Correlative surface imaging reveals chemical signatures for bacterial hotspots on plant roots
Wen
Liu†
ab,
Liuqin
Huang†
 ab,
Rachel
Komorek
c,
Pubudu P.
Handakumbura
a,
Yadong
Zhou
a,
Dehong
Hu
ab,
Rachel
Komorek
c,
Pubudu P.
Handakumbura
a,
Yadong
Zhou
a,
Dehong
Hu
 a,
Mark H.
Engelhard
a,
Mark H.
Engelhard
 a,
Hongchen
Jiang
a,
Hongchen
Jiang
 b,
Xiao-Ying
Yu
b,
Xiao-Ying
Yu
 c,
Christer
Jansson
a and
Zihua
Zhu
c,
Christer
Jansson
a and
Zihua
Zhu
 *a
*a
aEnvironmental Molecular Science Laboratory, Pacific Northwest National Laboratory, Richland, WA 99354, USA. E-mail: Zihua.zhu@pnnl.gov
bState Key Laboratory of Biogeology and Environmental Geology, China University of Geosciences, Wuhan, 430074, China
cEnergy and Environment Directorate, Pacific Northwest National Laboratory, Richland, WA 99354, USA
First published on 7th November 2019
Abstract
The rhizosphere is arguably the most complex microbial habitat on Earth, comprising an integrated network of plant roots, soil and a highly diverse microbial community (the rhizosphere microbiome). Understanding, predicting and controlling plant-microbe interactions in the rhizosphere will allow us to harness the plant microbiome as a means to increase or restore plant ecosystem productivity, improve plant responses to a wide range of environmental perturbations, and mitigate the effects of climate change by designing ecosystems for long-term soil carbon storage. To this end, it is imperative to develop new molecular approaches with high spatial resolution to capture interactions at the plant-microbe, microbe-microbe, and plant-plant interfaces. In this work, we designed an imaging sample holder that allows integrated surface imaging tools to map the same locations of a plant root-microbe interface with submicron lateral resolutions, providing novel in vivo analysis of root-microbe interactions. Specifically, confocal fluorescence microscopy, time-of-flight secondary ion mass spectrometry (ToF-SIMS), X-ray photoelectron spectroscopy (XPS), and scanning electron microscopy (SEM) were used for the first time for the correlative imaging of the Brachypodium distachyon root and its interaction with Pseudomonas SW25, a typical plant growth-promoting soil bacterium. Imaging data suggest that the root surface is inhomogeneous and that the interaction between Pseudomonas and Brachypodium roots was confined to only a few spots along the sampled root segments and that the bacterial attachment spots were enriched in Na- and S-related and high-mass organic species. We conclude that the attachment of the Pseudomonas cells to the root surface is outcompeted by strong root-soil mineral interactions but facilitated by the formation of extracellular polymeric substances (EPS).
Introduction
The rhizosphere, i.e., the thin region surrounding and including roots, is a well-known hotspot for microbial activity and abundance (the rhizosphere microbiome) due to the enrichment of root exudates and debris.1,2 Some members of the rhizosphere microbiome, referred to as plant growth-promoting bacteria (PGPB),3,4 can stimulate plant growth and productivity by reducing pathogenic infection,5 enhancing tolerance to abiotic stress such as drought,6 or increasing the provision of nutrients like P and N.7,8 The application of PGPB has proved to be promising as bio-fertilizers or pesticides, which can significantly reduce environmental pollution from the overuse of chemical fertilizers (e.g., N and P) and toxic pesticides in cropping systems.9,10 However, rhizosphere interactions are a complex function of biotic and abiotic factors, making it difficult to translate findings from PGPB inoculations performed under controlled laboratory conditions to the stochastic field environment.11,12 Thus, detailed, molecular and elemental understanding of plant-microbe interactions in the rhizosphere is vital for our ability to fully harness the rhizosphere microbiome for agricultural applications.13Brachypodium distachyon (Brachypodium) is a widely distributed annual monocot grass that has been proposed as a model organism for grasses, including bioenergy grasses, due to its suitable traits such as small genome, short lifetime, simple growth conditions and amenability to genetic modification.14–17 It has been widely used for many fundamental studies including plant-microbe interactions.18–21Pseudomonas sp. are commonly found in the rhizosphere and many isolates (e.g., Pseudomonas SW25 used in this study22 and P. fluorescens23) have been widely studied as model PGPB. However, the mechanisms of Pseudomonas on promoting plant growth are still under debate and may include pathogen suppression, P release, N fixation and hormonal regulation.23–25 Even less is known about the fundamental principles that control the interactions between plant roots and the Pseudomonas bacteria.
