Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Visualization of penicillin G acylase in bacteria and high-throughput screening of natural inhibitors using a ratiometric fluorescent probe†
Lu
Li‡
ab,
Lei
Feng‡
 bc,
Ming
Zhang‡
b,
Xin
He
b,
Shengqiao
Luan
b,
Chao
Wang
bc,
Ming
Zhang‡
b,
Xin
He
b,
Shengqiao
Luan
b,
Chao
Wang
 *b,
Tony D.
James
*b,
Tony D.
James
 *d,
Houli
Zhang
b,
Huilian
Huang
e and
Xiaochi
Ma
*d,
Houli
Zhang
b,
Huilian
Huang
e and
Xiaochi
Ma
 *a
*a
aJiangsu Key Laboratory of New Drug Research and Clinical Pharmacy, Xuzhou Medical University, Xuzhou 221004, China. E-mail: maxiaochi2019@163.com
bCollege of Pharmacy, Academy of Integrative Medicine, The National & Local Joint Engineering Research Center for Drug Development of Neurodegenerative Disease, Dalian Medical University, Dalian 116044, China. E-mail: wach_edu@sina.com
cState Key Laboratory of Fine Chemicals, Dalian University of Technology, Dalian 116024, China
dDepartment of Chemistry, University of Bath, Bath BA2 7AY, UK. E-mail: t.d.james@bath.ac.uk
eLaboratory of Modern Preparation of TCM, Ministry of Education, Jiangxi University of Traditional Chinese Medicine, Nanchang 330004, China
First published on 24th March 2020
Abstract
A ratiometric fluorescent probe (PNA) was developed to sense and image endogenous bacterial penicillin G acylase (PGA). Oleanolic acid was discovered as a potential natural inhibitor of PGA using high-throughput screening techniques based on PNA.
Penicillin G acylases (E.C. 3.5.1.11, PGA) mainly distribute in micro-organisms, belonging to the N-terminal hydrolase super family, which can catalyze the hydrolysis of β-lactam antibiotics producing 6-aminopenicillanic acid (6-APA), a key intermediate for the synthesis of various penicillins.1,2 Given the significant industrial importance of PGA, the expression of endogenous PGA in various bacteria has been evaluated using some common techniques, such as RT-PCR. In addition, penicillin acylases secreted by intestinal bacteria are one of the bile acid hydrolases, indicating the importance of penicillin acylases in the metabolism of bile acids, especially conjugated bile acids, by intestinal bacteria.3 Therefore, PGA belonging to penicillin acylases, not only mediates the production of 6-APA, but also contributes to the in vivo circulation of bile acid. Herein, on the basis of the hydrolase characteristics of PGA, we have developed an effective method for the real-time detection of endogenous bacterial PGA activity. In addition, we have investigated some potential inhibitors of PGA since such inhibitors could be used to interfere with the hydrolase activity of PGA in bacteria.
Fluorescent probes that are activated by biological enzymes with high selectivity and sensitivity, can be used for high-throughput analysis as a useful enzyme assay.4 1,8-Naphthalimide derivatives have been widely used to sense biological enzymes in cells, micro-organisms and even animals, due to excellent photophysicochemical properties and in many cases ratiometric fluorescence response which enables enhanced accuracy of measurement in complex systems.5 Recently, several fluorescent probes have been prepared to determine the PGA enzyme activity in vitro, based on deacylation of the phenylacetyl group from various fluorophores.6 However, few fluorescent probes have been developed to detect endogenous bacterial PGA activity, and image PGA in living bacterial cells.
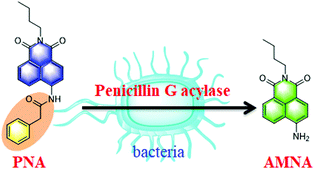
In the present work, we designed and developed a ratiometric fluorescent probe N-(n-butyl)-4-N-phenylacetamide-1,8-naphthalimide (PNA) for the real-time detection and imaging of bacterial PGA (Scheme 1). Additionally, a high-throughput screening system for PGA inhibitors has been established using PNA, and several inhibitors have been obtained from a natural compound library, in particular oleanolic acid with an IC50 value 15.35 μM, which could be used to block the hydrolysis of penicillins and conjugated bile acids by bacterial PGA.
PNA as a ratiometric fluorescent probe displayed fluorescence emission at 470 nm (Fig. S1, ESI†), after deacylation of the phenylacetyl moiety by PGA the N-(n-butyl)-4-amino-1,8-naphthalimide (AMNA) produced displays fluorescence emission at 540 nm (Fig. S1, ESI†). The hydrolysis of PNA mediated by PGA was determined by HPLC-DAD, and indicated the production of AMNA in a co-incubated solution of PNA with PGA (Fig. S2, ESI†). In addition, the interaction and hydrolysis mechanism between PNA and PGA was investigated using in silico docking analysis. As shown in Fig. S3 and S4 (ESI†), hydrogen bonds were observed between 4-NH and GLN B: 23 amino acid residue, the carbonyl group of the phenylacetyl moiety and the ALA B: 69 amino acid residue, which plays a key role in the hydrolysis of the amide bond of PNA. Importantly, PGA could efficiently catalyze the hydrolysis of PNA to produce AMNA.
