Open Access Article
Open Access ArticleFactors influencing the measurement of the secretion rate of extracellular vesicles†
Yi
Wen
a,
Yundi
Chen
a,
Guosheng
Wang
a,
Komal
Abhange
a,
Fei
Xue
a,
Zachary
Quinn
a,
Wenjun
Mao
*b and
Yuan
Wan
*a
aThe Pq Laboratory of Micro/Nano BiomeDx, Department of Biomedical Engineering, Binghamton University-SUNY, Binghamton, New York 13902, USA. E-mail: ywan@binghamton.edu; Fax: +1 607-777-5780; Tel: +1 607-777-5477
bDepartment of Cardiothoracic Surgery, The affiliated Wuxi People's Hospital of Nanjing Medical University, Wuxi, Jiangsu 214023, China. E-mail: maowenjun1@njmu.edu.cn; Tel: +86 510 82700775
First published on 10th July 2020
Abstract
Extracellular vesicles (EVs) are cell-derived vesicles which encapsulate a variety of molecules. Numerous studies have demonstrated EVs as signaling mediators of intercellular communication and are heavily involved under physiological and pathological conditions. In translational medicine, EVs have been used for disease diagnosis and treatment monitoring. EVs as natural nanocarriers for drug delivery and therapeutic EVs are also under intense investigation. While still in its infancy, relevant EV studies have been growing. For EV-centered research to thrive, a few fundamental unanswered questions, such as EV biogenesis, EV secretion rate (SR), EV content sorting mechanisms, etc. require further investigation. In this study, we measured the SR of EVs derived from 6 cancerous cell lines. Several factors that may interfere with EV secretion, isolation, and storage were also investigated. Our results show that the SR of EVs derived from various cancer cells was significantly different, indicating a heterogeneous EV secretion behavior among cell types. Moreover, 5 different drugs that interfere with cellular metabolism significantly influenced EV release. In addition, we found that (1) more EVs can be harvested at 24 h compared to 48 h of serum-free cell culture with a similar degree of FBS contamination; (2) filtration of the cell culture supernatant with a 0.22 μm filter causes ∼70% loss of EVs; (3) the isolation efficiency of EVs with the prevalent ultracentrifugation is only ∼14%; (4) storage at 4 °C for 3 days causes ∼21% loss of EVs. Overall, our findings provide a guideline for proper EV collection and storage in laboratory settings.
Introduction
Extracellular vesicles (EVs) are lipid bilayer-enclosed particles, which encapsulate cell-specific DNA, RNA, and proteins.1,2 EVs can be secreted by all eukaryotic cells and are found in various body fluids,3,4 however EV biogenesis is not fully understood yet. Based on the size and mechanism of release, EVs are generally categorized into three subsets: apoptotic bodies, microvesicles (MV), and exosomes.5 Apoptotic bodies (1–5 μm) are generated from apoptotic cells or from the fragmented membrane of dead cells.6,7 By contrast, MVs (100–1000 nm) and exosomes (30–120 nm) are derived from outward budding of the cell membrane and the endolysosomal pathway, respectively.8,9 The submicron-sized MVs and exosomes that hold promise for pathophysiologic and translational discoveries have recently experienced intense investigation.10 Previous studies have demonstrated that EVs play important roles in cell–cell communication by delivering cargos to recipient cells. In cancer research, growing evidence reveals that EVs regulate tumor immune responses, initiate the formation of pre-metastatic niches, determine organotropic metastasis, promote epithelial-to-mesenchymal transitions, and contribute to therapeutic resistance.11 In clinical translation, EVs as a useful addition to the liquid biopsy portfolio have been used for cancer diagnostics and treatment.12–14 In addition, EVs have been exploited for drug delivery and/or as therapeutic agents.15 In brief, EVs have become a hot topic in cancer research, diagnostics, and therapeutics. Nevertheless, a few fundamental characteristics of EVs still remain ambiguous, which may hinder future EV-centered research. Chief among these is the widely disputed secretion rate (SR) or magnitude of EV secretion. Similarly, the standard guidelines for EV preparation with cultured cells, handling of cell culture supernatant (CCS), and EV isolation with differential ultracentrifugation in laboratory settings have not been well studied either. Hence, the main objectives of this study are to clarify the aforementioned issues. In addition, we investigated the influence of five repurposed drugs previously used in cancer treatment research, including acetylsalicylic acid (ASA),16 celecoxib (CB),17 rapamycin (RAP),18 artemisinin (ART),19 and chloroquine (CQ), on EV secretion.20 Our results show that different cancer cells have varied respective SRs of EVs which can be significantly influenced by repurposed drugs. Moreover, we recommend only a 24 h serum-free culture for EV harvest. Replacement of 0.22 μm filtration with 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g centrifugation can preserve more EVs in CCS for further enrichment, although the isolation efficiency of EVs with differential ultracentrifugation is only ∼14%. Lastly, it is not recommended to preserve EVs in serum-free medium at 4 °C for more than 3 days as ∼21% of EVs would deteriorate.
000g centrifugation can preserve more EVs in CCS for further enrichment, although the isolation efficiency of EVs with differential ultracentrifugation is only ∼14%. Lastly, it is not recommended to preserve EVs in serum-free medium at 4 °C for more than 3 days as ∼21% of EVs would deteriorate.
