Open Access Article
Open Access ArticleDegrasyn exhibits antibiotic activity against multi-resistant Staphylococcus aureus by modifying several essential cysteines†
Kyu Myung
Lee
 a,
Philipp
Le
a,
Stephan A.
Sieber
a,
Philipp
Le
a,
Stephan A.
Sieber
 *a and
Stephan M.
Hacker
*a and
Stephan M.
Hacker
 *b
*b
aCenter for Integrated Protein Science, Department of Chemistry and Chair of Organic Chemistry II, Technische Universität München, Garching bei München, Germany. E-mail: stephan.sieber@tum.de
bDepartment of Chemistry, Technische Universität München, Garching bei München, Germany. E-mail: stephan.m.hacker@tum.de
First published on 10th February 2020
Abstract
Degrasyn inhibits deubiquitination enzymes and has anti-cancer activity. We here show that it also exhibits antimicrobial activity against multi-resistant Staphylococcus aureus. Structure activity relationship studies demonstrate an important role of the electrophilic α-cyanoacrylamide moiety as a Michael acceptor. A suite of chemical proteomic techniques unraveled binding of this moiety to various cysteine residues of essential proteins in a reversibly covalent manner.
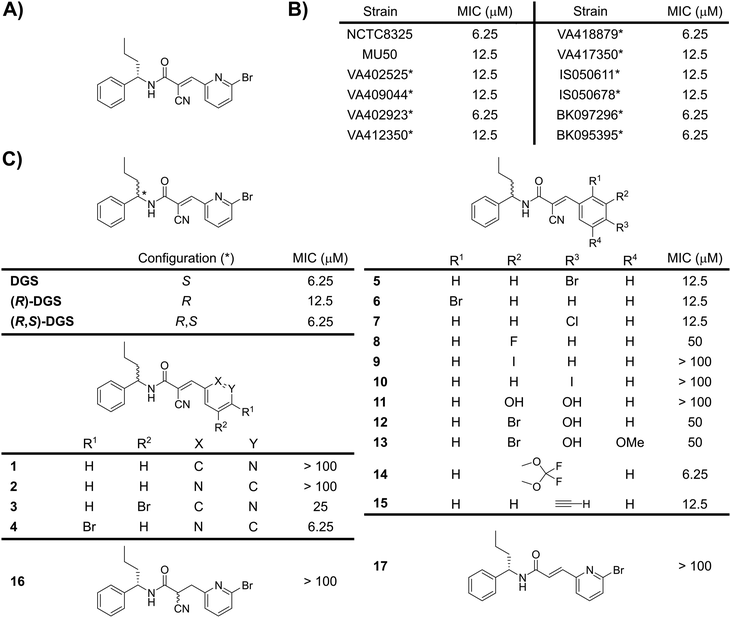
Rapid bacterial resistance development represents a major challenge for current antibacterial therapies.1,2 One strategy to identify novel antibacterials focuses on the repurposing of existing drugs originally developed against human proteins.3,4 In a recent drug-repurposing screen,5 we identified that the human deubiquitinating enzyme inhibitor degrasyn (also called WP1130) kills methicillin-sensitive S. aureus (MSSA) (Fig. 1A and B). Degrasyn induces apoptosis of leukemia cells.6,7 In addition, anti-infective effects, i.e. the reduction of intracellular replication of Listeria monocytogenes and viruses, were previously reported. These effects were mainly attributed to the inhibition of DUBs in macrophages.8–10 Direct antibiotic effects of degrasyn on isolated bacteria have to the best of our knowledge not been reported so far.
Here, we synthesized 19 degrasyn derivatives and analyzed their effects on bacterial growth. The nitrile-substituted Michael acceptor (α-cyanoacrylamide) turned out to be a signature moiety important for antibiotic activity through reversible covalent modification of cysteine residues in essential enzymes. Chemical proteomics revealed several target proteins of degrasyn belonging to important enzyme classes involved e.g. in cell wall, lipid and histidine biosynthesis indicating a polypharmacological mode-of-action.
Given the promising antibiotic effects of degrasyn against the MSSA strain NCTC8325 with a minimal inhibitory concentration (MIC) of 6.25 μM, we tested its activity against various methicillin-resistant S. aureus (MRSA) strains including several clinical isolates (Fig. 1B).5,11 Satisfyingly, the antibiotic activity was retained in all MRSA strains (MIC ≤ 12.5 μM) suggesting a different mode-of-action as compared to existing drugs.
