROsomes in the physiological salt solution. It should be noted that both asNEAT2 and asGL3 containing six T units at different positions in 21-mer clearly improved the PROsome stability, similar to polyT, suggesting that 6 T units in ASO should be enough for the photo-reaction with PRO having five CNVD moieties in the PIC membrane.
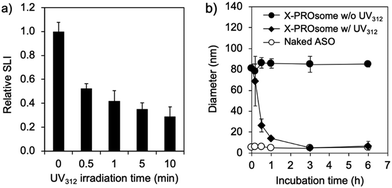
The cyclobutane structure formed by the [2+2] cycloaddition between CNVD and T in oligodeoxynucleotides can be de-crosslinked by photo-splitting reaction at UV312 irradiation.6b This de-crosslinking reaction makes PROsomes structurally unstable in the physiological environment. To confirm the reversibility of PROsomes, the time-dependent changes in SLI and hydrodynamic diameter of X-PROsomes in 150 mM NaCl solution were monitored under UV312 irradiation with a 15 W transilluminator (Fig. 4a and Fig. S5, ESI†). The relative SLI value of X-PROsomes rapidly decreased by only 0.5 min irradiation, associated with the considerable decrease in size of X-PROsomes. This result clearly demonstrates that the photo-splitting reaction of CNVD occurred by UV312 irradiation. The stability of X-PROsomes was additionally evaluated in fetal bovine serum (FBS)-containing media by fluorescence correlation spectroscopy (FCS) in terms of time-dependent changes in the hydrodynamic diameters of Alexa Fluor® 647-labeled ASO (AF647-ASO) (Fig. 4b). In this assay, two PROsome samples, X-PROsomes before and after UV312 irradiation for 0.5 min, were prepared and incubated with 10% FBS. The hydrodynamic diameter of X-PROsomes without UV312 irradiation was kept approximately 90 nm, indicating that AF647-ASO was stably confined in the nanovesicles even in the serum-containing medium. In contrast, the hydrodynamic diameter of X-PROsomes was reduced immediately after 0.5 min UV312 irradiation and reached the similar size to naked ASO, suggesting that the nanovesicles were dissociated to release free AF647-ASO. These results demonstrate the photo-switchable stability of PROsomes.
The feasibility of PROsomes as an ASO carrier was then investigated using cultured human lung cancer, A549 cells. The cellular uptake efficiency of PROsomes was evaluated by flow cytometry (Fig. 5a). Naked AF647-asNEAT2 exhibited a modest cellular uptake efficiency, presumably because chemically modified ASOs with phosphorothioate (PS)-backbone and locked nucleic acid (LNA)-wings (i.e., PS-gapmers) have significant tolerability against nucleases.11 The fluorescence intensity of cells treated with NX-PROsomes was comparable to that treated with naked AF647-ASO, suggesting that NX-PROsomes were unstable in the serum-containing medium and dissociated to free AF647-ASO. In contrast, X-PROsomes elicited a significantly enhanced cellular uptake efficiency, probably due to their higher tolerability in the serum-containing media. This enhanced cellular uptake of X-PROsomes can be explained by some reasons; (i) charge neutralization of ASO and (ii) larger surface area (curvature) of vesicular PICs compared with naked ASO, for facilitated adsorptive endocytosis.12 The additional uptake inhibition assays suggest that X-PROsomes were mainly uptaken by endocytic pathways, including macropinocytosis, because the uptake amount of X-PROsomes was significantly decreased by incubation at 4 °C and treatment with cytochalasin D (Fig. S6, ESI†).
Next, the intracellular ASO release from X-PROsomes was observed by confocal laser scanning microscopy (CLSM). In this experiment, UV312 was directly irradiated to the cells to destabilize X-PROsomes through de-crosslinking. Thus, the UV312 irradiation condition was first optimized to avoid the UV-induced cytotoxicity and DNA damage.13 The UV312-treated cells showed a negligible effect on their viability in the case of an irradiation time within 10 min (Fig. S7, ESI†). Also, the concentration of messenger RNA (mRNA) harvested from the cells was determined as an indicator of DNA damage after treatment with UV312 at different irradiation times (Fig. S8, ESI†). No significant reduction in mRNA concentration was observed for the cells treated with UV312 irradiation within 1 min. Thus, the UV312 irradiation for 0.5 min was selected for the intracellular ASO release assay. The X-PROsomes were prepared from AF647-ASOs and AF546-ASOs at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and incubated with cultured A549 cells for 24 h before UV312 irradiation. After UV312 irradiation, the cells were further incubated for 24 or 48 h to track the intracellular ASO release. The obtained CLSM images display that the yellow spots that result from the colocalization of AF647-ASOs and AF546-ASOs were reduced by UV312 irradiation (Fig. S9a, ESI†), which is more quantitatively shown by the calculated Pearson's correlation coefficients (Fig. S9b, ESI†). These results indicate that the UV312 irradiation triggered the time-dependent ASO release from X-PROsomes through de-crosslinking in the cells.
