Nanogel hybrid assembly for exosome intracellular delivery: effects on endocytosis and fusion by exosome surface polymer engineering†
Shin-ichi
Sawada‡
 ,
Yuko T.
Sato‡
,
Riku
Kawasaki
,
Jun-ichi
Yasuoka
,
Ryosuke
Mizuta
,
Yoshihiro
Sasaki
and
Kazunari
Akiyoshi
*
,
Yuko T.
Sato‡
,
Riku
Kawasaki
,
Jun-ichi
Yasuoka
,
Ryosuke
Mizuta
,
Yoshihiro
Sasaki
and
Kazunari
Akiyoshi
*
Department of Polymer Chemistry, Graduate School of Engineering, Kyoto University, Katsura, Nishikyo-ku, Kyoto 615-8510, Japan. E-mail: akiyoshi@bio.polym.kyoto-u.ac.jp
First published on 27th November 2019
Abstract
Surface polymer engineering was applied with a carrier of exosomes, namely, the amphiphilic cationic CHP (cCHP) nanogel, to improve the delivery of exosome content by forming complexes with the exosomes. Mouse macrophage cells were used to produce the exosomes, which were then mixed with the cCHP nanogel to form a hybrid. Transmission electron microscopy revealed that the surface of each exosome was coated with cCHP nanogel particles. Flow cytometry also showed a significant uptake of this exosome/nanogel hybrid by HeLa cells, with the main mechanism behind this internalization being endocytosis. A range of different molecules that inhibit different types of endocytosis were also applied to determine the particular pathway involved, with a caveolae-mediated endocytosis inhibitor being revealed to markedly affect the hybrid uptake. Next, we evaluated the fate of the internalized hybrid using fluorescent labeling, with the results suggesting fusion between endosomes and exosomes. Finally, revealing the functional efficacy of this approach, we showed that the nanogel system could successfully deliver functional exosomes into cells, as indicated by its ability to induce neuron-like differentiation in the recipient cells. Overall, our findings show the potential of using this hybrid nanocarrier system for transporting various contents in exosomes and ensuring their effective delivery in a functionally intact state.
Introduction
Recently, extracellular vesicles (EVs) secreted by various cells have attracted attention as a new system in cell-to-cell communication. In general, vesicles produced from the endocytic compartments called multivesicular bodies (MVBs) are referred to as exosomes (with sizes from 30 to 200 nm), while vesicles budding or shedding directly from the plasma membrane are referred to as microvesicles (with sizes from 100 to 1000 nm).1,2 Exosomes can contain proteins (cytosolic and membrane proteins) and nucleic acids (mRNA, siRNA, miRNA, and DNA), and play a key role in biological phenomena such as the immune system, inflammation, tumor metastasis, neurogenesis, aging, and infection.3,4 Several miRNAs inside exosomes released from disease-related cells have been used as biomarkers of diseases.5,6 Because of their ability to transport physiologically active substances to various tissues and cells, exosomes are considered to be effective natural nanocarriers.7,8 Recently, medical applications using exosomes for a drug delivery system (DDS) have progressed by loading miRNA/siRNA or hydrophobic drugs such as doxorubicin,9 curcumin,10 and paclitaxel.11 To effectively apply exosomes to DDS, surface modifications of exosomes produced by several methods have been reported because unmodified exosomes did not always show tissue targetability and were taken up by the reticuloendothelial system (RES) upon intravenous injection, similar to liposomes.12A typical method for the surface modification of exosomes is genetic engineering. To target brain cells, Erviti et al. successfully prepared dendritic cells to express membrane proteins fused with a neuron-specific peptide.13 Moreover, Kuroda et al. reported that exosomes expressing a peptide on their membrane to target the epidermal growth factor receptor (EGFR), which is highly expressed in cancer cells, were efficiently incorporated into cells.14 Tian et al. carried out a study in which the anticancer drug doxorubicin was complexed into exosomes secreted from immature mouse dendritic cells expressing the integrin-binding peptide fused with LAMP-2b proteins. It has also been reported from both in vitro and in vivo studies that doxorubicin-loaded exosomes specifically accumulate in cancer cells with high expression of integrin, resulting in a decrease in the number of cancer cells and suppression of their growth due to doxorubicin.9 In addition, the stabilization of the peptide fused to LAMP-2b proteins on the exosome surface was also reported. Glycosylation of the peptide-LAMP-2b-fused proteins was inhibited by degradation of the peptide.15 Furthermore, an exosome surface display technology achieved by expressing functional peptide-fused tetraspanins as well as LAMP-2b proteins in exosome-loaded cells was also established, which enabled the display of the peptide on both the outside and the inside of exosomes to design various chimeric tetraspanins.16 Kooijmans et al. also embedded anti-epidermal growth factor receptor nanobodies fused to glycosylphosphatidylinositol anchor signal peptides derived from decay-accelerating factor into the EV membrane.17 More recently, a new method has also been proposed for modifying nanobodies to phosphatidylserine (PS) on EV membrane surfaces using recombinant fusion proteins of nanobodies fused to phosphatidylserine (PS)-binding domains of lactadherin (C1C2).18
Other concepts have also been developed regarding new functionalization for exosomes using functional materials designed and synthesized artificially. For example, reports have been published on methods for labeling on the surface of exosomes using click chemistry based on the chemical modification of an alkyne as a membrane protein of exosomes or the incorporation of alkyne-modified lipids into the exosome membrane.19 Moreover, the modification of the exosome membrane with cationic lipids or polymers from outside could enhance incorporation into cells.20,21 In contrast, modified EVs were prepared by engineering to produce cells using membrane fusogenic liposomes that included drugs and functional lipids.22 A method of directly loading targeting ligands onto exosomes was also reported. Munagala et al. incorporated tumor-targeting ligands such as folic acid into exosomes by mixing the agent dissolved in organic solvents with exosome suspension.23 Moreover, Qi et al. developed an exosome delivery system using a magnetic field and transferrin-conjugated superparamagnetic nanoparticles.24
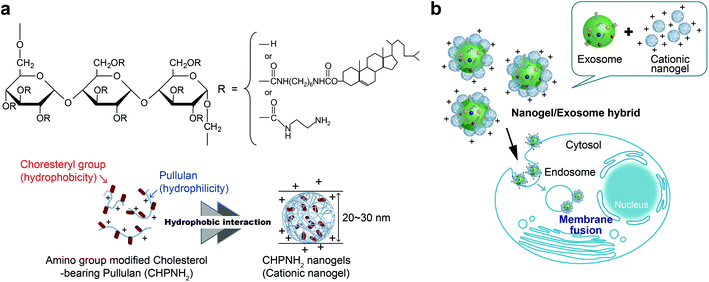
We propose here a new strategy for the effective delivery of exosomes into cells using functional macromolecular carriers such as amphiphilic nanogels. Specifically, we developed self-assembled nanogel engineering for a system for delivering biologics for application to nanomedicine and tissue engineering.25–27 Polysaccharide amphiphilic self-assembled nanogels applied to antigen delivery for a cancer vaccine (under clinical trial) and cationic nanogels act as effective nanocarriers for proteins25,28,29 and nucleic acids (plasmid or siRNA).30 In particular, the self-assembly of ethylenediamine-modified cholesteryl pullulan (cationic CHP) provided an effective intracellular protein delivery system with molecular chaperone-like activity, which enabled the capture of various protein and peptide molecules within the polymer matrix and their release in their native form.25 The cationic CHP (cCHP) nanogels were effective at penetrating the nasal mucosa and resulted in successful nasal vaccines.31,32 Furthermore, we reported that the liposome surface was coated with CHP nanogels mainly due to hydrophobic interaction between the cholesteryl group of CHP and the lipid domain of the liposome.33 In this report, we describe a novel exosome intracellular delivery system using cCHP nanogels (Fig. 1).
Experimental
Materials
Ethylenediamine-modified CHP (CHPNH2) (Fig. 1) was synthesized as reported previously.28 The degrees of substitution of the cholesteryl groups and amino groups were 1.2 and 15 per 100 anhydrous glucoside units. Anti-Hsc70 [anti-Hsc70 rat monoclonal (1B5)], anti-CD9 [anti-CD9 rabbit monoclonal (EPR2949)], anti-nerve-specific enolase (anti-NSE rabbit polyclonal), Alexa Fluor®488-conjugated anti-rabbit IgG, horseradish peroxidase (HRP)-conjugated anti-rat IgG, and HRP-conjugated anti-rabbit IgG were obtained from Enzo Life Sciences (NY, USA), Abcam (Cambridge, UK), Santa Cruz Biotechnology (CA, USA), and Biolegend (CA, USA). ECL western blotting detection reagent and PD SpinTrap G-25 were purchased from GE Healthcare (NY, USA). Transferrin receptor mouse mAb (H68.4), Lysotracker-Red, 3-octadecyl-2-[3-(3-octadecyl-2(3H)-benzoxazolylidene)-1-propenyl] perchlorate (DIO), 3,6-bis(diethylamino)-9-(2-octadecyloxy) carbonyl phenyl chloride (R18), and live cell imaging solution were purchased from Thermo Fisher Scientific (MA, USA). Stain buffer (FBS) was purchased from BD Biosciences (CA, USA). 5- or 6-(N-succinimidyloxycarbonyl)-fluorescein 3′,6′-diacetate (CFSE) was obtained from Dojindo Molecular Technologies, Inc. (Kumamoto, Japan). VECTASHIELD was obtained from Vector Laboratories Inc. (CA. USA).Cell culture
The mouse macrophage-like cell line Raw264.7 [RCB0535; Japanese Collection of Research Bioresources Cell Bank (JCRB Cell Bank)] was cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Thermo Fisher Scientific). HeLa cells (JCRB9004; JCRB Cell Bank) were cultured in minimal essential medium [MEM; Sigma-Aldrich (MO, USA)] supplemented with 10% FBS, 1% non-essential amino acids (NEAA; Sigma-Aldrich), and penicillin–streptomycin (Thermo Fisher Scientific). The rat pheochromocytoma cell line PC12HS (JCRB0266; JCRB Cell Bank), which is a clone highly sensitive to nerve growth factor (NGF), was cultured in DMEM supplemented with 10% heat-inactivated horse serum (Thermo Fisher Scientific) and penicillin–streptomycin. The human embryonic kidney cell line HEK293sus [CRL-1573; American Type Culture Collection (ATCC)] was cultured in 293 SFM II medium (Thermo Fisher Scientific) supplemented with 2% GlutaMAX™-I (Thermo Fisher Scientific). Human adipose-derived mesenchymal stem cells (hADSCs; Lonza) were cultured in ADSC basal medium (Lonza) with a poly-L-lysine-coated glass-bottomed dish. All cells were cultured in a 37 °C, 5% CO2, humidified incubator.Exosome isolation and lipid analysis
The cells were maintained in a medium with exosome-free FBS from which the FBS-derived exosomes had been removed by centrifugation at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 h. The culture supernatant was collected and then centrifuged at 1200g for 20 min and at 12
000g for 15 h. The culture supernatant was collected and then centrifuged at 1200g for 20 min and at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 min at 4 °C. After 0.22 μm filtration, it was ultra-centrifuged at 120
000g for 30 min at 4 °C. After 0.22 μm filtration, it was ultra-centrifuged at 120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 70 min at 4 °C. The pellets were rinsed with phosphate-buffered saline (PBS; Thermo Fisher Scientific), ultra-centrifuged again, and then dispersed in PBS. Protein concentrations were determined using BCA assay (Thermo Fisher Scientific). Protein concentrations of exosomes were adjusted to 300 μg mL−1. The amount of lipid was measured using Phospholipid C-Test Wako (Wako Pure Chemicals, Osaka, Japan).
000g for 70 min at 4 °C. The pellets were rinsed with phosphate-buffered saline (PBS; Thermo Fisher Scientific), ultra-centrifuged again, and then dispersed in PBS. Protein concentrations were determined using BCA assay (Thermo Fisher Scientific). Protein concentrations of exosomes were adjusted to 300 μg mL−1. The amount of lipid was measured using Phospholipid C-Test Wako (Wako Pure Chemicals, Osaka, Japan).
Lipids from Raw264.7 cells and Raw264.7-derived exosomes were extracted by the Bligh and Dyer method, namely, with chloroform/methanol/PBS (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.8, by vol.). Subsequently, chloroform/water (1
0.8, by vol.). Subsequently, chloroform/water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, by vol.) were added and mixed. The lower phase was collected, dried under nitrogen, and diluted in chloroform for analysis. Lipid extracts were analyzed by thin-layer chromatography (TLC) (Fig. S2†). These extracts were resolved on silica gel plates (0.25 mm thick; Merck), and lipid analysis was performed using a solvent system (chloroform/methanol/petroleum ether/acetone/acetic acid/water; 120
1, by vol.) were added and mixed. The lower phase was collected, dried under nitrogen, and diluted in chloroform for analysis. Lipid extracts were analyzed by thin-layer chromatography (TLC) (Fig. S2†). These extracts were resolved on silica gel plates (0.25 mm thick; Merck), and lipid analysis was performed using a solvent system (chloroform/methanol/petroleum ether/acetone/acetic acid/water; 120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 75
75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50
50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25
25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6.5
6.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, by vol.). The areas containing individual lipids were identified by comigration of standards and were visualized by staining with 10% copper acetate solution in 8% phosphoric acid and subsequent heating at 150 °C for 30 min. The relative amount of lipids was obtained through densitometry analysis by using JustTLC (Sweday, Södra Sandby, Sweden). Lipid analyses were conducted at the Toray Research Center, Inc. (Tokyo, Japan).
5, by vol.). The areas containing individual lipids were identified by comigration of standards and were visualized by staining with 10% copper acetate solution in 8% phosphoric acid and subsequent heating at 150 °C for 30 min. The relative amount of lipids was obtained through densitometry analysis by using JustTLC (Sweday, Södra Sandby, Sweden). Lipid analyses were conducted at the Toray Research Center, Inc. (Tokyo, Japan).
cCHP nanogel/exosome hybrid preparation
cCHP was suspended in PBS. The suspension was sonicated for 15 min with a probe sonicator (BRANSON, CT, USA) and centrifuged at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 min. The obtained supernatant was then filtered through a 0.22 μm filter (Merck Millipore, MA, USA). Exosome suspension (protein concentration of 300 μg mL−1) in PBS was added to an equal volume of cCHP nanogel suspension and mixed at various concentration ratios. The hybrid suspensions were kept for 30 min on ice.
000g for 30 min. The obtained supernatant was then filtered through a 0.22 μm filter (Merck Millipore, MA, USA). Exosome suspension (protein concentration of 300 μg mL−1) in PBS was added to an equal volume of cCHP nanogel suspension and mixed at various concentration ratios. The hybrid suspensions were kept for 30 min on ice.
Particle size and zeta-potential analysis
The particle size of cCHP nanogel/exosome hybrids with various mixture ratios in PBS (protein concentration of exosome fixed at 300 μg mL−1) was determined with a NanoSight LM10-HS instrument (NanoSight Ltd, Amesbury, UK) equipped with the nanoparticle tracking analysis (NTA) 2.3 analytical software. The exosome samples were measured with a 532 nm-wavelength laser at 25 °C. Particle suspensions were diluted with PBS to reach a concentration of 1–8 × 108 particles per milliliter for analysis. The particle size of the cCHP nanogel and the ζ-potential of hybrids were measured with a Zetasizer Nano ZS (Malvern Instruments, Worcestershire, UK). In the ζ-potential measurements, nanogel/exosome hybrids with various mixture ratios in PBS were prepared to have an exosome protein concentration of 3 μg mL−1. The detector angles were 173° for DLS measurement and 17° for ζ-potential measurement. The measured autocorrelation function was analyzed by a cumulant method. All measurements were performed at 25 °C. The results represent the means of three experiments.Immunoblotting analysis for exosome marker protein
An equal volume of sample buffer [50 mM Tris-HCl (pH 6.8), 2% SDS, 0.1% bromophenol blue, 10% glycerol, 100 mM DTT] was added to each of the samples. Proteins were separated by SDS-PAGE using a 12.5% polyacrylamide gel and transferred to PVDF membranes (ATTO, Tokyo, Japan). The membranes were treated with 0.5% skimmed milk in Tris-buffered saline with Tween-20 (TBST) [20 mM Tris-HCl (pH 7.4), 500 mM NaCl, 0.05% Tween 20] for blocking and were reacted with a primary antibody in TBST buffer. The following antibodies were used: anti-Hsc70 and anti-CD9. The membranes linked with the primary antibody were then reacted with anti-rat IgG-HRP or anti-rabbit IgG-HRP as the secondary antibody. The membrane was reacted with ECL western blotting detection reagents and bands were visualized using LAS-4000 (GE Healthcare).Transmission electron microscopy (TEM)
A total of 6 μg (protein) of exosome, 10 μg of cCHP nanogel, or 3.2 μg of cCHP/exosome hybrid (prepared at an equal weight ratio) with a volume ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.3 with 2% paraformaldehyde (PFA), or cCHP nanogel was applied on elastic carbon-coated grids (#100, ELS-C10; Okenshoji Co., Ltd, Japan) for 15 min. The samples were stained with 1% ammonium molybdate solution and observed using an HT7700 transmission electron microscope (Hitachi, Japan) at an accelerating voltage of 100 kV.
2.3 with 2% paraformaldehyde (PFA), or cCHP nanogel was applied on elastic carbon-coated grids (#100, ELS-C10; Okenshoji Co., Ltd, Japan) for 15 min. The samples were stained with 1% ammonium molybdate solution and observed using an HT7700 transmission electron microscope (Hitachi, Japan) at an accelerating voltage of 100 kV.
Flow cytometer analysis and inhibition of specific endocytic pathways
Regarding the cellular uptake of cCHP nanogel/exosome hybrids, purified Raw264.7-derived exosomes (30 μg as protein) were labeled with 62.8 μM CFSE for 30 min at 37 °C in a volume of 100 μL of PBS. Labeled exosomes were subjected to a PD Spin Trap G-25 to remove unreacted CFSE and filtered with a 0.22 μm PVDF filter. Raw264.7 exosomes were mixed with the desired amount of cCHP nanogel for 30 min. A total of 1.0 × 105 HeLa cells incubated with 5 μg (protein) of exosomes or cCHP nanogel/exosome (5 μg of each) hybrids for 4 h at 37 °C were washed twice with PBS, detached using trypsin, and resuspended in stain buffer. In an experiment using an endocytosis inhibitor, HeLa cells (1.0 × 105 cells per well) were pretreated with a clathrin-mediated endocytosis inhibitor (25 μM chlorpromazine), a macropinocytosis inhibitor [50 μM ethyl-isopropyl amiloride (EIPA)], a lysosome acidification inhibitor (50 μM chloroquine), a caveolae-mediated endocytosis inhibitor [2.5 mM methyl-β-cyclodextrin (MβCD)], or an actin polymerization inhibitor (5 μM cytochalasin D) for 30 min at 37 °C in 5% CO2. Then, the cells were cultured with fresh medium containing a hybrid of nanogel/exosome that was prepared at an equal weight ratio with one of the inhibitors for 4 h at 37 °C in 5% CO2. To investigate the energy-dependent endocytosis, low-temperature incubation (4 °C) was also carried out. The cells were washed with fresh medium, trypsinized, and suspended in stain buffer. Flow cytometry was performed on an LSR Fortessa cell analyzer with a 488 nm argon laser. The mean fluorescence of cells was measured by flow cytometry and is presented as the percentage relative to that at 37 °C or under inhibitor-free conditions.Confocal laser scanning fluorescence microscopy (CLSFM) observation
The intracellular localization of the cCHP nanogel/DIO-labeled exosome hybrids was observed using a confocal laser scanning fluorescence microscope (CLSFM). Raw264.7-derived exosomes (30 μg as protein) were labeled with 3 μM DIO for 1 h at 25 °C in a volume of 100 μL of PBS. Labeled exosomes were treated in the same way as CFSE-labeled exosomes. HeLa cells were plated in a glass-bottomed dish (Iwaki Glass, Japan) at a density of 1.0 × 105 cells per dish and cultured at 37 °C in 5% CO2 overnight. Then, 3 μg per protein of DIO-labeled exosome or cCHP nanogel/DIO-labeled exosome (3 μg of each) hybrids were added to HeLa cells, and the cells were cultured for a given period of time. Lysotracker Red was added to each dish 1 h before harvesting. The cells were fixed with 4% paraformaldehyde for 15 min, washed with PBS, and then covered with VECTASHIELD. The cells were observed using CLSFM equipped with a plan-APOCHROMAT 40× oil immersion objective (LSM780; Carl Zeiss, Germany) with excitation by using an argon laser (488 nm) and a He–Ne laser (543 nm).Fusion analysis
Purified Raw264.7-derived exosomes (30 μg as protein) were labeled with 3 μM DIO and/or R18 for 1 h at 25 °C. Labeled exosomes were subjected to a PD Spin Trap G-25 to remove unreacted dye and filtered with a 0.22 μm PVDF filter. HeLa cells were seeded on a glass-bottomed dish at a concentration of 1.0 × 105 cells per dish and cultured overnight. Subsequently, the cells were incubated with exosomes (3 μg as protein) or cCHP nanogel/exosome hybrids for 4 and 24 h at 37 °C, washed twice with live cell imaging solution, and then the medium was replaced with fresh live cell imaging solution. The specimens were observed under a CLSFM (LSM780) equipped with a plan-APOCHROMAT 40× oil immersion objective and the emission spectra of DIO and/or R18 were recorded using the lambda mode, with excitation at 488 nm (Ar laser) and detection at 495–688 nm (at intervals of 9 nm).Immunofluorescence staining
hADSCs were seeded on a poly-L-lysine-coated glass-bottomed dish at a concentration of 1.5 × 103 cells per dish and cultured overnight. PC12 cells were maintained in DMEM containing 1% penicillin–streptomycin, 2% horse serum, and 25 ng mL−1 nerve growth factor (NGF) to induce neuronal differentiation and then derived exosomes were isolated from the culture supernatant. The hADSCs were exposed to exosomes derived from differentiating PC12 cells (protein concentration of 10 or 40 μg mL−1) or cCHP nanogel/PC12-exosome hybrids [protein concentrations of exosome, 10 or 40 μg mL−1; cCHP, 25 or 100 μg mL−1; by preparing a weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5 (exosome
2.5 (exosome![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nanogel)]. After 24 h of incubation, the medium was changed to fresh DMEM, which was repeated every day. At 7 days, the cells were fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.5% Triton-X containing PBS (PBST) for 8 min. Anti-NSE antibody was added to the cells after a blocking procedure with 1% bovine serum albumin containing PBS for 1 h and the cells were incubated at 4 °C overnight. The secondary antibody (conjugated with Alexa Fluor488) was added and incubated for 1 h after washing with PBS. Nuclei were stained with Hoechst33342. The samples were observed by CLSFM (LSM780).
nanogel)]. After 24 h of incubation, the medium was changed to fresh DMEM, which was repeated every day. At 7 days, the cells were fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.5% Triton-X containing PBS (PBST) for 8 min. Anti-NSE antibody was added to the cells after a blocking procedure with 1% bovine serum albumin containing PBS for 1 h and the cells were incubated at 4 °C overnight. The secondary antibody (conjugated with Alexa Fluor488) was added and incubated for 1 h after washing with PBS. Nuclei were stained with Hoechst33342. The samples were observed by CLSFM (LSM780).
Acetylcholine esterase (AchE) assay
hADSCs were seeded on a 96-well plate at a concentration of 1.5 × 103 cells per well. The cells were exposed to HEK293sus-derived exosomes (protein concentration of 40 μg mL−1), PC12-derived exosomes (protein concentration of 40 μg mL−1), or cCHP nanogel/PC12-exosome hybrids [protein concentrations of exosome, 0–40 μg mL−1; CHPNH2, 0–100 μg mL−1; by preparing a weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5 (exosome
2.5 (exosome![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nanogel)] and cultured overnight. The medium was changed to DMEM, which was repeated every day. After 5 or 7 days of incubation, the cells were exposed to acetylcholine iodide (250 mM) and 5,5′-dithio-bis-(2-nitrobenzoic acid) (DTNB, 2 mM) with permeabilization using 0.5% PBST. The relative enzymatic activity of AchE was quantified by the initial slope of change in absorbance at 420 nm from the product, 2-nitro-5-mercaptobenzoic acid, by using a microplate reader. The enzyme activity of AchE was indicated as the absorbance value obtained by subtracting the blank value.
nanogel)] and cultured overnight. The medium was changed to DMEM, which was repeated every day. After 5 or 7 days of incubation, the cells were exposed to acetylcholine iodide (250 mM) and 5,5′-dithio-bis-(2-nitrobenzoic acid) (DTNB, 2 mM) with permeabilization using 0.5% PBST. The relative enzymatic activity of AchE was quantified by the initial slope of change in absorbance at 420 nm from the product, 2-nitro-5-mercaptobenzoic acid, by using a microplate reader. The enzyme activity of AchE was indicated as the absorbance value obtained by subtracting the blank value.
Statistical analysis
The data were analyzed for statistical significance using Student's t-test. The significance of differences was assessed with a two-sided test with an α level of 0.01 or 0.05.Results and discussion
Characterization of exosomes
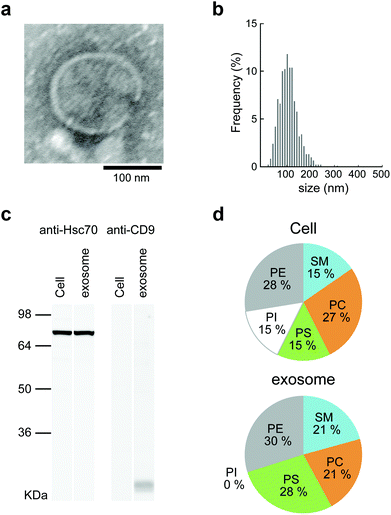
Mouse macrophage-like cells (Raw264.7) were selected as exosome-producing cells. The cells were incubated for 24 h in exosome-free fetal bovine serum (FBS), which had been centrifuged at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g before use. The culture supernatants obtained from each cell culture were collected and purified by differential centrifugation. For the relative quantitation of exosomes, the protein concentration of the purified exosomes was examined by BCA protein assay. The detailed results of analyzing the lipid compositions of cells and exosomes are shown in Fig. 2d. The lipid compositions of Raw264.7 cells and exosomes differed slightly, in that phosphatidylinositol (PI) was not present in the exosomes. The proportions of phosphatidylcholine (PC), phosphatidylserine (PS), phosphatidylethanolamine (PE), and sphingomyelin (SM) were 27
000g before use. The culture supernatants obtained from each cell culture were collected and purified by differential centrifugation. For the relative quantitation of exosomes, the protein concentration of the purified exosomes was examined by BCA protein assay. The detailed results of analyzing the lipid compositions of cells and exosomes are shown in Fig. 2d. The lipid compositions of Raw264.7 cells and exosomes differed slightly, in that phosphatidylinositol (PI) was not present in the exosomes. The proportions of phosphatidylcholine (PC), phosphatidylserine (PS), phosphatidylethanolamine (PE), and sphingomyelin (SM) were 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15
15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 28
28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 in Raw264.7 cells and 21
15 in Raw264.7 cells and 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 28
28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 21 in exosomes (Fig. 2d). The results show that Raw264.7 cells and exosomes contained much higher proportions of PE than the other cell strains.34 The lipid composition of Raw264.7 cells resembled that of the other macrophage cells.
21 in exosomes (Fig. 2d). The results show that Raw264.7 cells and exosomes contained much higher proportions of PE than the other cell strains.34 The lipid composition of Raw264.7 cells resembled that of the other macrophage cells.
The obtained exosomes were examined by transmission electron microscopy (TEM). Electron micrographs of the exosomes showed rounded structures similar to general exosomes (Fig. 2a). The sizes of the exosomes were measured by nanoparticle tracking analysis (NTA), and the average was approximately 150 nm (Fig. 2b). The ζ-potential of the exosomes was negative (−11.7 mV), as measured by using a Zetasizer Nano ZS. CD9, which is a 24 kDa member of the tetraspanins, known as exosome marker proteins, and heat shock cognate 71 kDa protein (Hsc70) were detected by western blotting analysis.
Preparation of nanogel/exosome hybrid
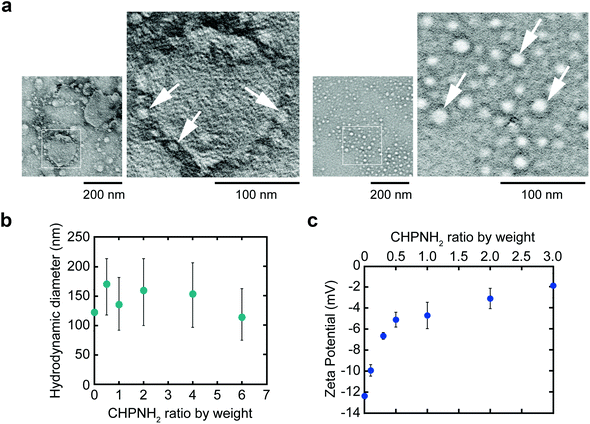
Ethylenediamine-modified CHP (CHPNH2) nanogels were prepared as cationic nanogels (cCHP nanogels) and then mixed with exosomes in PBS for 30 min on ice. The particle sizes of the complexes were measured by nanoparticle tracking analysis (NTA) (Fig. 3b). The size of the exosomes alone was 126 ± 35 nm. The size of the cCHP nanogels was about 30 nm, as determined by measuring dynamic light scattering (DLS). The interaction of exosomes with nanogels was estimated by measuring ζ-potential [cCHP (μg mL−1)/exosomes (μg mL−1, protein concentration) = 0 − 3]. The ζ-potential increased with increasing concentration of the nanogel (Fig. 3c). The anionic surfaces of the exosomes were encrusted with cationic nanogels. The evaluation of cCHP nanogel/Raw-exosome hybrid formation was also examined by agarose gel electrophoresis and polyanion addition. The results showed that almost no dissociation of the hybrids was observed under the condition where an excessive amount of polyanion was added (Fig. S3†). This suggests that the driving force for hybrid formation involved not only electrostatic interactions but also hydrophobic interactions via the cholesteryl groups of CHPNH2. We consider that the hybrid formation by multiple physical interactions is effective at improving the stability of the hybrid in a physiological environment. The cCHP nanogel/exosome hybrids were visually observed by TEM (Fig. 3a), with the images revealing that the surface of one exosome was coated with several cCHP nanogels. In addition, the size and ζ-potential of the nanogel/exosome hybrids were evaluated in PBS at 37 °C for 0, 4, and 24 h after preparation. The results revealed no change in the values (Table S1†). This suggests that the hybrid is stable under the experimental conditions.The driving force of the complexation between the liposome and the nanogel is hydrophobic interaction between the cholesteryl group of CHP and the lipid domain of the liposome.33 Therefore, it is considered that cationic nanogels can form complexes with exosomes by electrostatic or hydrophobic interactions and act as an exosome delivery system.
Cellular uptake of nanogel/exosome hybrid
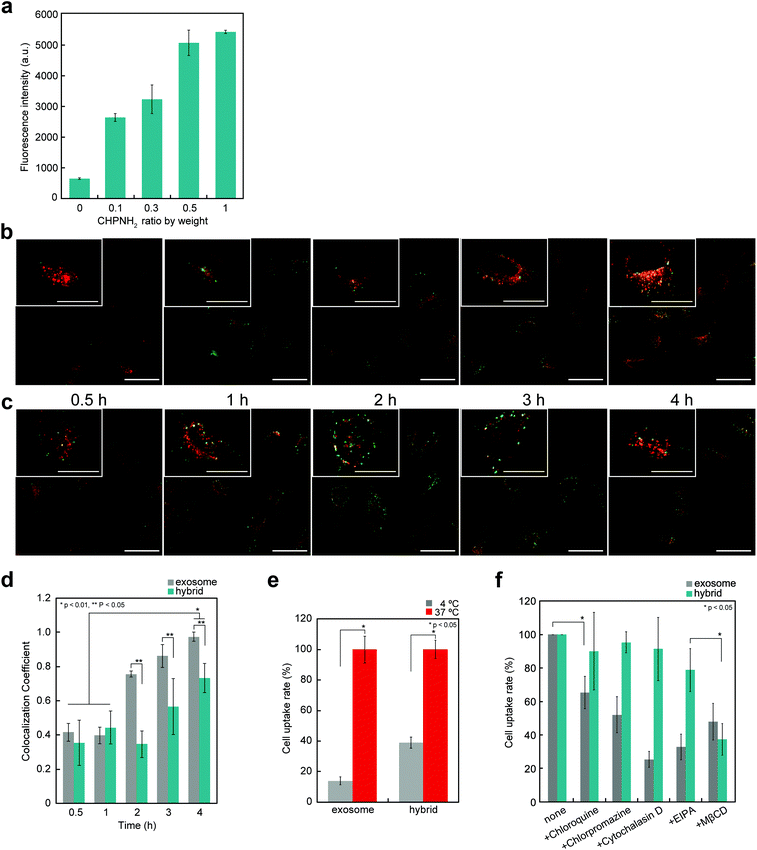
The cellular uptake of exosomes and nanogel/exosome hybrids by HeLa cells was investigated by flow cytometry. Carboxyfluorescein diacetate succinimidyl ester (CFSE) and 3-octadecyl-2-[3-(3-octadecyl-2(3H)-benzoxazolylidene)-1-propenyl] perchlorate (DIO) were selected as labeling reagents. CFSE is a membrane-permeable nonfluorescent compound that becomes fluorescent after the cleavage of its acetate groups by intracellular esterases. Exosomes are known to contain esterase, so CFSE can be used to label exosomes.35 DIO is a green-fluorescent lipophilic dye, which is used for exosome membrane labeling to examine the interaction with cells. CFSE- or DIO-treated exosomes were purified using a gel-filtration system to remove the fluorescent dye outside the exosomes. Significant fluorescence associated with the fluorescent-labeled exosomes was observed.The cellular uptake of exosomes drastically increased after the complexation of exosomes with nanogels. The efficiency depended on the concentrations of nanogels (Fig. 4a). The time-dependent uptake of exosomes was examined by confocal laser scanning fluorescence microscopy (CLSFM) (Fig. 4b and c). At 30 min after the addition of nanogel/exosome hybrids to cells, the hybrids adsorbed on the surface of the cells and gradually permeated into them. After 4 h, nanogel/exosome hybrids were efficiently internalized into the cells. Both exosomes and hybrids accumulated over time to the late endosome stage, which was revealed by an increase in the colocalization of the exosomes and late endosome marker (Lysotracker Red) (Fig. 4d). These results indicate that the main mechanism by which both exosomes and hybrids were internalized was endocytosis. However, under low-temperature conditions (4 °C), the cellular uptake of both exosomes and hybrids was significantly decreased, as revealed by flow cytometry (Fig. 4e).
The internalization of exosomes into cells occurs mainly via endocytosis,36 phagocytosis,37 and macropinocytosis,36,38 depending on the type of cell. To evaluate the mechanism of internalization of exosomes, the cells were pre-incubated for 30 min with several endocytic inhibitors, including a clathrin-mediated endocytosis inhibitor (chlorpromazine), a macropinocytosis inhibitor [ethyl-isopropyl amiloride (EIPA)], a lysosome acidification inhibitor (chloroquine), a caveolae-mediated endocytosis inhibitor [methyl-β-cyclodextrin (MβCD)], or an actin polymerization inhibitor (cytochalasin D), and then added to exosomes or hybrids. The inhibition efficiency was evaluated by flow cytometry (Fig. 4f). In the case of exosomes, every reagent tended to inhibit the uptake, indicating that the exosomes were internalized by multiple mechanisms. In contrast, the uptake of hybrids was significantly suppressed by the caveolae-mediated endocytosis inhibitor and slightly suppressed by the macropinocytosis inhibitor.
Fate of nanogel/exosome hybrid after internalization into cells
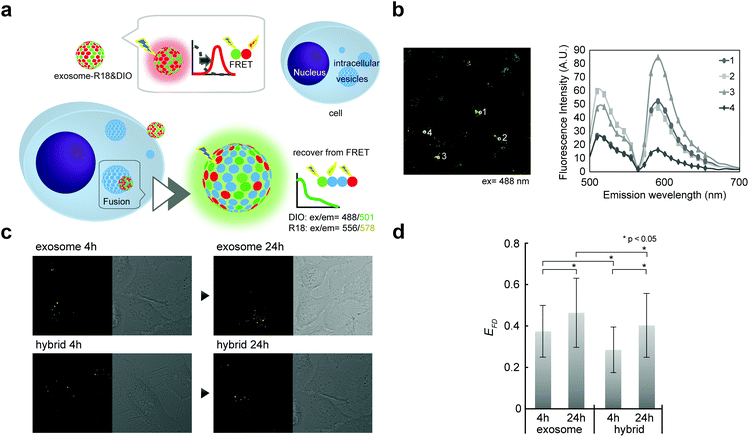
Although direct fusion with the plasma membrane of target cells39 or fusion with intercellular vesicles such as endosomes has been reported,40,41 the mechanisms involved in this are still not understood. We here investigated the possibility of fusion with membrane components (such as endosomes) after endocytosis. Fusion efficiencies between exosomes and membrane components were quantitatively evaluated as the lipid mixing ratio by FRET assay (Fig. 5a), which is widely utilized for fusion analysis, such as liposome–liposome, liposome–cell, and virus–cell fusion. Fluorescently labeled exosomes were prepared by two types of fluorescent lipids: DIO and R18 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio). Under these conditions, upon DIO excitation at 488 nm, the resulting solutions showed fluorescence emission at 504 nm (reflecting emissions from DIO), which was also observed at 583 nm due to fluorescence resonance energy transfer (FRET) between DIO and R18 (Fig. 5b, 1 and 3). If fusion/lipid mixing occurs, the fluorescence intensity at 504 nm should increase with a concomitant decrease of the intensity at 583 nm (Fig. 5b, 2 and 4). After the incubation of the fluorescently labeled exosome and hybrid with the cell for 4 and 24 h, fluorescence spectra inside the cell were analyzed. The emission spectra were recorded for each of the 100 bright spots in HeLa cells. The FRET restitution efficiency (EFD) of exosomes or nanogel/exosome hybrids was defined as follows:
1 molar ratio). Under these conditions, upon DIO excitation at 488 nm, the resulting solutions showed fluorescence emission at 504 nm (reflecting emissions from DIO), which was also observed at 583 nm due to fluorescence resonance energy transfer (FRET) between DIO and R18 (Fig. 5b, 1 and 3). If fusion/lipid mixing occurs, the fluorescence intensity at 504 nm should increase with a concomitant decrease of the intensity at 583 nm (Fig. 5b, 2 and 4). After the incubation of the fluorescently labeled exosome and hybrid with the cell for 4 and 24 h, fluorescence spectra inside the cell were analyzed. The emission spectra were recorded for each of the 100 bright spots in HeLa cells. The FRET restitution efficiency (EFD) of exosomes or nanogel/exosome hybrids was defined as follows: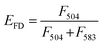 | (1) |
In both the exosome and hybrid, 30%–40% FRET restitution was observed (Fig. 5c and d). The results clearly showed that fluorescent lipids in exosomes were diluted by lipid molecules (or other membrane components such as membrane proteins) present in endosomes, probably due to the membrane fusion between the exosome and endosome. Cytosol delivery, for example, of microRNA, is indispensable for the appearance of activity due to its ability to avoid lysosomal degradation.
Manipulation of cellular function by nanogel/exosome hybrid delivery system
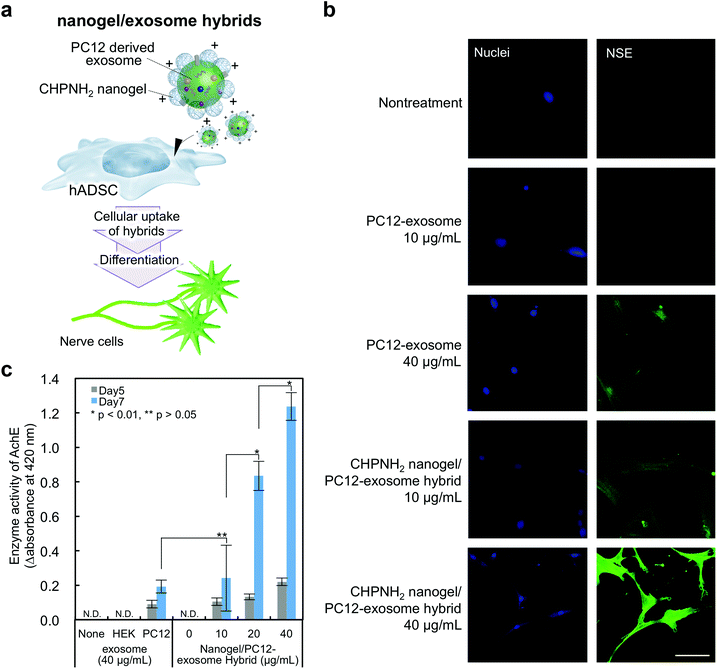
The availability of the current cationic nanogel system to manipulate cellular functions was evaluated by the induction of neuron-like differentiation toward adipocyte-derived stem cells (ADSCs) (Fig. 6a). It was revealed that neuron-like differentiation was induced by miR-125b, which was contained in exosomes isolated from differentiating rat pheochromocytoma (PC12) cells.42 Mesenchymal stem cells (MSCs) including ADSCs have become candidates for cell therapy due to their multiple potencies, including the abilities to self-renew and to differentiate into various cells such as muscle cells, osteoblasts, neurons, endothelial cells, and adipocytes.43 Therefore, the development of a way to establish engineered cells has been desired in the field of regenerative medicine.44In exosomes purified from the PC12 cell culture supernatant, CD9 and HSC70 were not detected by western blotting analysis, but CD81, known as another exosome marker protein, was confirmed (Fig. S4a†). The TEM image of the PC12-exosomes showed rounded and vesicle-like structures similar to typical exosomes (Fig. S4b†). The average size of the PC12-exosomes was approximately 150 nm, and the ζ-potential was negative (−11.5 ± mV) (Fig. S4c and Table S2†). The hybrids consisting of cCHP nanogels and PC12-exosomes were prepared; no significant change in size was observed in the case of Raw exosomes and nanogel hybrids, but an increase in ζ-potential was confirmed (Table S2†). Next, neuron-like differentiation toward ADSCs was investigated by detecting nerve differentiation marker proteins using immunofluorescence staining and quantifying the relative enzymatic activity of acetylcholine esterase. The PC12-exosome or cCHP nanogel/PC12-exosome hybrid was added to ADSCs and incubated for 7 days. The expression of nerve-specific enolase (NSE), which is known as a neuronal differentiation marker protein, was detected by immunofluorescence staining (Fig. 6b). Compared with the cells treated with PC12-exosome alone, stronger fluorescence signals were observed by treatment with the cCHP nanogel/PC12-exosome hybrid at the same concentration and neurite-like extensions were observed in NSE-positive cells. Therefore, the cationic nanogel system enhanced the differentiation toward ADSCs by achieving efficient intracellular delivery.
We next quantified the efficiency of neuron-like differentiation toward ADSCs by measuring the enzymatic activity of acetylcholine esterase (AchE). The differentiation was induced as well as immunofluorescence staining; we also used the HEK293sus-derived exosome as a control exosome, which should not induce differentiation. After 5 and 7 days of incubation, substrates (acetylcholine iodide, 250 mM; DTNB, 2 mM) were added to the cells and their relative enzymatic activities of AchE were estimated from the slope of the product absorbance changes at 420 nm within the first 15 min. The enzymatic activities of AchE were increased over time by treatment with the PC12-exosome or cCHP nanogel/PC12-exosome hybrid, in comparison with those in the group treated with the HEK293sus-exosome (Fig. 6c). Finally, the expression level of AchE in the cells treated with the complexes reached eight times higher than that upon treatment with the PC12-exosome alone.
Exosomes delivered by cationic nanogels enable the modulation of cellular function effectively despite the lower concentration. These results indicate that the newly developed cationic nanogel systems for exosome delivery are powerful tools to investigate the biological functions of exosomes.
Conclusions
We propose here a new form of exosome surface polymer engineering for effective intracellular exosome delivery by using self-assembled amphiphilic cationic nanogels. The cationic nanogel/exosome hybrid was effectively internalized into cells and acted as an effective delivery system for PC12-exosomes, resulting in the modulation of cellular functions such as the neuron-like differentiation of ADSCs. As the results indicate, the enhancement of cellular uptake and functional expression of exosomes by complexation with cationic nanogels is an effective method for analyzing their functions with a smaller number of exosomes. The value of exosomes as bio-shuttles was significantly increased by using a co-nanogel delivery system, although the efficiency of exosomes as cellular modulators was not always high. Polysaccharide self-nanogels themselves are effective nanocarriers for protein and nucleic acid (siRNA) delivery, as we previously reported. The nanogel/exosome hybrid should have potential for use as a dual nanocarrier system to enable the loading of various drugs to separate compartments in nanogels as well as exosomes. Moreover, we have developed stimulus-responsive self-nanogels by functionally associating polymers with responsivity to pH, light, and temperature. By simply mixing functional nanogels, new additional functions can be provided to exosomes. Exosome surface nanogel engineering offers a new option for the wide application of exosomes to drug delivery systems and nanomedicine.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by the Exploratory Research for Advanced Technology of the Japan Science and Technology Agency (JST-ERATO) and by Grant-in-Aid for JSPS Fellows (Grant Number 16H06313). We thank the Edanz Group (http://www.edanzediting.com/ac) for editing a draft of this manuscript.References
- G. van Niel, G. D'Angelo and G. Raposo, Shedding light on the cell biology of extracellular vesicles, Nat. Rev. Mol. Cell Biol., 2018, 19, 213–228 CrossRef CAS PubMed.
- M. Mathieu, L. Martin-Jaular, G. Lavieu and C. Théry, Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication, Nat. Cell Biol., 2019, 21, 9–17 CrossRef CAS PubMed.
- S. L. N. Maas, X. O. Breakefield and A. M. Weaver, Extracellular vesicles: unique intercellular delivery vehicles, Trends Cell Biol., 2017, 27, 172–188 CrossRef CAS PubMed.
- A. Takahashi, R. Okada, K. Nagao, Y. Kawamata, A. Hanyu, S. Yoshimoto, M. Takasugi, S. Watanabe, M. T. Kanemaki, C. Obuse and E. Hara, Exosomes maintain cellular homeostasis by excreting harmful DNA from cells, Nat. Commun., 2017, 8, 15287 CrossRef CAS PubMed.
- L. Barile and G. Vassalli, Exosomes: Therapy delivery tools and biomarkers of diseases, Pharmacol. Ther., 2017, 174, 63–78 CrossRef CAS PubMed.
- K. Boriachek, M. N. Islam, A. Möller, C. Salomon, N. T. Nguyen, M. S. A. Hossain, Y. Yamauchi and M. J. A. Shiddiky, Biological functions and current advances in isolation and detection strategies for exosome nanovesicles, Small, 2018, 14, 1702153 CrossRef PubMed.
- J. P. Armstrong, M. N. Holme and M. M. Stevens, Re-engineering extracellular vesicles as smart nanoscale therapeutics, ACS Nano, 2017, 11, 69–83 CrossRef CAS.
- O. G. de Jong, S. A. A. Kooijmans, D. E. Murphy, L. Jiang, M. J. W. Evers, J. P. G. Sluijter, P. Vader and R. M. Schiffelers, Drug delivery with extracellular vesicles: from imagination to innovation, Acc. Chem. Res., 2019, 52, 1761–1770 CrossRef CAS PubMed.
- Y. Tian, S. Li, J. Song, T. Ji, M. Zhu, G. J. Anderson, J. Wei and G. Nie, A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy, Biomaterials, 2014, 35, 2383–2390 CrossRef CAS PubMed.
- M. N. Oskouie, N. S. A. Moghaddam, A. E. Butler, P. Zamani and A. Sahebkar, Therapeutic use of curcumin-encapsulated and curcumin-primed exosomes, J. Cell. Physiol., 2019, 234, 8182–8191 CrossRef CAS PubMed.
- M. S. Kim, M. J. Haney, Y. Zhao, V. Mahajan, I. Deygen, N. L. Klyachko, E. Inskoe, A. Piroyan, M. Sokolsky and O. Okolie, Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells, Nanomedicine, 2016, 12, 655–664 CrossRef CAS PubMed.
- T. Smyth, M. Kullberg, N. Malik, P. Smith-Jones, M. W. Graner and T. J. Anchordoquy, Biodistribution and delivery efficiency of unmodified tumor-derived exosomes, J. Controlled Release, 2015, 199, 145–155 CrossRef CAS PubMed.
- L. Alvarez-Erviti, Y. Seow, H. Yin, C. Betts, S. Lakhal and M. J. Wood, Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes, Nat. Biotechnol., 2011, 29, 341–345 CrossRef CAS PubMed.
- S. Ohno, M. Takanashi, K. Sudo, S. Ueda, A. Ishikawa, N. Matsuyama, K. Fujita, T. Mizutani, T. Ohgi, T. Ochiya, N. Gotoh and M. Kuroda, Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells, Mol. Ther., 2013, 21, 185–191 CrossRef CAS PubMed.
- M. E. Hung and J. N. Leonard, Stabilization of exosome-targeting peptides via engineered glycosylation, J. Biol. Chem., 2015, 290, 8166–8172 CrossRef CAS PubMed.
- Z. Stickney, J. Losacco, S. McDevitt, Z. Zhang and B. Lu, A development of exosome surface display technology in living human cells, Biochem. Biophys. Res. Commun., 2016, 472, 53–59 CrossRef CAS PubMed.
- S. A. A. Kooijmans, C. G. Aleza, S. R. Roffler, W. W. van Solinge, P. Vader and R. M. Schiffelers, Display of GPI-anchored anti-EGFR nanobodies on extracellular vesicles promotes tumour cell targeting, J. Extracell. Vesicles, 2016, 5, 31053 CrossRef PubMed.
- S. A. A. Kooijmans, J. J. J. M. Gitz-Francois, R. M. Schiffelers and P. Vader, Recombinant phosphatidylserine-binding nanobodies for targeting of extracellular vesicles to tumor cells: a plug-and-play approach, Nanoscale, 2018, 10, 2413–2426 RSC.
- T. Tian, H. X. Zhang, C. P. He, S. Fan, Y. L. Zhu, C. Qi, N. P. Huang, Z. D. Xiao, Z. H. Lu, B. A. Tannous and J. Gao, Surface functionalized exosomes as targeted drug delivery vehicles for cerebral ischemia therapy, Biomaterials, 2018, 150, 137–149 CrossRef CAS PubMed.
- I. Nakase and S. Futaki, Combined treatment with a pH-sensitive fusogenic peptide and cationic lipids achieves enhanced cytosolic delivery of exosomes, Sci. Rep., 2015, 5, 10112 CrossRef.
- R. Tamura, S. Uemoto and Y. Tabata, Augmented liver targeting of exosomes by surface modification with cationized pullulan, Acta Biomater., 2017, 57, 274–284 CrossRef CAS PubMed.
- J. Lee, H. Lee, U. Goh, J. Kim, M. Jeong, J. Lee and J. H. Park, Cellular engineering with membrane fusogenic liposomes to produce functionalized extracellular vesicles, ACS Appl. Mater. Interfaces, 2016, 8, 6790–6795 CrossRef CAS PubMed.
- R. Munagala, F. Aqil, J. Jeyabalan and R. C. Gupta, Bovine milk-derived exosomes for drug delivery, Cancer Lett., 2016, 371, 48–61 CrossRef CAS PubMed.
- H. Qi, C. Liu, L. Long, Y. Ren, S. Zhang, X. Chang, X. Qian, H. Jia, J. Zhao, J. Sun, X. Hou, X. Yuan and C. Kang, Blood exosomes endowed with magnetic and targeting properties for cancer therapy, ACS Nano, 2016, 10, 3323–3333 CrossRef CAS.
- Y. Tahara and K. Akiyoshi, Current advances in self-assembled nanogel delivery systems for immunotherapy, Adv. Drug Delivery Rev., 2015, 95, 65–76 CrossRef CAS.
- Y. Tahara, S. Mukai, S. Sawada, Y. Sasaki and K. Akiyoshi, Nanocarrier-integrated microspheres: nanogel tectonic engineering for advanced drug delivery systems, Adv. Mater., 2015, 27, 5080–5088 CrossRef CAS PubMed.
- Y. Hashimoto, S. Mukai, S. Sawada, Y. Sasaki and K. Akiyoshi, Nanogel tectonic porous gel loading biologics, nanocarriers and cells for advanced scaffold, Biomaterials, 2015, 37, 107–115 CrossRef CAS PubMed.
- H. Ayame, N. Morimoto and K. Akiyoshi, Self-assembled cationic nanogels for intracellular protein delivery system, Bioconjugate Chem., 2008, 19, 882–890 CrossRef CAS PubMed.
- K. Watanabe, Y. Tsuchiya, Y. Kawaguchi, S. Sawada, H. Ayame, K. Akiyoshi and T. Tsubata, The use of cationic nanogels to deliver proteins to myeloma cells and primary T lymphocytes that poorly express heparan sulfate, Biomaterials, 2011, 32, 5900–5905 CrossRef CAS PubMed.
- Y. Sasaki, S. Toita and K. Akiyoshi, Cycloamylose-based nanocarriers as a nucleic acid delivery system, in Colloid and Interface Science in Pharmaceutical Research and Development, ed. H. Ohshima and K. Makino, Elsevier, Amsterdam, 1st edn, 2014, chapter 18, pp. 369–388 Search PubMed.
- T. Nochi, Y. Yuki, H. Takahashi, S. Sawada, M. Mejima, T. Kohda, N. Harada, I. G. Kong, A. Sato, N. Kataoka, D. Tokuhara, S. Kurokawa, Y. Takahashi, H. Tsukada, S. Kozaki, K. Akiyoshi and H. Kiyono, Nanogel antigenic protein-delivery system for adjuvant-free intranasal vaccines, Nat. Mater., 2010, 9, 572–578 CrossRef CAS PubMed.
- Y. Fukuyama, Y. Yuki, Y. Katakai, N. Harada, H. Takahashi, S. Takeda, M. Mejima, S. Joo, S. Kurokawa, S. Sawada, H. Shibata, E. J. Park, K. Fujihashi, D. E. Briles, Y. Yasutomi, H. Tsukada, K. Akiyoshi and H. Kiyono, Nanogel-based pneumococcal surface protein A nasal vaccine induces microRNA-associated Th17 cell responses with neutralizing antibodies against Streptococcus pneumoniae in macaques, Mucosal Immunol., 2015, 8, 1144–1153 CrossRef CAS PubMed.
- Y. Sekine, Y. Moritani, T. Ikeda-Fukazawa, Y. Sasaki and K. Akiyoshi, A hybrid hydrogel biomaterial by nanogel engineering: bottom-up design with nanogel and liposome building blocks to develop a multidrug delivery system, Adv. Healthcare Mater., 2012, 1, 722–728 CrossRef CAS PubMed.
- R. A. Haraszti, M. C. Didiot, E. Sapp, J. Leszyk, S. A. Shaffer, H. E. Rockwell, F. Gao, N. R. Narain, M. DiFiglia, M. A. Kiebish, N. Aronin and A. Khvorova, High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources, J. Extracell. Vesicles, 2016, 5, 32570 CrossRef PubMed.
- N. G. Van, I. Porto-Carreiro, S. Simoes and G. Raposo, Exosomes: a common pathway for a specialized function, J. Biochem., 2006, 140, 13–21 CrossRef PubMed.
- T. Tian, Y. L. Zhu, Y. Y. Zhou, G. F. Liang, Y. Y. Wang, F. H. Hu and Z. D. Xiao, Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery, J. Biol. Chem., 2014, 289, 22258–22267 CrossRef CAS PubMed.
- D. Feng, W. L. Zhao, Y. Y. Ye, X. C. Bai, R. Q. Liu, L. F. Chang, Q. Zhou and S. F. Sui, Cellular internalization of exosomes occurs through phagocytosis, Traffic, 2010, 11, 675–687 CrossRef CAS PubMed.
- D. Fitzner, M. Schnaars, D. van Rossum, G. Krishnamoorthy, P. Dibaj, M. Bakhti, T. Regen, U. K. Hanisch and M. Simons, Selective transfer of exosomes from oligodendrocytes to microglia by micropinocytosis, J. Cell Sci., 2011, 124, 447–458 CrossRef CAS PubMed.
- I. Parolini, C. Federici, C. Raggi, L. Lugini, S. Palleschi, A. De Milito, C. Coscia, E. Iessi, M. Logozzi, A. Molinari, M. Colone, M. Tatti, M. Sargiacomo and S. Fais, Microenvironmental pH is a key factor for exosome traffic in tumor cells, J. Biol. Chem., 2009, 284, 34211–34222 CrossRef CAS PubMed.
- T. Tian, Y. Wang, H. Wang, Z. Zhu and Z. Xiao, Visualizing of the cellular uptake and intracellular trafficking of exosomes by live-cell microscopy, J. Cell. Biochem., 2010, 111, 488–496 CrossRef CAS PubMed.
- S. Keerthikumar, L. Gangoda, M. Liem, P. Fonseka, I. Atukorala, C. Ozcitti, A. Mechler, C. G. Adda, C. S. Ang and S. Mathivanan, Proteogenomic analysis reveals exosomes are more oncogenic than ectosomes, Oncotarget, 2015, 6, 15375–15396 CrossRef PubMed.
- Y. S. Takeda and Q. Xu, Neuronal differentiation of human mesenchymal stem cells using exosomes derived from differentiating neuronal cells, PLoS One, 2015, 10, e0135111 CrossRef PubMed.
- A. Uccelli, L. Moretta and V. Pistoia, Mesenchymal stem cells in health and disease, Nat. Rev. Immunol., 2008, 8, 726–736 CrossRef CAS PubMed.
- D. A. Grande, Regenerative medicine in 2016: important milestones on the way to clinical translation, Nat. Rev. Rheumatol., 2017, 13, 67–68 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9bm01232j |
| ‡ These two authors contributed equally to this manuscript. |
| This journal is © The Royal Society of Chemistry 2020 |