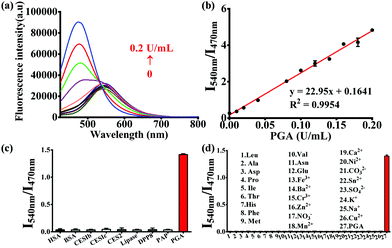
The optimal incubation temperature for the enzymatic hydrolysis of PNA by PGA, was found to be 37 °C (Fig. S5, ESI†). We then evaluated the fluorescence responses of PNA towards PGA as a function of PGA concentration (Fig. 1a). A good linear relationship was observed for the fluorescence intensity ratio at different PGA concentrations (0–0.2 U mL−1) (Fig. 1b), indicating that the intensity ratio was closely related to the PGA activity. Furthermore, the selectivity of PNA toward PGA was evaluated in the presence of various species. As shown in Fig. 1c, general proteins and enzymes including human serum albumin (HSA), bovine serum albumin (BSA), carboxylesterases (1b, 1c, and 2), lipase, dipeptidyl peptidase 8 (DPP8) and proline aminopeptidase (PAP) could not catalyze the hydrolysis of PNA. In addition, various amino acids and ions displayed no interference of the fluorescence intensity ratio (Fig. 1d). As an important parameter for this enzymatic reaction, the kinetic behavior of the hydrolysis of PNA mediated by PGA was investigated (Fig. S7, ESI†). A Michaelis–Menten kinetic model was observed for the hydrolysis experiment, with Vmax 6.44 nmol min−1 mg−1 and Km 9.12 μM, indicating a high affinity of PNA towards PGA.
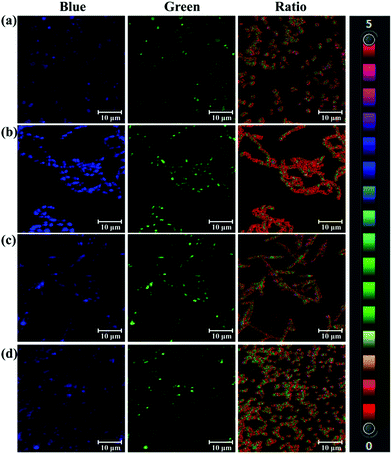
As a bacterial hydrolase, PGA has been reported to be expressed in several bacteria, such as lactobacillus.7 On the basis of hydrolase activity of endogenous bacterial PGA, PNA as a substrate of PGA was used to sense bacterial PGA and image bacteria. After screening several bacteria, Acinetobacter baumannii LMG 994 HAM, Bacillus cereus 994000168 LBK, Brevibacillus parabrevis 090915_03 LBK, and Staphylococcus aureus ssp. aureus DSM 799 DSM all expressed PGA. As shown in Fig. 2, the blue channel and green channel were monitored corresponding to the existence of PNA and AMNA in these bacteria. Importantly, the ratiometric nature of PNA, facilitates the precise determination of endogenous PGA activity for various bacterial species. According to the fluorescence intensity ratio values, it was found that Acinetobacter baumannii (ratio 1.995) expressed more PGA than other bacteria. Therefore, PNA as a ratiometric fluorescent probe could be used to sense and image endogenous bacterial PGA, which would be helpful for the characterisation of bacterial PGA and the investigation of the biological function of bacterial PGA.
Most kits for the activity assay and inhibitor screening of biological enzymes work on the basis of OD values at specific wavelengths for the designed chromogenic reagents, which show low sensitivity. Herein we used PNA with higher sensitivity to assay PGA activity and establish a high-throughput screening system to rapidly discover inhibitors for PNA.
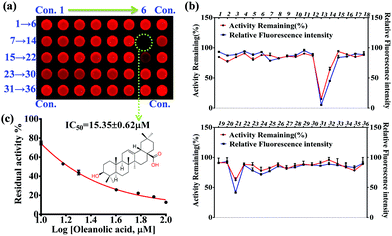
In the present work, 36 natural compounds with a variety of chemical skeletons (e.g. flavones, coumarins, monoterpenoids, diterpenoids, triterpenoids, steroids, alkaloids, and polyphenols) from a natural compound library have been evaluated for their inhibitory effects on PGA (Fig. S12 and Table S1, ESI†). Compared with the control group (Con.), weak fluorescence intensity was observed for the wells corresponding to compounds 13 and 21, indicating potential PGA inhibition (Fig. 3a). In comparison with the control group, the relative fluorescence intensity and remaining PGA activity of each well were determined as shown in Fig. 3b. As a result, compound 13 (oleanolic acid) was tested as a PGA inhibitor, and found to have an IC50 value of 15.35 ± 0.63 μM (Fig. 3c). Thus, using our high-throughput screening system, oleanolic acid was determined to be a potential PGA inhibitor from the natural compound library.