1 and incubated with cultured A549 cells for 24 h before UV312 irradiation. After UV312 irradiation, the cells were further incubated for 24 or 48 h to track the intracellular ASO release. The obtained CLSM images display that the yellow spots that result from the colocalization of AF647-ASOs and AF546-ASOs were reduced by UV312 irradiation (Fig. S9a, ESI†), which is more quantitatively shown by the calculated Pearson's correlation coefficients (Fig. S9b, ESI†). These results indicate that the UV312 irradiation triggered the time-dependent ASO release from X-PROsomes through de-crosslinking in the cells.
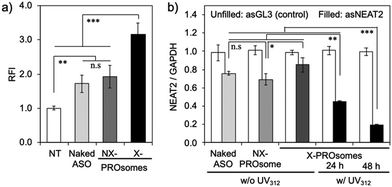
Eventually, the gene knockdown efficiency of PROsomes in cultured A549 cells was evaluated by quantitative reverse-transcription polymerase chain reaction (qRT-PCR). We designed the gene knockdown experiment as illustrated in Fig. S10, ESI,† where UV312 was irradiated for 0.5 min at 24 h post-transfection to induce de-crosslinking of X-PROsomes inside cells. Consistent with the cellular uptake study, the similar modest gene knockdown efficiencies (∼30%) were observed for naked asNEAT2 and asNEAT2-loaded NX-PROsomes (Fig. 5b). Despite the high cellular uptake efficiency, X-PROsomes without UV312 irradiation elicited only 10% decrease in NEAT2 level, suggesting inefficient ASO release from X-PROsomes presumably due to the covalent crosslinking between PRO and ASO. In contrast, the gene knockdown efficiency of X-PROsomes was significantly enhanced up to ∼55% and ∼80% in 24 h and 48 h post-incubation, respectively, by 0.5 min UV312 irradiation. Importantly, the X-PROsomes equipped with asGL3 as a control sequence induced almost no reduction in NEAT2 level. These results demonstrate the UV-triggered gene knockdown ability of X-PROsomes in an ASO sequence-specific manner. Of note, the X-PROsomes exhibited negligible cytotoxicity even at a high concentration of ASO (800 nM asGL3) (Fig. S11, ESI†).
In summary, the photo-responsive nanovesicle was successfully fabricated using PRO. The PROsomes were composed of anionic oligodeoxynucleotide mixtures and cationic block copolymers through their electrostatic interactions. While the UV365-crosslinking between CNVD-installed PRO and T-containing ASO provided the PROsomes with the structural durability in physiological milieu, the UV312-de-crosslinking destabilized the PROsomes directed toward the ASO release inside cells, resulting in the enhanced gene knockdown. This is the first study demonstrating the stimuli-responsive vesicular PIC for enhanced functionalities. The photo-reversible strategy can provide versatile applications of multifunctional polymeric nanovesicles for codelivery of various macromolecular drugs with their controlled release profiles. In the future study, we will further investigate the stability of the PIC membrane according to the number of photo-reactive moieties in the PRO strand. Also, other hydrophilic species will be simultaneously encapsulated into the hollow aqueous cavity of PROsomes for their cooperative functionalities.
This research was financially supported by the Center of Innovation (COI) Program (Grant Number: JPMJCE1305) and Research Complex Program from the Japan Science and Technology Agency (JST), Grant-in-Aid for Scientific Research of MEXT of Japan (JSPS KAKENHI Grant Number: 18K19900 for KM), and JSPS Core-to-Core Program (Grant Number: JPJSCCA20170007). We are grateful to Mr H. Hoshi, The University of Tokyo and the Research Hub for Advanced Nano Characterization at The University of Tokyo for the valuable support in TEM observation.
Conflicts of interest
There are no conflicts to declare.Notes and references
- (a) P. Tanner, P. Baumann, R. Enea, O. Onaca, C. Palivan and W. Meier, Acc. Chem. Res., 2011, 44, 1039 CrossRef CAS PubMed; (b) M. Marguet, C. Bonduelle and S. Lecommandoux, Chem. Soc. Rev., 2013, 42, 512 RSC.