The rhizosphere is a highly heterogeneous system, mainly composed of roots, mineral particles, organic matters and various microbes.1 Most commonly used analysis tools in this field such as total carbon analysis,26 FT-ICR-MS, NMR,27 and genomics28 are bulk analysis techniques, i.e., samples need to be extracted from the system using solvents. Although such approaches have provided key information for developing various models to explain mass transfer in the rhizosphere, they are associated with at least two intrinsic drawbacks. First, these approaches lack spatial information to describe the distribution of microbes or organic matter, e.g., along a root segment. Second, although some organic or bioorganic molecules are soluble, many molecular species may not be soluble and/or be firmly attached to mineral or root surfaces. Scanning electron microscopy (SEM)29 and transmission electron microscopy (TEM)30 have been introduced in the field, providing morphological information with good spatial resolution, albeit with mostly elemental and no or little molecular information. Traditionally, fluorescence microscopy has been widely used in this field, and recently nanoscale secondary ion mass spectrometry (NanoSIMS) has been employed to map the distribution of organic matter on mineral surfaces.31 These techniques can provide high spatial-resolution (down to tens of a nm) chemical maps, although, generally, only selected species with specially labelled fluorescent or isotope tags can be tracked.
Time-of-flight secondary ion mass spectrometry (ToF-SIMS) is a powerful surface analysis tool with several unique advantages.32 First, it can provide elemental, isotopic and molecular information simultaneously. Second, its information depth is very shallow (normally 1–3 nm), so surface-specific information can be collected. In addition, it has excellent sensitivity (ppm level) and very good spatial resolution (sub-micron).33 Therefore, it is a very useful tool in studies of the rhizosphere. Clearly, each technique has its own strength and weaknesses, and a single technique approach normally can provide only limited information. Therefore, a multi-technique approach is highly desirable for comprehensive interrogation of a complex system such as the rhizosphere.
In this work, confocal fluorescence microscopy, ToF-SIMS, X-ray photoelectron spectroscopy (XPS) and SEM were collectively used for the first time to image the Brachypodium root and root-microbe interactions.
Materials and methods
Four Brachypodium plants (named U1, U3, I1, and I2) were grown. The U1 and U3 plants were used as reference (without Pseudomonas treatment), and the I1 and I2 plants were treated with Pseudomonas. In brief, Brachypodium distachyon, reference line Bd21, was used as the plant system. Soil from the PNNL's field site in Prosser, WA, United States, was used as the growing medium. The Prosser soil is characterized as a Warden fine sandy/silt loam with 0.4% organic matter and 1 mg kg−1 ammonium N per dry weight. Brachypodium seeds were sown on sterilized (autoclaved) Prosser soil and placed in a Percival growth chamber with 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 h light
8 h light![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dark cycles with 22
dark cycles with 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 18 °C day
18 °C day![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) night temperatures, 60% RH, and ∼250 μmol m−2 s−1 light intensity. Pseudomonas fluorescens SW25 bacteria, which expressed the green fluorescence protein (GFP), were injected into the soil surrounding the I1 and I2 plants at 3 weeks post germination; the bacteria were grown to log phase OD600, pelleted and re-suspended to a specific OD = 0.6–0.8 before pipetting them into the soil/base of each plant. At the end of the experiment root samples were harvested for subsequent analysis.
night temperatures, 60% RH, and ∼250 μmol m−2 s−1 light intensity. Pseudomonas fluorescens SW25 bacteria, which expressed the green fluorescence protein (GFP), were injected into the soil surrounding the I1 and I2 plants at 3 weeks post germination; the bacteria were grown to log phase OD600, pelleted and re-suspended to a specific OD = 0.6–0.8 before pipetting them into the soil/base of each plant. At the end of the experiment root samples were harvested for subsequent analysis.