To elucidate the structure activity relationships (SAR) of degrasyn, we systematically altered its scaffold. Degrasyn (DGS) and its enantiomer ((R)-DGS) were prepared according to established synthetic procedures (Scheme 1). A change of the stereocenter resulted in a slight drop of the MIC from 6.25 μM for DGS to 12.5 μM for (R)-DGS suggesting only a minor role of the absolute configuration (Fig. 1C). Accordingly, a racemic mixture of (R,S)-DGS exhibited the same MIC value as DGS. Due to the minor role of the stereocenter and for the ease of synthesis, all further products were synthesized as racemic mixtures.
The synthesis of DGS variants with modifications at the pyridine ring (1–15, Fig. 1C) followed a modular blueprint as outlined in Scheme 2. While an unsubstituted pyridine ring led to a loss of antibiotic activity (1, 2), varying the positions of the bromine substituent and the nitrogen atom was tolerated with only a slight loss in activity (3, 4). Even the replacement of the pyridine ring with a phenyl ring retained considerable activity (5). Therefore, due to availability of building blocks and synthetic accessibility, we studied substituted phenyl rings at this position in more detail. Bromine- and chlorine-substituted phenyl rings were able to largely maintain antibiotic effects (5, 6, 7). In contrast, fluoro- and iodo-substituted phenyl rings resulted in a loss of activity (8, 9, 10). While di- and tri-substitutions with electron-donating groups (e.g. –OH, –OMe, etc.) were largely not tolerated (11, 12, 13), the introduction of a difluoroacetal moiety (14) retained full activity. Satisfyingly, substitution with an alkyne handle (15) only slightly increased the MIC, which is a prerequisite for subsequent target identification using conventional ABPP.
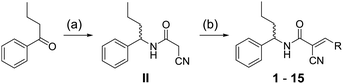 | ||
| Scheme 2 Synthesis of degrasyn derivatives (see Fig. 1 for specific R groups). (a) (i) NH2OH·HCl, NaOH, EtOH/H2O, 3 h (ii) H2, Pd/C, MeOH, 24 h (iii) 1-cyanoacetyl-3,5-dimethyl-1H-pyrazole, toluene, reflux, 2 h, 69%; (b) aryl aldehyde, piperidine, EtOH, reflux, 3 h, 33–75%. | ||
We next turned our synthetic efforts towards the reactive α-cyanoacrylamide moiety. Acrylamides are irreversible covalent inhibitors, which readily react with cysteine residues.12 Further fine-tuning with electron-withdrawing substituents such as nitriles yields reversibly covalent inhibitors, which have received great attention in drug development.13,14 As degrasyn bears this signature moiety, we investigated, if the ability to act as a Michael acceptor and the fine-tuning as α-cyanoacrylamide are both relevant for the antibiotic effect. The synthesis of 16, without the double bond of the Michael acceptor, was achieved by reduction of DGS using NaBH4 (Scheme S1, ESI†). The synthesis of 17, without the nitrile moiety, was performed using a Horner–Wadsworth–Emmons reaction (Scheme S1, ESI†). Interestingly, neither 16 nor 17 showed any antibiotic activity highlighting that both the Michael acceptor and the electron-withdrawing substituent are mandatory (Fig. 1C).
In order to better understand the mechanism-of-action, we performed target identification studies in S. aureus. We first applied conventional ABPP with 15 (Fig. S1, ESI†).15,16 Live bacterial cells were incubated with the probe, lysed, labeled proteins modified with biotin azide using copper-catalyzed azide–alkyne cycloaddition (CuAAC)17 and subsequently analyzed via liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS). The labeling was performed at three different concentrations (6.25 μM, 12.5 μM and 25 μM) and data analysis was performed with label-free quantification (LFQ).18 While we saw some labeling in a gel-based analysis, no prominent enrichment of protein targets was observed by mass spectrometry (Fig. S2 and Table S1, ESI†). In addition, competition studies with various concentrations of parent DGS did not lead to an obvious target profile (Fig. S3 and Table S1, ESI†). Thus, the reversibly covalent binding mechanism is not suitable for conventional ABPP studies as the transient target binding is incompatible with the enrichment procedure for MS.
We therefore performed competitive residue-specific proteomics using the recently developed isoDTB-ABPP method19 that is based on the isoTOP-ABPP (isotopic tandem orthogonal proteolysis-ABPP) platform12,20 as an alternative strategy. In this approach, S. aureus cells are lysed and the lysate is split into two samples. One sample is treated with DGS and the other one with DMSO as a control (Fig. 2A). Afterwards, the samples are both alkynylated with iodoacetamide-alkyne (IA-alkyne). Competitive cysteine binders such as DGS block this labeling at their specific binding sites in the proteome. In order to read out these differences in alkynylation, isotopically labeled desthiobiotin azide (isoDTB) tags19 are appended using CuAAC.17 The samples are combined, enriched on streptavidin, and digested. The resulting peptides are identified and quantified relative to each other using LC-MS/MS. The ratio R between the heavy-labeled (DMSO-treated) and light-labeled (DGS-treated) samples is a measure for the degree of modification of the specific cysteine with DGS.
 | ||
| Fig. 2 (A) Workflow for competitive, residue-specific proteomics with the isoDTB-ABPP technology. Competition of IA-alkyne labeling by reversibly binding DGS is preserved throughout the workflow by the irreversibly binding probe. DTB: desthiobiotin. (B) Volcano plot for the isoDTB-ABPP experiment with 100 μM DGS used as the competitor. Significantly competed cysteines that are discussed in the text are highlighted in red. Other liganded cysteines that are included in the concentration-dependent analysis (C) are highlighted in blue. The grey lines indicate cut-offs at −log10(p) = 1.3 and log2(R) = ±2 that were used as a criterion for hit selection. (C) Dependence of the degree of competition in isoDTB-ABPP experiments on the DGS concentration. Data points are measured values from isoDTB-ABPP experiments and lines are non-linear dose–response curve fits. The values in parentheses are the EC50 values of the competition for the respective cysteine. EC50 values with confidence intervals are given in Table S2 (ESI†). In (B) and (C), for each indicated cysteine, the name or the UniProt code of the respective protein and the residue number of the competed cysteine are given. All data results from duplicates. | ||
Besides the ability to obtain residue-specific information of binding with an unmodified covalent protein ligand, another important advantage of this approach is that also cysteines, which are reversibly engaged by DGS, are identified.14 Satisfyingly, treatment of S. aureus lysate with various concentrations of DGS ranging from 100 μM to 10 μM led to the identification of 151 cysteines in 114 proteins targeted by DGS (Fig. 2B, Fig. S4 and Table S1, ESI†). Interestingly, 96 of these cysteines were not liganded by any of 19 covalent α-chloro-acetamides that were previously used to profile cysteines in S. aureus.19 This indicates that the reversibly covalent α-cyano-acrylamide is able to interact with a unique set of cysteines. Many cysteines respond to DGS treatment in a clearly concentration-dependent manner (Fig. 2C and Tables S1, S2, ESI†). For the 98 cysteines that are liganded by DGS and quantified at all concentrations, the data could be fitted with a dose–response curve with a median R2 value of 0.92. This demonstrates that residue-specific proteomics with the isoDTB tags using a few different concentrations of a covalent protein ligand is able to deliver robust information on the binding affinity for many cysteines in parallel.
31 of the targeted cysteines belong to proteins with an essential function for the viability of S. aureus. DGSe.g. binds cysteine C100 of the bifunctional protein GlmU (EC50 = 22.3 μM, UniProt Code Q2G0S3), which is located close to the magnesium ion in the UDP-GlcNAc binding site of this protein.21 GlmU is an essential protein in the synthesis of UDP–GlcNAc and therefore important in e.g. cell wall biosynthesis.22DGS also binds cysteine C38 of the ribulose-5-phosphate reductase TarJ (EC50 = 33.4 μM, UniProt Code Q2G1B9), which is involved in the binding of the catalytic zinc ion.21 TarJ is essential in the synthesis of poly(ribitol phosphate)teichoic acid, which are the main cell wall teichoic acids of S. aureus.23 Furthermore, modification of the catalytic nucleophile C165 of the 3-oxoacyl-[acyl-carrier-protein] synthase FabF (EC50 = 35.3 μM, UniProt Code Q2FZR9), an enzyme essential for lipid biosynthesis, was detected.21,24 In this way, interaction of DGS with cysteine residues in functionally relevant binding sites of several essential proteins could be a major driver of its antibiotic activity.
To further complement these results, we performed a global analysis of cellular protein levels in response to DGS treatment. Cells were cultivated with ½MIC concentration of DGS (in order to prevent premature cell lysis) and investigated by LC-MS/MS via LFQ. In line with the multiple targets observed by isoDTB-ABPP, a large fraction of 33 proteins was upregulated and only nine proteins were downregulated (Fig. 3A and Table S1, ESI†). Of the dysregulated proteins seven are essential for S. aureus survival, five of which were down-regulated ribosomal proteins. Analysis of the Gene Ontology terms of the upregulated proteins revealed a strong enrichment of terms associated with histidine biosynthesis (Fig. 3B). Together with the isoDTB-ABPP data these results suggest a polypharmacological mode-of-action for DGS leading to the alteration of multiple essential pathways, which is reflected by complex changes in the proteome.
In summary, we identified degrasyn as a novel antibiotically active compound against S. aureus including clinically isolated MRSA strains. In line with the essential role of the α-cyanoacrylamide as Michael acceptor covalent capture of target proteins failed due to limited stability of the linkage throughout the chemoproteomic protocol. While reversible covalent inhibitors are very promising, this is one technological drawback. Competitive residue-specific proteomics using the isoDTB-ABPP method turned out as an excellent complementary strategy to identify the targeted cysteines of degrasyn in vitro. 31 of these cysteines were labelled in proteins essential for viability of S. aureus highlighting a polypharmacological mode-of-action which was further corroborated by the up- or down-regulation of several proteins in a whole proteome study. While for degrasyn significant human toxicity is a challenge (IC50 = 3.4 ± 0.3 μM in an MTT assay in A549 cells), the high antibacterial potency obtained by screening a small compound library encourages the repurposing of other human inhibitors or drugs for exploiting their antibacterial potential.
SAS acknowledges funding by the Center for Integrated Protein Science (CIPSM) and the European Research Council (grant agreement No. 725085, CHEMMINE, ERC consolidator grant), SMH by the Fonds der Chemischen Industrie (Liebig Fellowship) and the TUM Junior Fellow Fund and KML by the Korea Research Institute of Chemical Technology (KRICT).
Conflicts of interest
There are no conflicts to declare.Notes and references
- A. Cassini, et al. , Lancet. Infect. Dis., 2019, 19, 56–66 CrossRef PubMed.
- M. Lakemeyer, W. Zhao, F. A. Mandl, P. Hammann and S. A. Sieber, Angew. Chem., Int. Ed., 2018, 57, 14440–14475 CrossRef CAS PubMed.
- A. Miro-Canturri, R. Ayerbe-Algaba and Y. Smani, Front. Microbiol., 2019, 10, 41 CrossRef PubMed.
- L. Peyclit, S. A. Baron and J. M. Rolain, Front. Cell. Infect. Microbiol., 2019, 9, 193 CrossRef PubMed.
- P. Le, et al. , Nat. Chem. DOI:10.1038/s41557-019-0378-7.
- L. F. Peterson, et al. , Blood, 2015, 125, 3588–3597 CrossRef CAS PubMed.
- G. A. Bartholomeusz, et al. , Blood, 2007, 109, 3470–3478 CrossRef CAS PubMed.
- M.-E. Charbonneau, et al. , PLoS One, 2014, 9, e104096–e104096 CrossRef PubMed.
- K. M. Burkholder, et al. , Infect. Immun., 2011, 79, 4850–4857 CrossRef CAS PubMed.
- K. D. Passalacqua, et al. , Antimicrob. Agents Chemother., 2016, 60, 4183–4196 CrossRef CAS PubMed.
- J. Vomacka, et al. , Chemistry, 2016, 22, 1622–1630 CrossRef CAS PubMed.
- K. M. Backus, et al. , Nature, 2016, 534, 570–574 CrossRef CAS PubMed.
- J. M. Bradshaw, et al. , Nat. Chem. Biol., 2015, 11, 525–531 CrossRef CAS PubMed.
- K. Senkane, et al. , Angew. Chem., Int. Ed., 2019, 58, 11385–11389 CrossRef CAS PubMed.
- M. J. Evans and B. F. Cravatt, Chem. Rev., 2006, 106, 3279–3301 CrossRef CAS PubMed.
- P. P. Geurink, L. M. Prely, G. A. van der Marel, R. Bischoff and H. S. Overkleeft, Top. Curr. Chem., 2012, 324, 85–113 CrossRef CAS PubMed.
- V. V. Rostovtsev, L. G. Green, V. V. Fokin and K. B. Sharpless, Angew. Chem., Int. Ed., 2002, 41, 2596–2599 CrossRef CAS PubMed.
- J. Cox, et al. , Mol. Cell. Proteomics, 2014, 13, 2513–2526 CrossRef CAS PubMed.
- P. R. A. Zanon, L. Lewald and S. M. Hacker, Angew. Chem., Int. Ed. DOI:10.1002/anie.201912075.
- S. M. Hacker, et al. , Nat. Chem., 2017, 9, 1181–1190 CrossRef CAS PubMed.
- The UniProt Consortium, Nucleic Acids Res., 2019, 47, D506–D515 CrossRef PubMed.
- M. Wang, et al. , RSC Adv., 2017, 7, 13858–13867 RSC.
- E. W. Sewell and E. D. Brown, J. Antibiot., 2014, 67, 43–51 CrossRef CAS PubMed.
- Y. Wang and S. Ma, ChemMedChem, 2013, 8, 1589–1608 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Supporting scheme, figures and tables, experimental procedures and compound characterization. See DOI: 10.1039/c9cc09204h |
| This journal is © The Royal Society of Chemistry 2020 |