- (a) O. Onaca, R. Enea, D. W. Hughes and W. Meier, Macromol. Biosci., 2009, 9, 129 CrossRef CAS PubMed; (b) C. G. Palivan, R. Goers, A. Najer, X. Zhang, A. Car and W. Meier, Chem. Soc. Rev., 2016, 45, 377 RSC.
- (a) F. Meng, Z. Zhong and J. Feijen, Biomacromolecules, 2009, 10, 197 CrossRef CAS PubMed; (b) X. Hu, Y. Zhang, Z. Xie, X. Jing, A. Bellotti and Z. Gu, Biomacromolecules, 2017, 18, 649 CrossRef CAS PubMed.
- (a) C. Alvarez-Lorenzo, L. Bromberg and A. Concheiro, Photochem. Photobiol., 2009, 85, 848 CrossRef CAS PubMed; (b) F. Ercole, T. P. Davis and R. A. Evans, Polym. Chem., 2010, 1, 37 RSC.
- B. S. Kim, S. Chuanoi, T. Suma, Y. Anraku, K. Hayashi, M. Naito, H. J. Kim, I. C. Kwon, K. Miyata, A. Kishimura and K. Kataoka, J. Am. Chem. Soc., 2019, 141, 3699 CrossRef CAS PubMed.
- (a) Y. Yoshimura and K. Fujimoto, Org. Lett., 2008, 10, 3227 CrossRef CAS PubMed; (b) T. Sakamoto, Y. Tanaka and K. Fujimoto, Org. Lett., 2015, 17, 936 CrossRef CAS PubMed.
- (a) K. Fujimoto, S. Matsuda, Y. Yoshimura, T. Matsumura, M. Hayashi and I. Saito, Chem. Commun., 2006, 3223 RSC; (b) K. Fujimoto, T. Sakamoto, K. Konishi-Hiratsuka and Y. Yoshimura, ChemBioChem, 2010, 11, 1661 CrossRef CAS PubMed; (c) T. Sakamoto, A. Shigeno, Y. Ohtaki and K. Fujimoto, Biomater. Sci., 2014, 2, 1154 RSC.
- T. Gutschner, M. Hämmerle, M. Eissmann, J. Hsu, Y. Kim, G. Hung, A. Revenko, G. Arun, M. Stentrup, M. Gross, M. Zörnig, A. R. MacLeod, D. L. Spector and S. Diederichs, Cancer Res., 2013, 73, 1180 CrossRef CAS PubMed.
- (a) W.-F. Dong, A. Kishimura, Y. Anraku, S. Chuanoi and K. Kataoka, J. Am. Chem. Soc., 2009, 131, 3804 CrossRef CAS PubMed; (b) S. Chuanoi, A. Kishimura, W.-F. Dong, Y. Anraku, Y. Yamasaki and K. Kataoka, Polym. J., 2014, 46, 130 CrossRef CAS.
- Y. Anraku, A. Kishimura, Y. Yamasaki and K. Kataoka, J. Am. Chem. Soc., 2013, 135, 1423 CrossRef CAS PubMed.
- (a) J. Kurreck, E. Wyszko, C. Gillen and V. A. Erdmann, Nucleic Acids Res., 2002, 30, 1911 CrossRef CAS PubMed; (b) X. H. Liang, H. Sun, W. Shen and S. T. Crooke, Nucleic Acids Res., 2015, 43, 2927 CrossRef CAS PubMed.
- (a) W. Jiang, B. Y. Kim, J. T. Rutka and W. C. Chan, Nat. Nanotechnol., 2008, 3, 145 CrossRef CAS PubMed; (b) K. Ezzat, Y. Aoki, T. Koo, G. McClorey, L. Benner, A. Coenen-Stass, L. O'Donovan, T. Lehto, A. Garcia-Guerra, J. Nordin, A. F. Saleh, M. Behlke, J. Morris, A. Goyenvalle, B. Dugovic, C. Leumann, S. Gordon, M. J. Gait, S. El-Andaloussi and M. J. Wood, Nano Lett., 2015, 15, 4364 CrossRef CAS PubMed.
- K. Imoto, N. Kobayashi, S. Katsumi, Y. Nishiwaki, T. A. Iwamoto, A. Yamamoto, Y. Yamashina, T. Shirai, S. Miyagawa, Y. Dohi, S. Sugiura and T. Mori, J. Invest. Dermatol., 2002, 119, 1177 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0cc01750g |
| This journal is © The Royal Society of Chemistry 2020 |