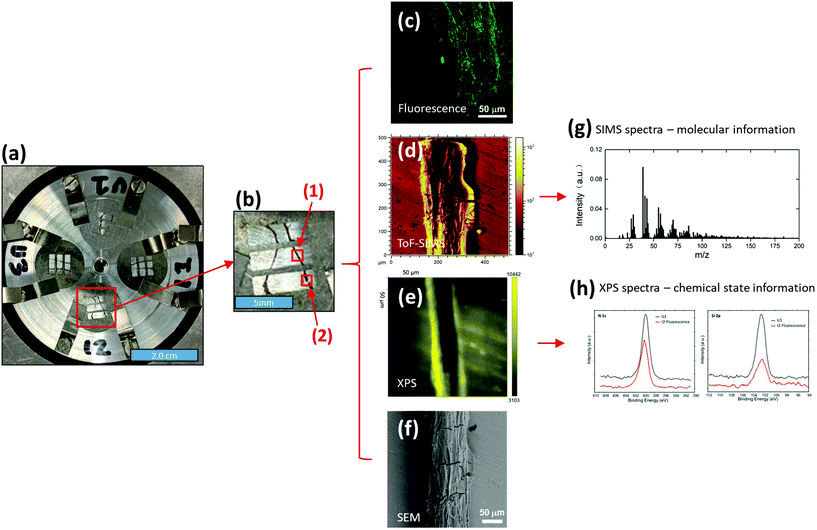
Two root segments (about 12–15 mm each) close to the bacterial inoculation areas were selected and excised from each of the I1 and I2 plants. Similarly, two roots from the corresponding areas were selected and cut from each of the U1 and U3 plants. The roots were gently rinsed with deionized water to remove loosely attached soil particles and then immobilized onto a special sample holder (Fig. 1a) that was developed as part of this study. In brief, eight stainless steel pins were used to immobilize four molybdenum (Mo) masks on a ∼5.0 cm diameter aluminum (Al) sample holder (thickness ∼ 6.0 mm). Four Mo masks (12.7 mm diameter and 0.10 mm thick each) were used to press root samples to make them flat. Sample flatness was critical to ensure high quality data from surface analysis tools such as ToF-SIMS and XPS. On each Mo mask, three 5.0 × 1.5 mm2 windows were open for imaging analysis. Such a design allowed us to easily determine accurate locations for multi-imaging analysis. For example, as shown in Fig. 1b, two root samples were immobilized under a Mo mask, and six selected locations, including locations (1) and (2), could be easily located and imaged using different imaging tools.
The initial analysis of fresh root segments was performed by fluorescence microscopy using an upright confocal fluorescence microscope (Zeiss LSM 710) under ambient conditions. The segments were excited using an Ar ion laser with a 488 nm wavelength. The objective was 40× NA 0.75. The fluorescence detection had a wavelength range of 498 nm to 550 nm for GFP. A 3D Z stack was acquired for every sample. The locations of fluorescence images were recorded for subsequent ToF-SIMS, XPS and SEM imaging analysis.
Fluorescence microscopy was followed by ToF-SIMS imaging using a TOF-SIMS5 instrument (IONTOF GmbH, Műnster, Germany). Before SIMS analysis, the sample holder was put into the introduction chamber of the SIMS instrument, and the root samples were dried under vacuum. After drying, the sample holder was introduced into the analysis chamber for SIMS imaging analysis. A 25 keV Bi3+ beam was used as the analysis beam to collect SIMS spectra and images. The Bi3+ beam was focused to be ∼0.5 μm diameter and scanned over 200 × 200 μm2 to 500 × 500 μm2 areas. The current of the Bi3+ beam was about 0.36 pA with 10 kHz pulse frequency, and the data collection time was 600 s per imaging testing. The total ion dose was under the static limit so only surface information (<2 nm) was collected. While collecting data, a low energy electron flood gun (10 eV, ∼1.0 μA current) was used to compensate for surface charging. The pressure in the analysis chamber was about 2 × 10−8 mbar. A positive ion imaging testing and a negative ion imaging testing were performed on each selected location (totally 24 locations, 6 on each Mo mask, as shown in Fig. 1a and b). Due to the complexity of ToF-SIMS spectral data, principal component analysis (PCA)34,35 was used to extract effective information, following procedures described in our previous work.36–38 Because only SIMS signals on the root surface were of interest for PCA analysis, data reconstruction was required. SIMS imaging capability showed its importance here. During data reconstruction, only root surface areas were selected based on SIMS images (e.g., Fig. 1d). Thus, SIMS signals on root surfaces were shown in reconstructed mass spectra for further PCA analysis, and interference signals from the substrate could be removed.