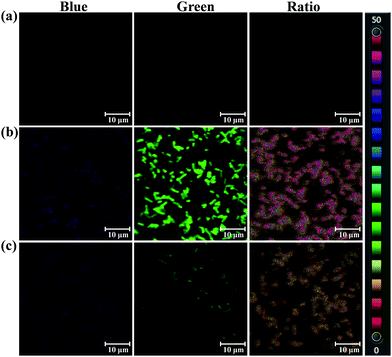
The selectivity and specificity of sensing endogenous bacterial PGA by PNA has been elucidated by fluorescence imaging experiments using PGA expression regulation and the inhibitor oleanolic acid. In the imaging experiment of bacteria, the PGA plasmid was constructed and transfected into E. coli, which led to the preparation of an E. coli strain with high expression of PGA (Escherichia coli–PGA+). As shown in Fig. 4, a strong fluorescence intensity in the green channel was observed for E. coli–PGA+, which suggests that the fluorescence imaging of E. coli was PGA activity dependent. Additionally, the co-incubation of oleanolic acid (50 μM) and PNA in E. coli–PGA+ culture inhibited the production of AMNA according to the weak fluorescence intensity from the green channel. The ratio images confirmed high expression of PGA in E. coli–PGA+ and the strong inhibitory effect of oleanolic acid towards PGA.
In summary, based on the hydrolase properties of PGA, a ratiometric fluorescent probe PNA has been successfully designed and developed for sensing and imaging bacterial PGA. Using PNA for PGA activity assay, a high-throughput screening system has been established for the discovery of new PGA inhibitors, and oleanolic acid was discovered as a potential PGA inhibitor from a natural compound library, capable of interfering in the hydrolysis of penicillins and conjugated bile acids by bacterial PGA.
This work was supported financially by the National Natural Science Foundation of China (No. 81872970, 81930112), National Key R&D Program of China (2018YFC1705900), National Science and Technology Major Project of the Ministry of Science and Technology of China (No. 2018ZX09735005), Liaoning Provincial Key R&D Program (2019JH2/10300022), Liaoning Revitalization Talents Program (XLYC1907017), and Support Program for Innovative Talents in the Universities of Liaoning Province and Program for High-level Talents of Dalian City (2017RQ119). TDJ wishes to thank the Royal Society for a Wolfson Research Merit Award.
Conflicts of interest
There are no conflicts to declare.Notes and references
- (a) V. S. Avinash, A. V. Pundle, S. Ramasamy and C. G. Suresh, Crit. Rev. Biotechnol., 2016, 36, 303–316 CrossRef CAS PubMed; (b) P. Rathinaswam, S. M. Gaikwad, C. G. Suresh, A. A. Prabhune, J. A. Brannigan, G. G. Dodson and A. V. Pundle, Int. J. Biol. Macromol., 2012, 50, 25–30 CrossRef PubMed.
- (a) S. Deng, X. Ma, M. Sun, D. Wei and E. Su, Catal. Commun., 2016, 79, 31–34 CrossRef CAS; (b) X. Pan, L. Wang, J. Ye, S. Qin and B. He, Appl. Microbiol. Biotechnol., 2018, 102, 1749–1758 CrossRef CAS PubMed.
- R. S. Kumar, J. A. Brannigan, A. A. Prabhune, A. V. Pundle, G. G. Dodson, E. J. Dodson and C. G. Suresh, J. Biol. Chem., 2006, 281, 32516–32525 CrossRef CAS PubMed.