Experimental section
Cell culture and EV collection
Breast cancer cell line MDA-MB-231 cells, lung adenocarcinoma cell lines NCI-H441, NCI-H661 and NCI-H2228, ovarian carcinoma cell line SKOV3, and colon adenocarcinoma cell line HT-29 were purchased from the ATCC. All cells passed testing for mycoplasma contamination during all experiments. MDA-MB-231 cells and NCI-H441 cells were cultured in DMEM (Corning, USA). SKOV3 cells and HT29 cells were maintained in McCoy's 5a Modified Medium (Gibco, USA). NCI-H661 cells and NCI-H2228 cells were grown in RPMI 1640 medium with 25 mM HEPES and L-glutamine (GE Healthcare, USA). All culture media were supplemented with 5% (v/v) exosome-depleted FBS (A2720801, Thermo Fisher), 100 units per ml penicillin, 100 μg ml−1 streptomycin and 10% non-essential amino acid. Cells were maintained in 95% humidified atmosphere of 5% CO2 at 37 °C. Once cell confluence reached 40%, the culture media were removed and rinsed with PBS thrice followed by cell culture with non-FBS media for 48 h (the final cellular confluence was ∼90%). First, we determined whether EVs from the filtered, undiluted CCS could be measured using Nanosight. CCS was filtered with 0.22 μm low protein binding polyethersulfone filter (Whatman). In the other group, CCS was filtered and ultracentrifuged at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g at 4 °C for 4 h. The EV pellet was resuspended in 100 μl of PBS followed by appropriate dilution. Two EV samples were measured with Nanosight to determine EV size and concentration. Next, we investigated the influence of final cellular confluence (after 48 h FBS-free culture) on the measurement of EV SR. CCS derived from cells at 45%, 70%, and 90% confluence was filtered and diluted 10-fold followed by Nanosight measurement. Next, we studied the influence of filtration on the measurement of SR. CCS derived from 70% confluence was filtered in one group and centrifuged at 20
000g at 4 °C for 4 h. The EV pellet was resuspended in 100 μl of PBS followed by appropriate dilution. Two EV samples were measured with Nanosight to determine EV size and concentration. Next, we investigated the influence of final cellular confluence (after 48 h FBS-free culture) on the measurement of EV SR. CCS derived from cells at 45%, 70%, and 90% confluence was filtered and diluted 10-fold followed by Nanosight measurement. Next, we studied the influence of filtration on the measurement of SR. CCS derived from 70% confluence was filtered in one group and centrifuged at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 min at 4 °C in the other group. Following 10-fold dilution, EVs from CCS samples were measured with Nanosight. Afterwards, we forewent filtration, using only 20
000g for 15 min at 4 °C in the other group. Following 10-fold dilution, EVs from CCS samples were measured with Nanosight. Afterwards, we forewent filtration, using only 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g centrifugation to remove intact cells and cellular debris. Subsequently, cells were cultured in FBS-free media for 24 h or 48 h. CCS was collected and centrifuged followed by Nanosight measurement. Lastly, we studied the influence of ultracentrifugation and EV storage on the measurement of EV SR.
000g centrifugation to remove intact cells and cellular debris. Subsequently, cells were cultured in FBS-free media for 24 h or 48 h. CCS was collected and centrifuged followed by Nanosight measurement. Lastly, we studied the influence of ultracentrifugation and EV storage on the measurement of EV SR.
Treatment of cells with repurposed drugs in gradient doses
ASA (Acros Organics) was dissolved in ethanol to prepare 1.25 mM and 2.5 mM solutions. CB (TCI America) was dissolved in DMSO to prepare 5 μM, 10 μM, and 20 μM solutions. RAP (Adipogen) was dissolved in DMSO to prepare 5 nM, 10 nM, 20 nM, 40 nM and 80 nM solutions. ART (TCI America) was dissolved in DMSO to prepare 10 μM, 20 μM, 40 μM, 80 μM and 160 μM solutions. CQ (Chem-Impex) was directly dissolved in DMEM to prepare 5 μM, 10 μM, 20 μM and 40 μM solutions. MDA-MB-231 cells were treated with each gradient concentration of drug. Equal amounts of solvent in each respective group was used to treat cells as a control. Cell viability was determined with CCK-8 assay following the manufacturer's instruction. Once the drug treated MDA-MB-231 cells reached ∼60% confluence, the media were removed and cells were rinsed thrice with PBS. Cells were further cultured with serum-free media for 24 h before measurement of EV concentration and size distribution. CCS was centrifuged at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 min at 4 °C followed by Nanosight measurement.
000g for 15 min at 4 °C followed by Nanosight measurement.
EV characterization
10 μl of concentrated EVs in PBS were loaded on the 400-mesh Formvar-coated copper grid and allowed to incubate for 3 min at room temperature. Excess samples were drained with filter paper and stained with 1% filtered uranyl acetate solution for 1 min. Prepared samples were imaged with a Hitachi TEM at an acceleration voltage of 100 Kv.21 Size distribution and concentration of EVs were measured with Nanosight NS300 according to manufacturer's instructions followed by automated analysis with NTA software (NTA version: 3.4 Build 3.4.003) to calculate the size distribution and concentration. Three 30 second videos were taken. Data were represented as mean±SD. EV protein markers, CD9 and TSG101, were identified with western blot. Briefly, the supernatant was centrifuged at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 min at 4 °C, filtered with a 0.22 μm filter, and then lyophilized followed by RIPA lysis of the contained EVs. In parallel, parental cells were lysed with RIPA. The respective EV lysate and cellular lysate were analyzed with Micro BCA Protein assay. Protein samples were processed with acrylamide gels and then transferred onto PVDF membranes. The protein blot was blocked for 1 hour at room temperature with 5% nonfat dry milk in PBS/0.05% Tween 20 and incubated overnight at 4 °C with Santa Cruz Biotechnology antibodies against TSG101 (sc-7964) and CD9 (sc13118). Afterward, secondary antibodies were incubated for 1 hour at room temperature. Samples were washed with PBS/0.05% Tween 20 for 10 minutes thrice. Blots were developed with chemiluminescence.