XPS measurements were conducted on the same samples after ToF-SIMS analysis. A Physical Electronics Quantera Scanning X-ray Microprobe was used. This system used a focused monochromatic Al Kα X-ray (1486.7 eV) source for excitation and a spherical section analyzer. The instrument had a 32-element multichannel detection system. The X-ray beam was incident normal to the sample and the photoelectron detector was at 45° off-normal. High energy resolution spectra were collected on root surfaces using a pass-energy of 69.0 eV with a step size of 0.125 eV. For the Ag 3d5/2 line, these conditions produced a FWHM of 0.92 eV ± 0.05 eV. The binding energy (BE) scale was calibrated using the Cu 2p3/2 feature at 932.62 ± 0.05 eV and the Au 4f7/2 feature at 83.96 ± 0.05 eV. The sample experienced variable degrees of charging. Low energy electrons at ∼1 eV, 19 μA and low energy Ar+ ions were used to minimize this charging.
SEM imaging was performed after the XPS measurements. A Hitachi TM-1000 scanning electron microscope (Chiyoda, Tokyo, Japan) was used with an accelerating voltage of 15.0 kV. Before SEM imaging, about 2 nm Au was coated on top of the sample to reduce charging. The imaging areas were ranging from 100 × 75 μm2 to 500 × 375 μm2.
Results and discussion
Fluorescence was used to assess whether or not Pseudomonas cells were evenly distributed on the Brachypodium root surface. Interestingly, we found that the attachment of Pseudomonas was obvious at only two locations on the root, locations (1) and (2) of the I2 plant (Fig. 1b).The fluorescence microscopy results prompted the question of how the surface properties of the attachment spots differed from the rest of the root. Here, ToF-SIMS spectra (Fig. 2) can provide valuable chemical information, including elemental, isotopic and molecular information, to elucidate the difference. However, ToF-SIMS spectra are complex, and each spectrum may be composed of hundreds of ion signals. Thus, statistical analysis was used to distinguish features that differentiate the regions with and without Pseudomonas.
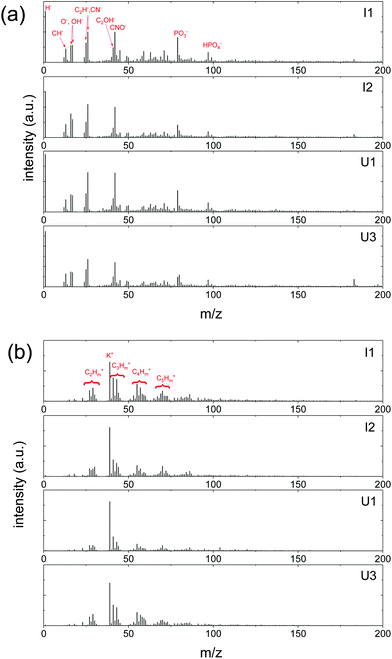 | ||
| Fig. 2 Representative ToF-SIMS spectra of I1, I2, U1 and U3 samples. (a) Negative ion spectra and (b) positive ion spectra. | ||
Principal Component Analysis (PCA) has been widely used in ToF-SIMS data analysis for over a decade. Fig. 3 shows the PCA analysis results of negative ion spectra collected from the four sets of Brachypodium root samples. The PC1 scores plot (Fig. 3a) revealed that only two spots were clearly separated. It should be noted that after PCA analysis, each spectrum has its own PC1 score value, that is to say, each data spot in Fig. 3a corresponds to a spectrum. Thus, based on the PC1 score values, these two separated data spots are found to be corresponding to the spectra from the locations (1) and (2) of the I2 sample (Fig. 1b). The two separated data spots are close to each other, and the major difference between them and the remaining data spots is the decrease in PC1 scores. The PC1 loadings plot (Fig. 3b) shows that the positive loadings of PC1 are mainly CN and POx-related species, as well as low-mass organic species, while the negative PC1 loadings are mainly Cl and organic SOx species, as well as high-mass organic species. The loadings plot data suggest that the surface of attachment spots for the Pseudomonas cells had relatively more Cl, organic SOx and high-mass organic species, but less CN, POx-related species and low-mass organic species. Moreover, the PC1 scores of the remaining data spots are close together, suggesting that all Pseudomonas-free root surfaces exhibited a similar micro-chemical environment.
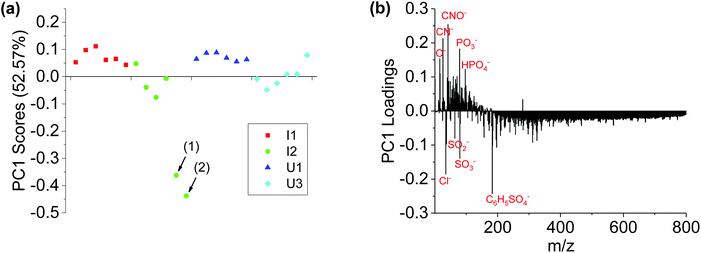 | ||
| Fig. 3 PCA analysis results of negative ion spectra of I1, I2, U1 and U3 samples. (a) PC1 scores plot and (b) PC1 loadings plot. It should be noted that the data spots (1) and (2) in (a) correspond to the two locations (1) and (2) in Fig. 1b, where Pseudomonas attachment was observed. | ||
Fig. 4 shows the PCA analysis results of positive ion spectra collected from the four sets of Brachypodium root samples. The PCA scores plot (Fig. 4a) is qualitatively consistent with the negative ion results. The PC1 scores of locations (1) and (2) are well separated from the remaining data spots. The loadings plot (Fig. 4b) shows that the major positive loadings of PC1 are Na-related species and high-mass organic species, and the negative loadings are K+, NH4+ and low-mass organic N species. The data suggest that the root surface of the Pseudomonas attachment spots had relatively more Na-related species and high-mass organic species, but less K+, NH4+ and low-mass organic N species.
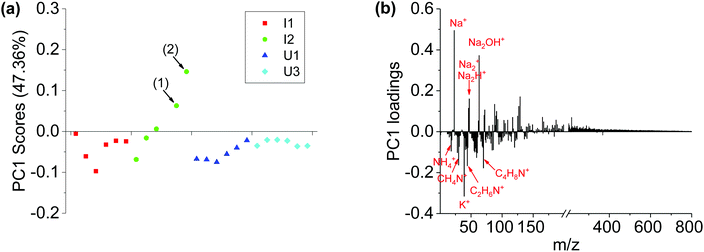 | ||
| Fig. 4 PCA analysis results of positive ion spectra of I1, I2, U1 and U3 samples. (a) PC1 scores plot and (b) PC1 loadings plot. It should be noted that the data spots (1) and (2) in (a) are corresponding to the two locations (1) and (2) in Fig. 1b, where Pseudomonas was observed. | ||
Importantly, PCA analysis results from negative ion spectra and positive ion spectra are consistent with each other. First, PC1 scores could separate Pseudomonas attachment spots from Pseudomonas-free root segments. Second, both positive ion results and negative ion results show that small N-contained organic species are enriched on the Pseudomonas-free root surface, while high-mass organic species are enriched on the surface of Pseudomonas attachment spots.
Fig. 5 shows XPS data from two representative locations on the roots of the I2 and U1 plants. More N, K and Si were observed on the Pseudomonas-free root surface, while more S was observed on Pseudomonas attachment spots. These results are well consistent with the results from ToF-SIMS/PCA analysis. For example, both XPS S spectra and PCA analysis results of ToF-SIMS negative ion spectra show more –SOx groups on the surface of Pseudomonas attachment spots. Such a consistency is very reasonable, because both techniques are surface sensitive, sharing similar information depth (a few nanometers) during analysis. It should be noted that XPS can provide chemical state information, which cannot be obtained from fluorescence, SIMS and SEM analysis. A notable observation in S 2p spectra (Fig. 5b) is two chemical states of S (S6+ and S0/S2−) on the Pseudomonas-free root surface, but only one dominant chemical state of S (S6+) on the Pseudomonas attachment spots. One possible explanation for such an observation is that there was some (∼1 mM) MgSO4 in the growth medium for Pseudomonas, so that some SO42− might stay in the extracellular polymeric substances of the Pseudomonas biofilm.
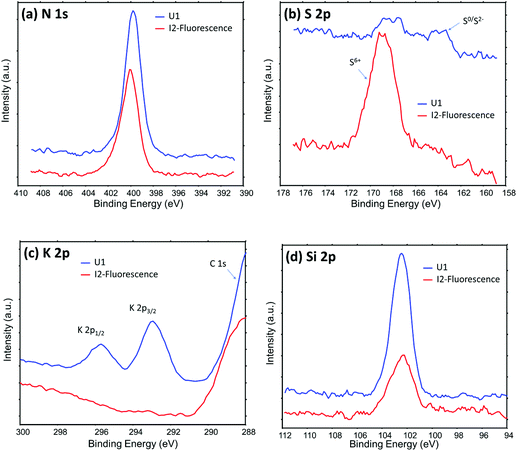 | ||
| Fig. 5 High energy resolution XPS data showing the chemical difference between the Pseudomonas-free root surface (U1 plant) and the root surface of Pseudomonas attachment spots (I2 plant, location (1) in Fig. 2b). | ||
Fig. 6 displays the SEM images of roots with and without Pseudomonas attachment. The root surface with Pseudomonas attachment looks smoother, almost free of soil particles. In contrast, many small soil particles were observed on the Pseudomonas-free root surface. This situation is in agreement with the ToF-SIMS and XPS data that there are more Si and K (from soil mineral particles) on the Pseudomonas-free root surface.
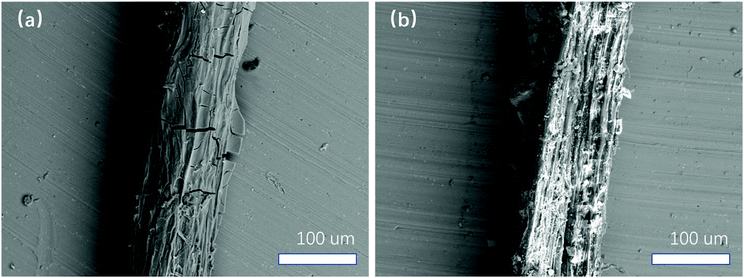 | ||
| Fig. 6 SEM images of root surfaces obtained after ToF-SIMS measurements. (a) Root surface from the I2 plant at location (1) with Pseudomonas attachment shown in Fig. 1b. (b) Pseudomonas-free root surface on a root segment from the U1 plant. | ||
The above SEM observation is very interesting. From the literature, biofilms are usually implicated in aggregation processes because soil particles stick to them.39 If so, a possible explanation for the above observation is that Pseudomonas could only attach on soil particle-free areas. However, SEM images show that all root surfaces from U1 and U3 are with some considerable amount of soil particles, indicating that soil particle-free root surfaces are rare before Pseudomonas treatment. If so, another possibility is that Pseudomonas attachment might reduce direct interactions between root and soil particles.
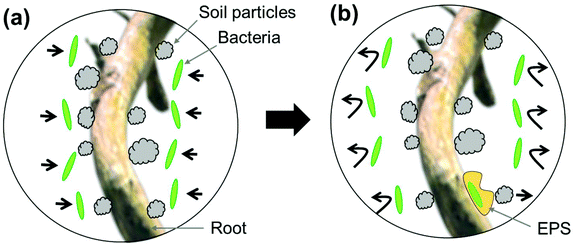
It is notable that Pseudomonas attachment was observed only on two locations along the four root segments from two Pseudomonas-inoculated Brachypodium plants (I1 and I2), i.e., locations (1) and (2) of the I2 plant. One possible explanation for such an observation is that the Pseudomonas treatment process was not uniform. However, this is doubtful since a large amount of Pseudomonas bacterial solution was added to the soil around the I1 and I2 plants, and root segments for imaging were selected close to the inoculation site. A more plausible explanation is that direct interactions between Pseudomonas cells and Brachypodium roots is weak and outcompeted by root-mineral interactions (Fig. 7a). Such a weak interaction between Pseudomonas cells and Brachypodium roots has been confirmed by a separate research study in our lab.40 In that work, the Brachypodium roots grew in liquid media with glass beads (not in soil), in which Pseudomonas seemed to have difficulty in attaching to the free root surface in the media, but could aggregate on the glass bead-root interface.40 Therefore, we tend to believe that Pseudomonas may produce extracellular polymeric substances to form a biofilm on a small amount of root surfaces and that the biofilm can physically separate roots and soil particles (Fig. 7b).
Conclusions
Our data suggest that the root surface is a very inhomogeneous system, and chemical imaging tools with sufficient spatial resolution (from nm to mm) are valuable in elucidating the complex interactions at the plant-microbe, microbe-microbe, and plant-plant interfaces. Using a correlative imaging approach comprised of fluorescence microscopy, ToF-SIMS, high-energy resolution XPS, and SEM, and enabled by the development of a universal sample holder, we found that the interaction between Pseudomonas fluorescens SBW25 and Brachypodium distachyon roots was weak and confined to only a few spots along the sampled root segments. Chemical imaging supported by PCA analysis suggests that the bacterial attachment spots were enriched in Na- and S-related and high-mass organic species, whereas the bacterial-free root surface was enriched in N, K and Si species. We hypothesize that (1) the enrichment of N, K and Si on the Pseudomonas-free root surface indicates the presence of soil particles and (2) the attachment of the Pseudomonas cells to the root surface is outcompeted by strong root-soil mineral interactions but facilitated by the formation of EPS, as reflected by the accumulation of high-mass organic species at the attachment spots. Given the considerable interest in harnessing the plant microbiome for mitigating climate change and improving plant productivity and health,41–45 it is critical that we obtain a comprehensive insight into the mechanisms that govern plant-microbe interactions. We argue that the correlative surface imaging strategy presented here offers great potential for our ability to understand, predict and control rhizosphere interactions for desirable outputs.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the Department of Energy (DOE) Office of Biological and Environmental Research (BER) and was conducted at the Environmental Molecular Sciences Laboratory (EMSL), a DOE user facility, as a contribution to the EMSL Strategic Science Area (project 22142, user proposal 50170). The authors thank for helpful discussions with Professor Rene Boiteau at Oregon State University.References
- A. H. Ahkami, R. A. White III, P. P. Handakumbura and C. Jansson, Rhizosphere engineering: Enhancing sustainable plant ecosystem productivity, Rhizosphere, 2017, 3, 233–243 CrossRef.
- H. P. Bais, T. L. Weir, L. G. Perry, S. Gilroy and J. M. Vivanco, The role of root exudates in rhizosphere interactions with plants and other organisms, Annu. Rev. Plant Biol., 2006, 57, 233–266 CrossRef CAS PubMed.
- M. Ahemad and M. Kibret, Mechanisms and applications of plant growth promoting rhizobacteria: Current perspective, J. King Saud Univ., Sci., 2014, 26(1), 1–20 CrossRef.
- J. W. Kloepper, J. Leong, M. Teintze and M. N. Schroth, Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria, Nature, 1980, 286(5776), 885–886 CrossRef CAS.
- C. Pieterse, S. Van Wees, E. Hoffland, J. A. Van Pelt and L. C. Van Loon, Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression, Plant Cell, 1996, 8(8), 1225–1237 CAS.
- J. Yang, J. W. Kloepper and C.-M. Ryu, Rhizosphere bacteria help plants tolerate abiotic stress, Trends Plant Sci., 2009, 14(1), 1–4 CrossRef CAS PubMed.
- A. Zaidi, M. Khan, M. Ahemad and M. Oves, Plant growth promotion by phosphate solubilizing bacteria, Acta Microbiol. Immunol. Hung., 2009, 56(3), 263–284 CrossRef CAS PubMed.
- N. Otieno, R. D. Lally, S. Kiwanuka, A. Lloyd, D. Ryan, K. J. Germaine and D. N. Dowling, Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates, Front. Microbiol., 2015, 6, 745 Search PubMed.
- G. Berg, Plant–microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture, Appl. Microbiol. Biotechnol., 2009, 84(1), 11–18 CrossRef CAS PubMed.
- P. Vejan, R. Abdullah, T. Khadiran, S. Ismail and A. N. Boyce, Role of Plant Growth Promoting Rhizobacteria in Agricultural Sustainability—A Review, Molecules, 2016, 21(5), 573 CrossRef PubMed.
- R. Dey, K. K. Pal, D. M. Bhatt and S. M. Chauhan, Growth promotion and yield enhancement of peanut (Arachis hypogaea L.) by application of plant growth-promoting rhizobacteria, Microbiol. Res., 2004, 159(4), 371–394 CrossRef CAS PubMed.
- S. Compant, C. Clément and A. Sessitsch, Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization, Soil Biol. Biochem., 2010, 42(5), 669–678 CrossRef CAS.
- R. d. Souza, A. Ambrosini and L. M. Passaglia, Plant growth-promoting bacteria as inoculants in agricultural soils, Genet. Mol. Biol., 2015, 38(4), 401–419 CrossRef PubMed.
- J. Brkljacic, E. Grotewold, R. Scholl, T. Mockler, D. F. Garvin, P. Vain, T. Brutnell, R. Sibout, M. Bevan, H. Budak, A. L. Caicedo, C. Gao, Y. Gu, S. P. Hazen, B. F. Holt, S.-Y. Hong, M. Jordan, A. J. Manzaneda, T. Mitchell-Olds, K. Mochida, L. A. J. Mur, C.-M. Park, J. Sedbrook, M. Watt, S. J. Zheng and J. P. Vogel, Brachypodium as a Model for the Grasses: Today and the Future, Plant Physiol., 2011, 157(1), 3–13 CrossRef CAS PubMed.