- (a) H. W. Liu, L. Chen, C. Xu, Z. Li, H. Zhang, X. B. Zhang and W. Tan, Chem. Soc. Rev., 2018, 47, 7140–7180 RSC; (b) J. Zhang, X. Chai, X. P. He, H. J. Kim, J. Yoon and H. Tian, Chem. Soc. Rev., 2019, 48, 683–722 RSC; (c) L. Feng, J. Ning, X. X. Tian, C. Wang, L. Y. Zhang, X. C. Ma and T. D. James, Coord. Chem. Rev., 2019, 399, 213026 CrossRef CAS; (d) X. F. Wu, W. Shi, X. H. Li and H. M. Ma, Acc. Chem. Res., 2019, 52, 1892–1904 CrossRef CAS PubMed; (e) J. Ning, T. Liu, P. P. Dong, W. Wang, G. B. Ge, B. Wang, Z. L. Yu, L. Shi, X. G. Tian, X. K. Huo, L. Feng, C. Wang, C. P. Sun, J. N. Cui, T. D. James and X. C. Ma, J. Am. Chem. Soc., 2019, 141, 1126–1134 CrossRef CAS PubMed; (f) K. Dou, G. Chen, F. B. Yu, Y. X. Liu, L. X. Chen, Z. P. Cao, T. Chen, Y. L. Li and J. M. You, Chem. Sci., 2017, 8, 7851–7861 RSC; (g) J. Ning, W. Wang, G. B. Ge, P. Chu, F. D. Long, Y. L. Yang, Y. L. Peng, L. Feng, X. C. Ma and T. D. James, Angew. Chem., Int. Ed., 2019, 221, 47–57 Search PubMed.
- (a) X. F. Wu, L. H. Li, W. Shi, Q. Y. Gong, X. H. Li and H. M. Ma, Anal. Chem., 2016, 88, 1440–1446 CrossRef CAS PubMed; (b) B. C. Zhu, C. C. Gao, Y. Z. Zhao, C. Y. Liu, Y. M. Li, Q. Wei, Z. M. Ma, B. Du and X. L. Zhang, Chem. Commun., 2011, 47, 8656–8658 RSC; (c) L. Feng, P. Li, J. Hou, Y. L. Cui, X. G. Tian, Z. L. Yu, J. N. Cui, C. Wang, X. K. Huo, J. Ning and X. C. Ma, Anal. Chem., 2018, 90, 13341–13347 CrossRef CAS PubMed; (d) R. M. Duke, E. B. Veale, F. M. Pfeffer, P. E. Kruger and T. Gunnlaugsson, Chem. Soc. Rev., 2010, 39, 3936–3953 RSC; (e) X. F. Hou, Q. X. Yu, F. Zeng, C. M. Yu and S. Z. Wu, Chem. Commun., 2014, 50, 3417–3420 RSC; (f) Z. R. Dai, L. Feng, Q. Jin, H. L. Cheng, Y. Li, J. Ning, Y. Yu, G. B. Ge, J. N. Cui and L. Yang, Chem. Sci., 2017, 8, 2795–2803 RSC; (g) X. X. Zhang, H. Wu, P. Li, Z. J. Qu, M. Q. Tan and K. L. Han, Chem. Commun., 2016, 52, 8283–8286 RSC; (h) L. Feng, Q. S. Yan, B. J. Zhang, X. G. Tian, C. Wang, Z. L. Yu, J. N. Cui, D. A. Guo, X. C. Ma and T. D. James, Chem. Commun., 2019, 55, 3548–3551 RSC; (i) G. J. Scerri, J. C. Spiteri, C. J. Mallia and D. C. Magri, Chem. Commun., 2019, 55, 4961–4964 RSC; (j) Y. H. Lee, P. Verwilst, H. S. Kim, J. Ju, J. S. Kim and K. Kim, Chem. Commun., 2019, 55, 12136–12139 RSC; (k) C. Ma, M. Ma, Y. Zhang, X. Zhu, L. Zhou, R. Fang, X. Liu and H. Zhang, Spectrochim. Acta, Part A, 2019, 212, 48–54 CrossRef CAS PubMed; (l) F. Ye, N. Wu, P. Li, Y. L. Liu, S. J. Li and Y. Fu, Spectrochim. Acta, Part A, 2019, 222, 117242 CrossRef CAS PubMed; (m) Q. Ma, C. Wang, Y. Bai, J. Xu and X. Guo, Spectrochim. Acta, Part A, 2019, 223, 117334 CrossRef CAS PubMed.
- (a) Y. Meyer, J. A. Richard, B. Delest, P. Noack, P. Y. Renard and A. Romieu, Org. Biomol. Chem., 2010, 8, 1777–1780 RSC; (b) Y. Meyer, J. A. Richard, M. Massonneau, P. Y. Renard and A. Romieu, Org. Lett., 2008, 10, 1517–1520 CrossRef CAS PubMed; (c) J. A. Richard, M. Massonneau, P. Y. Renard and A. Romieu, Org. Lett., 2008, 10, 4175–4178 CrossRef CAS PubMed.
- (a) J. M. Lambert, R. S. Bongers, W. M. de Vos and M. Kleerebezem, Appl. Environ. Microbiol., 2008, 4719–4726 CrossRef CAS PubMed; (b) S. W. Deng, E. Z. Su, X. Q. Ma, S. L. Yang and D. Z. Wei, J. Biotechnol., 2015, 199, 62–68 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0cc00197j |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: