Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Conformationally rigid pyrazoloquinazoline α-amino acids: one- and two-photon induced fluorescence†
Jonathan D.
Bell
 a,
Alexander H.
Harkiss
a,
David
Nobis
a,
Alexander H.
Harkiss
a,
David
Nobis
 a,
Eilidh
Malcolm
a,
Astrid
Knuhtsen
a,
Christopher R.
Wellaway
a,
Eilidh
Malcolm
a,
Astrid
Knuhtsen
a,
Christopher R.
Wellaway
 b,
Andrew G.
Jamieson
b,
Andrew G.
Jamieson
 a,
Steven W.
Magennis
a,
Steven W.
Magennis
 a and
Andrew
Sutherland
a and
Andrew
Sutherland
 *a
*a
aWestCHEM, School of Chemistry, The Joseph Black Building, University of Glasgow, Glasgow, G12 8QQ, UK. E-mail: Andrew.Sutherland@glasgow.ac.uk
bMedicinal Chemistry, GlaxoSmithKline Medicines Research Centre, Gunnels Wood Road, Stevenage, SG1 2NY, UK
First published on 13th January 2020
Abstract
The synthesis and photophysical properties of a new class of α-amino acid bearing a rigid pyrazoloquinazoline chromophore are described. Confromational constraint of the amino acid side-chains resulted in high emission quantum yields, while the demonstration of two-photon-induced fluorescence via near-IR excitation signifies their potential for sensitive bioimaging applications.
Fluorescence spectroscopy is a powerful technique for studying biological structure and function and for visualizing molecular interactions and intracellular processes.1 This is due to its high sensitivity and information content, its multiplexing capability, the ability to probe processes over a wide time range, and the availability of an array of versatile fluorescent probes. The investigation of cellular functions such as enzyme mechanisms and protein–protein interactions have often utilized extrinsic fluorescent labels such as green fluorescent protein (GFP) and its derivatives2 or intrinsic labels such as the proteinogenic fluorescent α-amino acids, tyrosine or tryptophan.3 However, the attachment of a fluorescent protein can alter the stability and functionality of the fusion partner, while tyrosine and tryptophan have poor optical properties and the occurrence of multiple residues in different environments can complicate their analysis.3
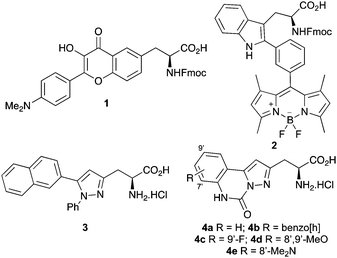
In recent years these limitations have been overcome by the development of unnatural fluorescent α-amino acids.4 The photophysical properties of such amino acids can be tuned for a particular application and they can be incorporated into proteins and peptides by solid phase peptide synthesis (SPPS) or by unnatural amino acid mutagenesis.5 Of particular interest are amino acids with side-chain chromophores that can be easily incorporated into peptides and proteins with minimal disruption to structure and function.4,6 While some of these fluorescent α-amino acids have found application in biological and medicinal imaging, an inherent issue of small chromophores is that ultraviolet (UV) light is required for excitation. Prolonged exposure to UV light causes cell and tissue damage through photo-bleaching. In recent years, this limitation has been overcome by the use of two-photon fluorescence microscopy, which permits excitation of the chromophore at longer wavelength, allowing long-term imaging of biological specimens, deeper tissue penetration, with minimization of photo-bleaching and photo-toxicity.7 While this technique has been utilized with a wide range of fluorophores and applied to the imaging of various biological processes and diseases,8 there are relatively few reports with unnatural α-amino acids. Examples include the seminal work of Mély and co-workers who used two-photon excitation of peptides containing 3-hydroxyflavone derived α-amino acids (e.g.1, Fig. 1) for monitoring interactions with cell membranes and intracellular RNA,9 while the Vendrell group used two-photon excitation of a cyclic peptide bearing a tryptophan-BODIPY conjugate (2) for the visualization of fungal infections.10
We previously reported the synthesis of 5-arylpyrazole-derived α-amino acids such as 3 (Fig. 1).11 These compounds possessed some interesting photophysical properties however, the rotational flexibility between the aryl and pyrazole rings resulted in low quantum yields and brightness. As conformationally rigid, molecular structures permit more efficient π-overlap and subsequently brighter chromophores,12 we proposed that conformational restriction of these 5-arylpyrazole motifs would lead to α-amino acids with enhanced photophysical properties. Here we describe a new class of α-amino acid (4), bearing planar pyrazoloquinazoline chromophores. We also report the photophysical properties of these compounds, demonstrating how conformational restraint leads to fluorophores with high quantum yields and spectroscopic properties that are sensitive to environment polarity. We also show the application of these α-amino acids to two-photon excitation, for potential use in imaging UV sensitive, biological systems.
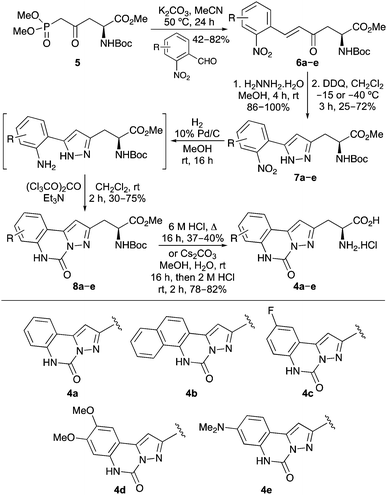
A synthetic route for the preparation of pyrazoloquinazoline-containing α-amino acids 4 was developed from L-aspartic acid. Initially, L-aspartic acid was converted to phosphononorvaline derivative 5 in five steps and 84% overall yield using previously described methods.13,14 A range of 2-nitrobenzaldehydes was reacted with phosphononorvaline 5via a Horner–Wadsworth–Emmons reaction, which gave the E-isomer of enones 6a–e in 42–82% yields (Scheme 1).14,15 The enone moiety was used to construct the pyrazole ring by reaction of 6a–e with hydrazine via a one-pot condensation/aza-Michael process. Low temperature oxidation of the resulting 2-pyrazolines with DDQ gave 5-(2-nitroaryl)pyrazoles 7a–e.11 Carbonylation was then achieved by reduction of the nitroaryl moiety, followed by reaction with triphosgene under basic conditions.16 This gave pyrazoloquinazolines 8a–e in 30–75% yields over the two steps. Deprotection to the parent amino acids was either conducted in one step under acidic conditions (4b, 37%; 4c, 40%) or more efficiently by a two-step strategy involving ester hydrolysis, followed by removal of the Boc-protecting group under milder acidic conditions (4a, 80%; 4d, 82%; 4e, 78%).
The UV/Vis absorption and emission spectra of the amino acids were recorded in methanol at a concentration of 0.5 × 10−5 M (Fig. 2 and Table 1).17 As expected for chromophores that are planar in the ground state, the amino acids, particularly 4a–d, showed structured absorption spectra with well-resolved vibrational bands (Fig. 2a).18 The emission spectra of amino acids 4a–e, following one-photon excitation at the longest wavelength absorption band, showed strong fluorescence ranging from 342–414 nm, with a vibrational progression which mirrored that of the absorption spectra (Fig. 2b).19 Amino acid 4e bearing the electron rich dimethylamino substituent gave a red-shifted, structureless emission maximum at 414 nm, which is indicative of a combination of locally excited and internal charge transfer states.20 More importantly, the quantum yields of 4a–e were characteristic of conformational restriction and showed a ten-fold increase in comparison to the non-rigid pyrazole-derived amino acid 3. As a result, these α-amino acids are substantially brighter than the corresponding non-rigid systems.
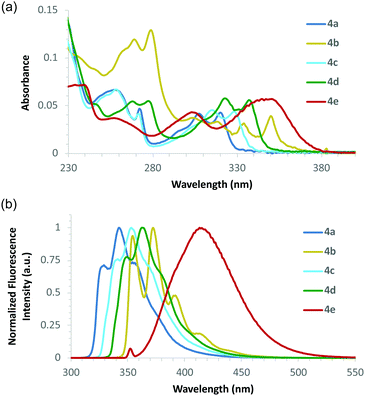 | ||
| Fig. 2 (a) Absorption spectra of amino acids 4a–4e recorded at 0.5 × 10−5 M in MeOH. (b) Emission spectra of 4a–4e at 0.5 × 10−5 M in MeOH. | ||
| Amino acid | λ Abs (nm) | ε (cm−1 M−1) | λ Em (nm) | Φ F | Brightnessc (cm−1 M−1) |
|---|---|---|---|---|---|
| a All spectra were recorded at 0.5 × 10−5 M in MeOH. b Quantum yields (ΦF) were determined in MeOH using anthracene and L-tryptophan as standards. c Brightness was calculated as the product of the QY and the molar absorptivity at the wavelength maximum specified. | |||||
| 3 | 249 | 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 700 |
356 | 0.04 | 950 |
| 4a | 321 | 8300 | 342 | 0.42 | 3500 |
| 4b | 350 (279) | 7700 (24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800) 800) |
372 (373) | 0.47 (0.27) | 3600 (6700) |
| 4c | 328 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
353 | 0.47 | 4700 |
| 4d | 337 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 200 |
363 | 0.56 | 6300 |
| 4e | 349 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 200 |
414 | 0.48 | 5900 |
As amino acid 4e showed the most interesting photophysical properties, this compound was further investigated. The fluorescence decay was measured at an emission wavelength of 430 nm in methanol using time-correlated single-photon counting, following excitation at 390 nm (see ESI†). Three decay components were fitted with lifetimes of 0.79, 2.17 and 7.90 ns, and A-factors of 0.60, 0.36 and 0.04, respectively. This suggests the molecule exists predominantly in two states, as has been observed previously for dimethylamino-substituted fluorophores.21 The amplitude-weighted average lifetime is 1.57 ns, which is comparable to fluorophores typically used in fluorescence lifetime imaging microscopy (FLIM).22
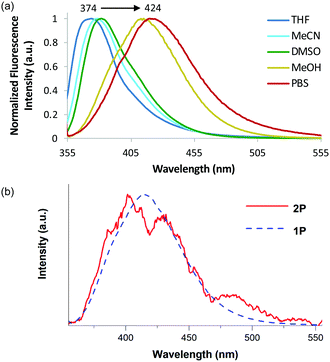
To assess the environmental sensitivity of amino acid 4e, its photophysical properties were examined in a range of solvents. The absorption maxima (342–348 nm) were found to be independent of polarity (see ESI†), indicating negligible intramolecular interaction between the electron-rich dimethylamino and electron-deficient N-acylpyrazole moieties in the ground state. In contrast, the emission spectra were found to be highly sensitive to the solvent used, with increasing polarity leading to concomitantly broadened, structureless emission spectra at longer wavelengths (Fig. 3a).23,24 For example, in THF, an emission maximum at 374 nm was observed, while in phosphate-buffered saline (PBS), the emission maximum was found at 424 nm. These spectral features and strong solvatochromism are likely due to increasing solvent stabilization of both the locally excited and intramolecular charge transfer states.
Application of amino acid 4e for two-photon spectroscopy was next investigated. Two- and three-photon excitation has recently been shown to be a promising approach for the ultrasensitive analysis of fluorescent nucleobases, suggesting a similar approach may be applicable to α-amino acids.25 Two-photon absorption was confirmed with a quadratic dependence of the emitted intensity on the excitation intensity (see ESI†). Furthermore, the molecule has no one-photon absorption in the near IR, precluding linear excitation. The emission spectra of amino acid 4e in methanol after one- and two-photon excitation have a similar profile (Fig. 3b). The two-photon cross section was measured to be 0.38 ± 0.06 GM using rhodamine B as a reference compound (see ESI†). This value is comparable to other molecules of similar size.8d,e Thus, 4e can undergo two-photon absorption using near-IR excitation, thereby avoiding the use of UV excitation and the issues of photobleaching.
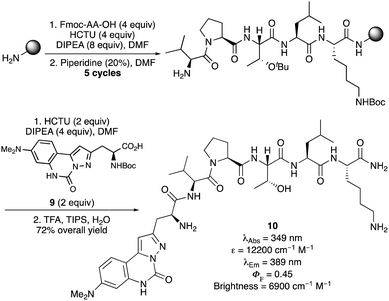
The potential of these amino acids for incorporation into peptides via SPPS methodology was also investigated. A model pentapeptide (Val-Pro-Thr-Leu-Lys), based on the Bax-binding domain of Ku70, a multifunctional protein involved in DNA repair and cell-death regulation was chosen.26 This pentapeptide and similar analogues have been shown to have low cytotoxicity and are effective at penetrating living cells.26b The pentapeptide was prepared in an automated, peptide synthesizer using TentaGelTM S Rink Amide (RAM) resin as the polymer support27 and routine SPPS methodology (Scheme 2). Coupling of each Fmoc-protected amino acid was performed by HCTU28 activation, followed by piperidine-mediated N-deprotection, which gave the N-terminal unprotected pentapeptide. Boc-Protected pyrazoloquinazoline-derived amino acid 9 (see ESI† for preparation) was then coupled manually onto the polymer-supported pentapeptide. A TFA cleavage cocktail was used to remove the protecting groups and release the hexapeptide from the polymer support. Purification by reverse phase-HPLC gave hexapeptide 10 in 72% yield with >95% purity. Analysis of the photophysical properties of hexapeptide 10 showed that apart from a small hypsochromic shift in the emission spectra (Fig. 4), the key photophysical properties of α-amino acid 4e were retained, including a high quantum yield (0.45) and brightness value (6900 cm−1 M−1).
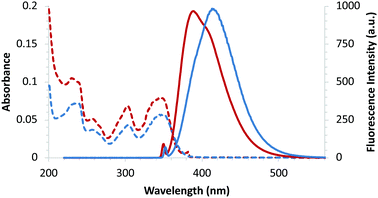 | ||
| Fig. 4 Absorption (dotted lines) and emission (solid lines) spectra of amino acid 4e (blue) and hexapeptide 10 (red), recorded at 0.5 × 10−5 M in MeOH. | ||
In summary, a new class of conformationally rigid, unnatural α-amino acid has been synthesized using a highly regioselective olefination reaction to introduce side-chain diversity, followed by a one-pot condensation/aza-Michael reaction and carbonylation process to form the key pyrazoloquinazoline ring system. Although containing relatively small side-chains, these compounds were found to be strongly fluorescent with high quantum yields (42–56%). The potential of amino acid 4e as a sensitive bio-imaging tool was demonstrated with successful excitation of this compound at 700 nm using two-photon spectroscopy. Furthermore, amino acid 4e was efficiently incorporated into a biologically relevant peptide as part of a SPPS process. Based on the exciting optical properties of this new class of α-amino acid, current work is investigating the introduction of these minimally invasive compounds into other biologically relevant peptides for in vitro and in vivo spectroscopic and microscopic applications.
Financial support from EPSRC (studentships to J. D. B., EP/N509176/1 and A. H. H., EP/K503058/1), University of Glasgow (studentship to D. N.) and GlaxoSmithKline is gratefully acknowledged.
Conflicts of interest
There are no conflicts to declare.Notes and references
- For some recent reviews, see: (a) L.-Y. Niu, Y.-Z. Chen, H.-R. Zheng, L.-Z. Wu, C.-H. Tung and Q.-Z. Yang, Chem. Soc. Rev., 2015, 44, 6143 RSC; (b) A. B. Chinen, C. M. Guan, J. R. Ferrer, S. N. Barnaby, T. J. Merkel and C. A. Mirkin, Chem. Rev., 2015, 115, 10530 CrossRef CAS PubMed; (c) X. Chen, F. Wang, J. Y. Hyun, T. Wei, J. Qiang, X. Ren, I. Shin and J. Yoon, Chem. Soc. Rev., 2016, 45, 2976 RSC.
- (a) R. Y. Tsien, Annu. Rev. Biochem., 1998, 67, 509 CrossRef CAS PubMed; (b) R. Heim, A. B. Cubitt and R. Y. Tsien, Nature, 1995, 373, 663 CrossRef CAS PubMed.
- R. W. Sinkeldam, N. J. Greco and Y. Tor, Chem. Rev., 2010, 110, 2579 CrossRef CAS PubMed and references therein.
- For recent reviews, see: (a) A. T. Krueger and B. Imperiali, ChemBioChem, 2013, 14, 788 CrossRef CAS PubMed; (b) R. Kubota and I. Hamachi, Chem. Soc. Rev., 2015, 44, 4454 RSC; (c) A. H. Harkiss and A. Sutherland, Org. Biomol. Chem., 2016, 14, 8911 RSC.
- For examples, see: (a) A. Chatterjee, H. Xiao, P.-Y. Yang, G. Soundararajan and P. G. Shultz, Angew. Chem., Int. Ed., 2013, 52, 5106 CrossRef CAS; (b) S. Chen, N. E. Fahmi, L. Wang, C. Bhattacharya, S. J. Benkovic and S. M. Hecht, J. Am. Chem. Soc., 2013, 135, 12924 CrossRef CAS PubMed; (c) L. C. Speight, A. K. Muthusamy, J. M. Goldberg, J. B. Warner, R. F. Wissner, T. S. Willi, B. F. Woodman, R. A. Mehl and E. J. Petersson, J. Am. Chem. Soc., 2013, 135, 18806 CrossRef CAS PubMed.
- For recent examples, see: (a) P. Cheruku, J.-H. Huang, H.-J. Yen, R. S. Iyer, K. D. Rector, J. S. Martinez and H.-L. Wang, Chem. Sci., 2015, 6, 1150 RSC; (b) M. R. Hilaire, I. A. Ahmed, C.-W. Lin, H. Jo, W. F. DeGrado and F. Gai, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 6005 CrossRef CAS PubMed; (c) M. Arribat, E. Rémond, S. Clément, A. Van Der Lee and F. Cavelier, J. Am. Chem. Soc., 2018, 140, 1028 CrossRef CAS PubMed; (d) A. H. Harkiss, J. D. Bell, A. Knuhtsen, A. G. Jamieson and A. Sutherland, J. Org. Chem., 2019, 84, 2879 CrossRef CAS.
- A. Diaspro, G. Chirico and M. Collini, Q. Rev. Biophys., 2005, 38, 97 CrossRef CAS.
- (a) R. Yuste and W. Denk, Nature, 1995, 375, 682 CrossRef CAS PubMed; (b) C. Xu, W. Zipfel, J. B. Shear, R. M. Williams and W. W. Webb, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 10763 CrossRef CAS PubMed; (c) W. R. Zipfel, R. M. Williams, R. Christie, A. Y. Nikitin, B. T. Hyman and W. W. Webb, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 7075 CrossRef CAS PubMed; (d) R. S. K. Lane and S. W. Magennis, RSC Adv., 2012, 2, 11397 RSC; (e) R. S. K. Lane, R. Jones, R. W. Sinkeldam, Y. Tor and S. W. Magennis, ChemPhysChem, 2014, 15, 867 CrossRef CAS PubMed.
- (a) V. Y. Postupalenko, O. M. Zamotaiev, V. V. Shvadchak, A. V. Strizhak, V. G. Pivovarenko, A. S. Klymchenko and Y. Mély, Bioconjugate Chem., 2013, 24, 1998 CrossRef CAS PubMed; (b) M. Sholokh, O. M. Zamotaiev, R. Das, V. Y. Postupalenko, L. Richert, D. Dujardin, O. A. Zaporozhets, V. G. Pivarenko, A. S. Klymenchenko and Y. Mély, J. Phys. Chem. B, 2015, 119, 2585 CrossRef CAS.
- L. Mendive-Tapia, C. Zhao, A. R. Akram, S. Preciado, F. Alberico, M. Lee, A. Serrels, N. Kielland, N. D. Read, R. Lavilla and M. Vendrell, Nat. Commun., 2016, 7, 10940 CrossRef CAS PubMed.
- L. Gilfillan, R. Artschwager, A. H. Harkiss, R. M. J. Liskamp and A. Sutherland, Org. Biomol. Chem., 2015, 13, 4514 RSC.
- (a) M. Nepraš, N. Almonasy, F. Bureš, J. Kulhánek, M. Dvorák and M. Michl, Dyes Pigm., 2011, 91, 466 CrossRef; (b) M. S. Michie, R. Götz, C. Franke, M. Bowler, N. Kumari, V. Magidson, M. Levitus, J. Loncarek, M. Sauer and M. J. Schnermann, J. Am. Chem. Soc., 2017, 139, 12406 CrossRef CAS PubMed; (c) J. C. Anderson, C.-H. Chang, A. P. Jathoul and A. J. Syed, Tetrahedron, 2019, 75, 347 CrossRef CAS.
- D. E. Rudisill and J. P. Whitten, Synthesis, 1994, 851 CrossRef CAS.
- (a) L. S. Fowler, D. Ellis and A. Sutherland, Org. Biomol. Chem., 2009, 7, 4309 RSC; (b) L. S. Fowler, L. H. Thomas, D. Ellis and A. Sutherland, Chem. Commun., 2011, 47, 6569 RSC.
- F. Gosselin and W. D. Lubell, J. Org. Chem., 1998, 63, 7463 CrossRef CAS PubMed.
- F. Varano, D. Catarzi, V. Colotta, G. Filacchioni, A. Galli, C. Costagli and V. Carlà, J. Med. Chem., 2002, 45, 1035 CrossRef CAS.
- The absorption and emission spectra for each amino acid are presented in the ESI†.
- (a) I. B. Berlman, J. Phys. Chem., 1970, 74, 3085 CrossRef CAS; (b) N. I. Nijegorodov and W. S. Downey, J. Phys. Chem., 1994, 98, 5639 CrossRef CAS.
- It should be noted that while excitation of the absorption band at longest wavelength gave the most intense emission spectra for most of the amino acids, this was not the case for 4b. Excitation of the peak at 279 nm gave the most intense fluorescence and highest brightness value (Table 1).
- V. A. Galievsky, S. I. Druzhinin, A. Demeter, S. A. Kovalenko, T. Senyushkina, P. Mayer and K. A. Zachariasse, J. Phys. Chem. A, 2011, 115, 10823 CrossRef CAS PubMed.
- S. R. Meech and D. Phillips, J. Chem. Soc., Faraday Trans. 2, 1987, 83, 1941 RSC.
- P. Sarder, D. Maji and S. Achilefu, Bioconjugate Chem., 2015, 26, 963 CrossRef CAS PubMed.
- S. Achelle, I. Nouira, B. Pfaffinger, Y. Ramondenc, N. Plé and J. Rodríguez-López, J. Org. Chem., 2009, 74, 3711 CrossRef CAS PubMed.
- J. R. Lakowicz, Principles of Fluorescence Spectroscopy, Springer, New York, 2006 Search PubMed.
- (a) D. Nobis, R. S. Fisher, M. Simmermacher, P. A. Hopkins, Y. Tor, A. C. Jones and S. W. Magennis, J. Phys. Chem. Lett., 2019, 10, 5008 CrossRef CAS PubMed; (b) R. S. Fisher, D. Nobis, A. F. Füchtbauer, M. Bood, M. Grøtli, L. M. Wilhelmsson, A. C. Jones and S. W. Magennis, Phys. Chem. Chem. Phys., 2018, 20, 28487 RSC.
- (a) J. A. Downs and S. P. Jackson, Nat. Rev. Mol. Cell Biol., 2004, 5, 367 CrossRef CAS; (b) J. A. Gomez, J. Chen, J. Ngo, D. Hajkova, I.-J. Yeh, V. Gama, M. Miyagi and S. Matsuyama, Pharmaceuticals, 2010, 3, 3594 CrossRef CAS.
- W. Li and B. Yan, J. Org. Chem., 1998, 63, 4092 CrossRef CAS.
- HCTU: O-(1-H-6-Chlorobenzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures, characterisation data for compounds, photophysical data for amino acids and, NMR spectra for all novel compounds. See DOI: 10.1039/c9cc09064a |
| This journal is © The Royal Society of Chemistry 2020 |