000g for 15 min at 4 °C, filtered with a 0.22 μm filter, and then lyophilized followed by RIPA lysis of the contained EVs. In parallel, parental cells were lysed with RIPA. The respective EV lysate and cellular lysate were analyzed with Micro BCA Protein assay. Protein samples were processed with acrylamide gels and then transferred onto PVDF membranes. The protein blot was blocked for 1 hour at room temperature with 5% nonfat dry milk in PBS/0.05% Tween 20 and incubated overnight at 4 °C with Santa Cruz Biotechnology antibodies against TSG101 (sc-7964) and CD9 (sc13118). Afterward, secondary antibodies were incubated for 1 hour at room temperature. Samples were washed with PBS/0.05% Tween 20 for 10 minutes thrice. Blots were developed with chemiluminescence.
Cell counting
All cells in flasks were fixed with 4% paraformaldehyde at 4 °C for 15 min followed by nuclear staining with 1 μg ml−1 DAPI (Invitrogen) at 4 °C refrigeration for 30 min. The fluorescence images were captured using a Zeiss Axiophot 373 microscope. The number of nuclei was counted with ImageJ.Mass spectrometry
Protein contents of EVs were detected by NanoLC-MS/MS analysis. After RIPA lysis, the protein amount in lysate was first determined via Bradford assay. Samples were also processed for in-solution digestion and solid phase extraction. In brief, samples were resuspended in 20 μl of 50% trifluoroethanol in 100 mM Tris HCL, pH 8.0. The peptides were reduced by adding 0.5 M TCEP and incubated for 45 min at room temperature. Subsequently, the processed peptides were alkylated by adding 1.6 μl 1 M iodoacetamide and incubated at room temperature for 1 h in a dark environment. The samples were continuously diluted by 10-fold using 100 mM Tris buffer pH 8.0, followed by the addition of 1 μg of trypsin and digestion at 37 °C for 18 h. Label-free quantification was employed for the analysis. For database searching, all MS spectra were converted into DTA files. The protein-level FDR was also calculated and restricted to lower than 5%. Data were presented as abundance in this report.Statistics analysis
The results are presented as mean ± standard deviation (SD). Statistical comparisons were performed by two-tailed T-test and two-tailed ANOVA test. A p-value <0.05 was considered statistically significantly.Results and discussion
Sample preparation
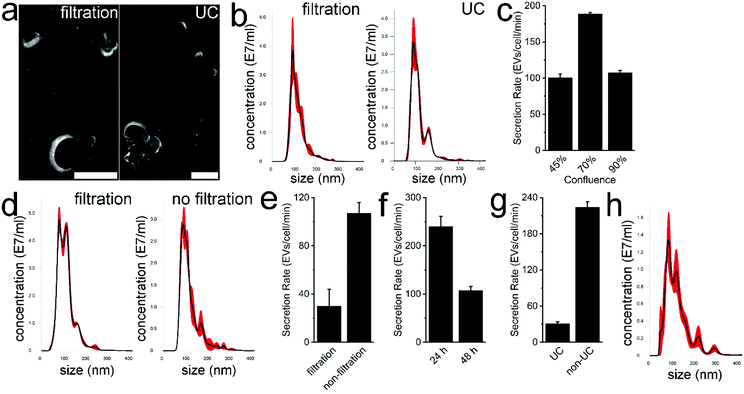
In previous studies, highly concentrated EV samples prepared by ultracentrifugation were used for Nanosight measurement to determine the EV concentration and size distribution.22,23 The total amount of EVs can then be calculated using the volume of EV resuspension and measured EV concentration. Subsequently, the EV amount is divided by cell numbers and culture time to generate the SR of EVs. Some concerns as to the accuracy of the data arose in such measurements. First, the isolation efficiency of EVs with ultracentrifugation may not be included for data processing. Second, even if this factor is involved, the isolation efficiency may be subject to fluctuation due to the variant specimen volume, processing time, centrifugation force, EV adsorption to the container surfaces, etc.24 Particularly, the high concentration of EV resuspension may require hundreds or thousands of dilutions to accommodate into the recommended concentration range of the Nanosight measurement.25 The agglomeration of EVs and protein aggregation caused by ultracentrifugation poses another challenge to Nanosight measurement. The EV adsorption to surfaces such as micropipette tips and microtubes in a serial dilution would significantly influence the measurement accuracy. Therefore, the determined SR of EVs in previous studies may be different by orders of magnitude.To eliminate, or at least alleviate, these shortcomings, we directly measured the EV concentration in serum-free medium. In one group, the CCS of MDA-MB-231 cells was simply filtered followed by characterization. In the control group, CCS was processed with filtration, ultracentrifugation, resuspension, and dilution before measurement. Serum-free medium was used as a blank control in Nanosight measurements. Both TEM images show the characteristic saucer-shaped morphology of MDA-MB-231 EVs (Fig. 1a). The respective average size of EVs is 118.4 nm and 120.1 nm (Fig. 1b). No significant difference was found in size distribution (t-test, p < 0.001). Of note, the average concentration of EVs in filtered, undiluted CCS was determined as 1.84 × 109 particles per ml. These results indicate that it is feasible to directly measure EVs from serum-free medium. It also implies a high concentration of EVs in undiluted CCS, which exceeds the recommended concentration range for Nanosight, i.e., 106 to 109 particles per ml. Thus, in the following studies, the serum-free CCS was routinely diluted 10 times to avoid potential overranging. Of note, since cells are cultured in serum-free media for 48 hours following culture with 5% exosome-depleted FBS, the scarce remaining FBS does not interfere with measurement.
Next the effect of cellular confluence on the SR of EVs was studied. After 48 h serum-free culture, MDA-MB-231 three separate cultures reached ∼45%, ∼70%, and ∼90% confluence. Following routine filtration with a 0.22 μm filter, the average size of EVs in each group was 114, 117 and 113 nm, respectively, showing good repeatability of our measurement. The calculated average SR of EVs in each group was 100, 189, and 108 EVs per cell per min, respectively (Fig. 1c). The SR of EVs at ∼70% cell confluence was significantly higher than that at ∼45% and ∼90% cell confluence (t-test, p < 0.001). This shows that high confluence and cell amount are not proportional to the measured SR of EVs. First, in the stationary phase, the number of cells in the active cell cycle can drop to 10% or more. Second, the secreted fresh EVs may be quickly taken up by surrounding cells. Meanwhile, the high cell amount as the denominator in SR calculation also influences the calculated value. On the other hand, low confluence is not preferred either. Theoretically, at ∼45% confluence, cells should still be in the logarithmic growth phase, which is important to EV secretion as endocytosis is heavily involved in exosome genesis. We speculate that the relatively small amount of secreted EVs in serum-free medium may easily adsorb to culture flask surfaces and thus lower the value of the measured SR. Nonetheless, 70% confluence as the end point is recommended for EV harvest and was thus used in the following studies. In the case of MDA-MB-231 cells, when cell confluence reaches 20% or 40%, the FBS supplemented medium can be replaced with serum-free medium and culture can be continued for an additional 48 or 24 hours, respectively.
Successively, we investigated the influence of filtration on SR measurement. To efficiently remove MDA-MB-231 MVs and large cellular debris, 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g centrifugation was used to process CCS. In the control group, CCS was filtered with a 0.22 μm filter. The morphology of MDA-MB-231 EVs in centrifuged CCS and typical EV markers, TSG101 and CD9, was characterized (Fig. S1†). The findings indicated the presence of EVs in centrifuged CCS. The average size of MDA-MB-231 EVs in centrifuged CCS and filtered CCS was 122.8 and 113.2 nm, respectively (Fig. 1d). The size of measured EVs in centrifuged CCS was comparatively higher, 90% of EVs had a size of 185.7 ± 6.5 nm or smaller. By contrast, 90% of EVs in filtered CCS has a size of 163.3 ± 3.9 nm or smaller. Although a significant difference was found between the two groups (t-test, p < 0.0001), the majority of EVs in both groups were smaller than 400 nm which is suitable for Nanosight measurement. We also found filtration can significantly impair the measurement accuracy of the SR (Fig. 1e). The calculated average SR of EVs in filtered CCS and in non-filtered CCS was 30 and 108 EVs per cell per min (t-test, p < 0.001). The physical entrapment of EVs within the 0.22 μm filter leads to the distortion of the measured SR. The EVs might also be lacerated or ruptured under the extremely high driving pressure within the filter. It is worth noting that very low protein binding polyethersulfone filters were used for CCS filtration. The trapping of EVs in filters made of cellulose nitrate, nylon, or regenerated cellulose may further worsen the SR measurement due to the nature of low/very high protein adsorption. On the other hand, we also speculate EVs can be seriously damaged during isolation with the ultrafiltration approach. Given the significant decrease of EVs after filtration and slight size increase of EVs without filtration, we decided to use 20
000g centrifugation was used to process CCS. In the control group, CCS was filtered with a 0.22 μm filter. The morphology of MDA-MB-231 EVs in centrifuged CCS and typical EV markers, TSG101 and CD9, was characterized (Fig. S1†). The findings indicated the presence of EVs in centrifuged CCS. The average size of MDA-MB-231 EVs in centrifuged CCS and filtered CCS was 122.8 and 113.2 nm, respectively (Fig. 1d). The size of measured EVs in centrifuged CCS was comparatively higher, 90% of EVs had a size of 185.7 ± 6.5 nm or smaller. By contrast, 90% of EVs in filtered CCS has a size of 163.3 ± 3.9 nm or smaller. Although a significant difference was found between the two groups (t-test, p < 0.0001), the majority of EVs in both groups were smaller than 400 nm which is suitable for Nanosight measurement. We also found filtration can significantly impair the measurement accuracy of the SR (Fig. 1e). The calculated average SR of EVs in filtered CCS and in non-filtered CCS was 30 and 108 EVs per cell per min (t-test, p < 0.001). The physical entrapment of EVs within the 0.22 μm filter leads to the distortion of the measured SR. The EVs might also be lacerated or ruptured under the extremely high driving pressure within the filter. It is worth noting that very low protein binding polyethersulfone filters were used for CCS filtration. The trapping of EVs in filters made of cellulose nitrate, nylon, or regenerated cellulose may further worsen the SR measurement due to the nature of low/very high protein adsorption. On the other hand, we also speculate EVs can be seriously damaged during isolation with the ultrafiltration approach. Given the significant decrease of EVs after filtration and slight size increase of EVs without filtration, we decided to use 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g centrifugation to replace filtration, which still can efficiently exclude EVs larger than 500 nm.
000g centrifugation to replace filtration, which still can efficiently exclude EVs larger than 500 nm.
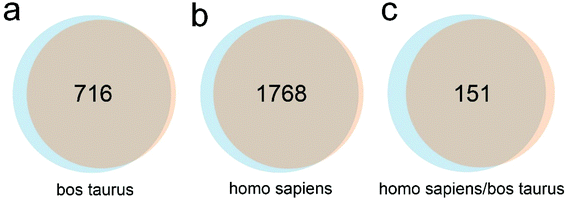
Subsequently, we examined the influence of culture time on the SR of EVs. Serum-free CCS of MDA-MB-231 cells was harvested at 24 h and 48 h time points followed by measurement (Fig. 1f). The average SR of EVs was 240 and 107 EVs per cell per min, respectively (t-test, p < 0.01). This strongly indicates SR determination can be significantly influenced by sample preparation. In addition, in the 24 h serum-free culture, the scarcity of residual FBS may drive down the cell metabolic rate and proliferation and thus impair the SR of EVs. Alternatively, overcrowd cells after 48 h culture may efficiently uptake the released EVs from adjacent cells, leading to relatively small amounts of EVs that can be secreted into the CCS. We also speculate that the two scenarios may co-exist. NanoLC-MS/MS analysis was performed to identify the FBS contaminants in EVs collected with ultracentrifugation at 24 h and 48 h time points. In the 24 h group, 3039 proteins were identified, including 831 proteins derived from Bos Taurus, 2026 proteins derived from Homo sapiens, and 182 proteins noted as “Homo sapiens/Bos taurus”. In the 48 h group, 2723 proteins were identified, including 738 proteins derived from Bos Taurus, 1824 proteins derived from Homo sapiens, and 161 proteins marked as “Homo sapiens/Bos taurus”. The percentage of Bos taurus-derived proteins in total identified proteins was 27.34% (831 out of 3039) and 27.1% (738 out of 2723), respectively (n = 3, t-test, p > 0.05). In the Venn diagrams, values represent protein overlap between the two groups (Fig. 2), showing ∼90% similarity. In addition, 1936 out of 2026 (95.6%) and 2613 out of 2723 (96%) homo sapiens-derived proteins can be identified in the public database Vesiclepedia. Furthermore, 90 out of the top 100 proteins from Vesiclepedia were identified in both EVs isolated at 24 and 48 h time point. The top 25 mutual proteins derived from FBS and their corresponding abundance in the 48 h group indicated that an additional 24 h of serum-free culture has a limited effect on further FBS elimination (Table S1†). MS analysis confirmed that vimentin, cytokeratin, EGFR, and mammary cancer stem cell marker CD44, appear in both EV samples, which agrees with the phenotype of this triple-negative breast cancer cell line. In addition, markers routinely used for EV characterization, such as CD63, CD9, CD81, TSG101, ADAM10, Syntenin-1, Annexin II, and GAPDH were also identified via MS. Notably, due to limited fractions, the amount of identified protein is less than that of 10 fractions we analyzed in a previous study.12 Altogether, 24 h serum-free culture is recommended for EV harvest. It shortens the EV collection period, but also offers higher amounts of EVs in comparison with 48 h culture. We also only maintained cells in serum-free medium for 24 hours before EV harvest in the following studies.
Moreover, EV isolation with ultracentrifugation and EV storage practices influences SR measurement. Based on the EV amount isolated with ultracentrifugation, the average SR of MDA-MB-231 EVs was determined to be 31 EVs per cell per min. The reduced SR value observed could be caused by low isolation efficiency and EV damage during pelleting. By contrast, without processing with ultracentrifugation, the average EV SR was 224 EVs per cell per min (Fig. 1g), and the average size of isolated EVs was 99.5 nm. The significant difference between the SR (t-test, p < 0.001) demonstrated that the performance of EV isolation approaches must be included in SR measurements, otherwise the characterization may cause an order of magnitude difference. On the other hand, based on the EV amount, we determined that the average isolation efficiency of EVs with ultracentrifugation is only 14%, which is in line with reported values.26,27 As for storage, based on the measured EV concentration in fresh CCS and CCS preserved at 4 °C for 72 h, we found that ∼21% of EVs could degrade. The average size of preserved EVs slightly increased to 129 nm, and 90% of EVs had a size of 210.9 ± 13 nm or smaller (Fig. 1h). This alteration may be caused by the degradation of small EVs and/or fusion between EVs. Altogether, it is not recommended to store EVs at 4 °C for more than 3 days.
Influence of drugs and cell type on the SR of EVs
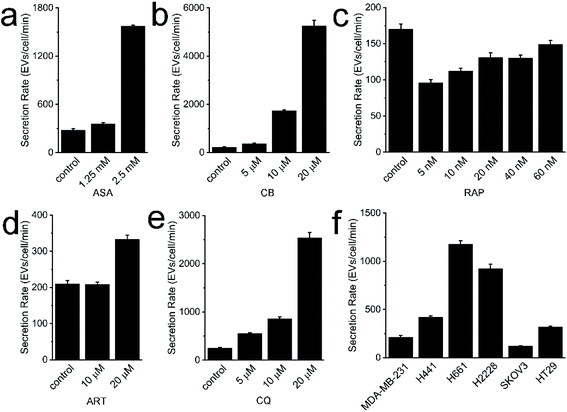
In translational studies, EVs have been explored as drug delivery nanocarriers and therapeutic agents for disease treatment.28,29 Clinical-grade EVs for the treatment of pancreatic cancer have been investigated and produced on a large-scale, employing good manufacturing practice standards.30,31 Accordingly, large quantities of EVs are required for drug delivery. Engineered hybrid EV/liposomes have also been reported.32 This strategy could alleviate this scalability issue to a certain extent. However, EVs are still inherently harvested in small quantities from culture cells. It was reported that only ∼3.4 × 1010 EVs can be harvested from ∼6.75 × 107 cells cultured in FBS-free media for 48 hours.33 To expand cells from 2.5 × 106 to 6.75 × 107 would take additional ∼6 days. The relatively low yield and long timescale may be obstacles for relevant studies and applications. Therefore, the development of methods that can improve the yield of EVs secreted by parental cells is desired. Studies have reported that EV yields could be manipulated by the stimulation of cells, plasma receptors, plasma membranes, and even intracellular electrolytes.34–36 Therefore, we speculate certain “stimulant” chemicals may promote EV generation; however, relevant studies have not been performed yet. On the other hand, cancer cell derived EVs contain oncogenic payloads inherited from parental cells. They can easily and extensively disseminate through the bloodstream whereupon they may then influence recipient cell activities, induce immune tolerance, and promote the formation of pre-metastatic niche. Some repurposed drugs have been used for cancer treatment, however, it is unclear whether these drugs can influence EV secretion and the relevant significance thereof. In this study, five repurposed drugs, i.e. ASA, CB, RAP, ART, and CQ were incubated with MDA-MB-231 cells in gradient doses. We studied whether these drugs could promote EV secretion, which may provide an alternative way for improving EV yield. Meanwhile, potential findings may also inspire our future investigation into the functions of these released EVs.Following the well-established protocol, cell viability and EV SR were determined. Since low doses were used to treat cells for only 24 hours,37–44 cell viability was not significantly impaired (Fig. S2†). The lowest cell viability of ∼93.4% was found in the 60 nM RAP group. In other groups, cell viability ranged from 95.6% to 105.1%. In the ASA study, the average SR of EVs in the control, 1.25 mM, and 2.5 mM groups was 279, 357, and 1573 EVs per cell per min, respectively (Fig. 3a). In the CB study, the average SR of EVs in the control, 5 μM, 10 μM, and 20 μM groups was 222, 364, 1636, and 5252 EVs per cell per min, respectively (Fig. 3b). In the RAP study, the average SR of EVs in the control, 5 nM, 10 nM, 20 nM, 40 nM, and 60 nM groups was 117, 96, 112, 131, 130, and 149 EVs per cell per min (Fig. 3c). In the ART study, the average SR of EVs in the control, 10 μM and 20 μM groups was 210, 207, and 333 EVs per cell per min (Fig. 3d). In the CQ study, the average SR of EVs in the control, 5 μM, 10 μM, and 20 μM groups was 248, 556, 856, and 2536 EVs per cell per min, respectively (Fig. 3e). High doses of repurposed drugs, including ASA, CB, ART and CQ, may promote the SR of EVs by stimulating cellular endocytosis, cellular budding, or other associated mechanisms. MDA-MB-231 cells, a well-known drug resistant cell line, may release drug loaded EVs in order to maintain cellular hemostasis.45 The detachment of drug treated cells from the flask followed by their removal with centrifugation caused decreased cell amounts when counting. It may also contribute to the high measured SR value. We also speculate that the enhanced SR might be caused by apoptotic or necrotic MDA-MB-231 cells after drug treatment. However, given cell viability was not significantly affected by these repurposed drugs, the corresponding influence of dead cells may be very limited. Of note, the measured SR in the group of MDA-MB-231 cells treated with high dose of RAP did not significantly increase in comparison with that of other groups. It may be caused by the inhibition of endocytosis and cytoskeleton reorganization, which would further inhibit EV generation.46,47 However, the relevant mechanism of inhibition is still unclear.48 Nevertheless, the involved mechanisms and functions of released EVs deserve further investigation.
In addition to MDA-MB-231 cells, the SR of EVs from NCI-H441, NCI-H771, NCI-H2226, SKOV3, and HT-29 cells was 210, 589, 462, 60, and 160 EVs per cell per min (Fig. 3f). As expected, the SR of EVs from different cancerous cell lines are significantly different, with an overall dynamic range of 1 log. We speculate the difference may stem from the relative malignancy of each cell line. The mode of cell proliferation, i.e., aggregated or dispersed growth, may also significantly influence the SR of EVs. The agglomerative growth of cells and mucus secretion, such as HT29, may not allow EVs to be easily released to extracellular space. Instead, the released EVs could be rapidly and efficiently taken up by the abutting cells. Moreover, it might be possible to use the SR of EVs as a marker for the prediction of the malignancy of parental cells.
Conclusion
Overall, the order of magnitude difference of the SR of EVs ranges from 1–3 logs. The measurement of SR of EVs can be influenced by sample pretreatment, cell confluence, culture time, isolation approach, storage, viability, cell types, and many other unidentified factors. Based on our findings, the use of a minimal amount of FBS to ensure normal cell proliferation and proper cell confluence, 24 h serum-free culture, and efficient isolation approaches would facilitate EV harvest with less FBS-derived contaminants.Conflicts of interest
All authors declare that they have no conflicts of interest and no competing interest.Acknowledgements
This work was partially supported by Binghamton University Faculty Startup Fund 910252-35 and Binghamton University S3IP award ADLG195.References
- M. Colombo, G. Raposo and C. Théry, Annu. Rev. Cell Dev. Biol., 2014, 30, 255–289 CrossRef CAS PubMed.
- E. R. Abels and X. O. Breakefield, Cell. Mol. Neurobiol., 2016, 36, 301–312 CrossRef CAS PubMed.
- A. Gallo, M. Tandon, I. Alevizos and G. G. Illei, Plos One, 2012, 7(3), e30679 CrossRef CAS PubMed.
- C.-X. Wu and Z.-F. Liu, J. Invest. Dermatol., 2018, 138, 89–97 CrossRef CAS PubMed.
- E. Van der Pol, A. N. Böing, P. Harrison, A. Sturk and R. Nieuwland, Pharmacol. Rev., 2012, 64, 676–705 CrossRef CAS PubMed.
- E. Willms, H. J. Johansson, I. Mäger, Y. Lee, K. E. M. Blomberg, M. Sadik, A. Alaarg, C. I. E. Smith, J. Lehtiö, S. E. L. Andaloussi, M. J. A. Wood and P. Vader, Sci. Rep., 2016, 6, 22519 CrossRef CAS PubMed.
- V. R. Minciacchi, M. R. Freeman and D. Di Vizio, Semin. Cell Dev. Biol., 2015, 40, 41–51 CrossRef CAS PubMed.
- K. Rilla, S. Pasonen-Seppänen, A. J. Deen, V. V. Koistinen, S. Wojciechowski, S. Oikari, R. Kärnä, G. Bart, K. Törrönen, R. H. Tammi and M. I. Tammi, Exp. Cell Res., 2013, 319, 2006–2018 CrossRef CAS PubMed.
- C. Théry, M. Ostrowski and E. Segura, Nat. Rev. Immunol., 2009, 9, 581–593 CrossRef PubMed.
- B.-T. Pan, K. Teng, C. Wu, M. Adam and R. M. Johnstone, J. Cell Biol., 1985, 101, 942–948 CrossRef CAS PubMed.
- H. Valadi, K. Ekström, A. Bossios, M. Sjöstrand, J. J. Lee and J. O. Lötvall, Nat. Cell Biol., 2007, 9, 654–659 CrossRef CAS PubMed.
- Y. Wan, G. Cheng, X. Liu, S. J. Hao, M. Nisic, C. D. Zhu, Y. Q. Xia, W. Q. Li, Z. G. Wang, W. L. Zhang, S. J. Rice, A. Sebastian, I. Albert, C. P. Belani and S. Y. Zheng, Nat. Biomed. Eng., 2017, 1, 0058 CrossRef CAS PubMed.
- Y. Wan, B. Liu, H. Lei, B. Zhang, Y. Wang, H. Huang, S. Chen, Y. Feng, L. Zhu, Y. Gu, Q. Zhang, H. Ma and S. Y. Zheng, Ann. Oncol., 2018, 29, 2379–2383 CrossRef CAS PubMed.
- Y. Wan, M. Maurer, H. Z. He, Y. Q. Xia, S. J. Hao, W. L. Zhang, N. S. Yee and S. Y. Zheng, Lab Chip, 2019, 19, 2346–2355 RSC.
- S. A. Kooijmans, P. Vader, S. M. van Dommelen, W. W. van Solinge and R. M. Schiffelers, Int. J. Nanomed., 2012, 7, 1525–1541 CAS.
- E. Piazuelo, P. Esquivias, A. De Martino, C. Cebrián, B. Conde, S. Santander, S. Emperador, M. A. García-González, P. Carrera-Lasfuentes and A. Lanas, Dig. Dis. Sci., 2016, 61, 2896–2907 CrossRef CAS PubMed.
- Z.-J. Dai, X.-B. Ma, H.-F. Kang, J. Gao, W.-L. Min, H.-T. Guan, Y. Diao, W.-F. Lu and X.-J. Wang, Cancer Cell Int., 2012, 12, 53 CrossRef CAS PubMed.
- H. Carraway and M. Hidalgo, Breast Cancer Res., 2004, 6, 219 CrossRef CAS PubMed.
- M. P. Crespo-Ortiz and M. Q. Wei, J. Biomed. Biotechnol., 2011, 2012, 247597 Search PubMed.
- Y. Zhang, Y. Cao, X. Sun, Y. Feng, Y. Du, F. Liu, C. Yu and F. Jin, Int. Immunopharmacol., 2017, 42, 100–107 CrossRef PubMed.
- Y. Wan, G. Cheng, X. Liu, S. J. Hao, M. Nisic, C. D. Zhu, Y. Q. Xia, W. Q. Li, Z. G. Wang, W. L. Zhang, S. J. Rice, A. Sebastian, I. Albert, C. P. Belani and S. Y. Zheng, Nat. Biomed. Eng., 2017, 1, 0058 CrossRef CAS PubMed.
- S. M. Davidson, J. A. Riquelme, Y. Zheng, J. M. Vicencio, S. Lavandero and D. M. Yellon, Sci. Rep., 2018, 8, 15885 CrossRef PubMed.
- F. J. Verweij, M. P. Bebelman, C. R. Jimenez, J. J. Garcia-Vallejo, H. Janssen, J. Neefjes, J. C. Knol, R. de Goeij-de Haas, S. R. Piersma, S. R. Baglio, M. Verhage, J. M. Middeldorp, A. Zomer, J. van Rheenen, M. G. Coppolino, I. Hurbain, G. Raposo, M. J. Smit, R. F. G. Toonen, G. van Niel and D. M. Pegtel, J. Cell Biol., 2018, 217, 1129–1142 CrossRef CAS PubMed.
- S. Yokota, H. Kuramochi, K. Okubo, A. Iwaya, S. Tsuchiya and T. Ichiki, PLoS One, 2019, 14, e0224091 CrossRef CAS PubMed.
- V. Filipe, A. Hawe and W. Jiskoot, Pharm. Res., 2010, 27, 796–810 CrossRef CAS PubMed.
- J. Caradec, G. Kharmate, E. Hosseini-Beheshti, H. Adomat, M. Gleave and E. Guns, Clin. Biochem., 2014, 47, 1286–1292 CrossRef CAS PubMed.
- M. Oksvold, Arch. Med., 2015, 6, 18 Search PubMed.
- D. Ha, N. Yang and V. Nadithe, Acta Pharm. Sin. B, 2016, 6, 287–296 CrossRef PubMed.
- R. S. Conlan, S. Pisano, M. I. Oliveira, M. Ferrari and I. M. Pinto, Trends Mol. Med., 2017, 23, 636–650 CrossRef CAS PubMed.
- M. Mendt, S. Kamerkar, H. Sugimoto, K. M. McAndrews, C. C. Wu, M. Gagea, S. Yang, E. V. R. Blanko, Q. Peng, X. Ma, J. R. Marszalek, A. Maitra, C. Yee, K. Rezvani, E. Shpall, V. S. LeBleu and R. Kalluri, JCI Insight, 2018, 3(8), e99263 CrossRef PubMed.
- H. G. Lamparski, A. Metha-Damani, J. Y. Yao, S. Patel, D. H. Hsu, C. Ruegg and J. B. Le Pecq, J. Immunol. Methods, 2002, 270, 211–226 CrossRef CAS PubMed.
- Y. T. Sato, K. Umezaki, S. Sawada, S. A. Mukai, Y. Sasaki, N. Harada, H. Shiku and K. Akiyoshi, Sci. Rep., 2016, 6, 21933 CrossRef CAS PubMed.
- Y. Wan, L. Wang, C. Zhu, Q. Zheng, G. Wang, J. Tong, Y. Fang, Y. Xia, G. Cheng, X. He and S. Y. Zheng, Cancer Res., 2018, 78, 798–808 CrossRef CAS PubMed.
- A. Savina, C. M. Fader, M. T. Damiani and M. I. Colombo, Traffic, 2005, 6, 131–143 CrossRef CAS PubMed.
- G. Lachenal, K. Pernet-Gallay, M. Chivet, F. J. Hemming, A. Belly, G. Bodon, B. Blot, G. Haase, Y. Goldberg and R. Sadoul, Mol. Cell. Neurosci., 2011, 46, 409–418 CrossRef CAS PubMed.
- G. Raposo and W. Stoorvogel, J. Cell Biol., 2013, 200, 373–383 CrossRef CAS PubMed.
- M. Sambi, V. Samuel, B. Qorri, S. Haq, S. V. Burov, E. Markvicheva, W. Harless and M. R. Szewczuk, Drug Des., Dev. Ther., 2020, 14, 1995–2019 CrossRef PubMed.
- G. T. Ma, S. K. Lee, K.-K. Park, J. Park, S. H. Son, M. Jung and W.-Y. Chung, Cell. Physiol. Biochem., 2018, 49, 1460–1475 CrossRef CAS PubMed.
- G. Wang, J. Li, L. Zhang, S. Huang, X. Zhao and X. Zhao, Biochem. Biophys. Res. Commun., 2017, 490, 969–976 CrossRef CAS PubMed.
- E. Seront, R. Boidot, C. Bouzin, O. Karroum, B. Jordan, B. Gallez, J.-P. Machiels and O. Feron, Br. J. Cancer, 2013, 109, 2597–2606 CrossRef CAS PubMed.
- K. Kumari, S. Keshari, D. Sengupta, S. C. Sabat and S. K. Mishra, BMC Cancer, 2017, 17, 858 CrossRef PubMed.
- N. Sharma, S. Thomas, E. B. Golden, F. M. Hofman, T. C. Chen, N. A. Petasis, A. H. Schonthal and S. G. Louie, Cancer Lett., 2012, 326, 143–154 CrossRef CAS PubMed.
- Z. J. Dai, X. B. Ma, H. F. Kang, J. Gao, W. L. Min, H. T. Guan, Y. Diao, W. F. Lu and X. J. Wang, Cancer Cell Int., 2012, 12, 53 CrossRef CAS PubMed.
- W. C. Noh, W. H. Mondesire, J. Peng, W. Jian, H. Zhang, J. Dong, G. B. Mills, M. C. Hung and F. Meric-Bernstam, Clin. Cancer Res., 2004, 10, 1013–1023 CrossRef CAS PubMed.
- A. Takahashi, R. Okada, K. Nagao, Y. Kawamata, A. Hanyu, S. Yoshimoto, M. Takasugi, S. Watanabe, M. T. Kanemaki, C. Obuse and E. Hara, Nat. Commun., 2017, 8, 15287 CrossRef CAS PubMed.
- L. Liu, Y. Luo, L. Chen, T. Shen, B. Xu, W. Chen, H. Zhou, X. Han and S. Huang, J. Biol. Chem., 2010, 285, 38362–38373 CrossRef CAS PubMed.
- L. Liu, L. Chen, J. Chung and S. Huang, Oncogene, 2008, 27, 4998–5010 CrossRef CAS PubMed.
- A. W. Thomson, H. R. Turnquist and G. Raimondi, Nat. Rev. Immunol., 2009, 9, 324–337 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0an01199a |
| This journal is © The Royal Society of Chemistry 2020 |