- K.-B. G. Scholthof, S. Irigoyen, P. Catalan and K. K. Mandadi, Brachypodium: A Monocot Grass Model Genus for Plant Biology, Plant Cell, 2018, 30(8), 1673–1694 CrossRef CAS PubMed.
- M. Opanowicz, P. Vain, J. Draper, D. Parker and J. H. Doonan, Brachypodium distachyon: making hay with a wild grass, Trends Plant Sci., 2008, 13(4), 172–177 CrossRef CAS PubMed.
- T. Douché, H. S. Clemente, V. Burlat, D. Roujol, B. Valot, M. Zivy, R. Pont-Lezica and E. Jamet, Brachypodium distachyon as a model plant toward improved biofuel crops: Search for secreted proteins involved in biogenesis and disassembly of cell wall polymers, Proteomics, 2013, 13(16), 2438–2454 CrossRef PubMed.
- F. P. Do Amaral, V. C. Pankievicz, A. C. M. Arisi, E. M. de Souza, F. Pedrosa and G. Stacey, Differential growth responses of Brachypodium distachyon genotypes to inoculation with plant growth promoting rhizobacteria, Plant Mol. Biol., 2016, 90(6), 689–697 CrossRef CAS PubMed.
- S. Zhong, S. Ali, Y. Leng, R. Wang and D. F. Garvin, Brachypodium distachyon–Cochliobolus sativus pathosystem is a new model for studying plant–fungal interactions in cereal crops, Phytopathology, 2015, 105(4), 482–489 CrossRef PubMed.
- T. L. Fitzgerald, J. J. Powell, K. Schneebeli, M. M. Hsia, D. M. Gardiner, J. N. Bragg, C. L. McIntyre, J. M. Manners, M. Ayliffe and M. Watt, Brachypodium as an emerging model for cereal–pathogen interactions, Ann. Bot., 2015, 115(5), 717–731 CrossRef CAS PubMed.
- M. Figueroa, S. Alderman, D. F. Garvin and W. F. Pfender, Infection of Brachypodium distachyon by formae speciales of Puccinia graminis: early infection events and host-pathogen incompatibility, PLoS One, 2013, 8(2), e56857 CrossRef CAS PubMed.
- A. Adesemoye, M. Obini and E. Ugoji, Comparison of plant growth-promotion with Pseudomonas aeruginosa and Bacillus subtilis in three vegetables, Braz. J. Microbiol., 2008, 39(3), 423–426 CrossRef CAS PubMed.
- P. B. Rainey, Adaptation of Pseudomonas fluorescens to the plant rhizosphere, Environ. Microbiol., 1999, 1(3), 243–257 CrossRef CAS PubMed.
- O. Martínez-Viveros, M. Jorquera, D. Crowley, G. Gajardo and M. Mora, Mechanisms and practical considerations involved in plant growth promotion by rhizobacteria, J. Soil Sci. Plant Nutr., 2010, 10(3), 293–319 Search PubMed.
- V. T. Pham, H. Rediers, M. G. Ghequire, H. H. Nguyen, R. De Mot, J. Vanderleyden and S. Spaepen, The plant growth-promoting effect of the nitrogen-fixing endophyte Pseudomonas stutzeri A15, Arch. Microbiol., 2017, 199(3), 513–517 CrossRef CAS PubMed.
- B. Zhu, J. L. M. Gutknecht, D. J. Herman, D. C. Keck, M. K. Firestone and W. Cheng, Rhizosphere priming effects on soil carbon and nitrogen mineralization, Soil Biol. Biochem., 2014, 76, 183–192 CrossRef CAS.
- D. I. Kaplan, C. Xu, S. Huang, Y. Lin, N. Tolić, K. M. Roscioli-Johnson, P. H. Santschi and P. R. Jaffé, Unique organic matter and microbial properties in the rhizosphere of a wetland soil, Environ. Sci. Technol., 2016, 50(8), 4169–4177 CrossRef CAS PubMed.
- S. Shi, E. E. Nuccio, Z. J. Shi, Z. He, J. Zhou and M. K. Firestone, The interconnected rhizosphere: high network complexity dominates rhizosphere assemblages, Ecol. Lett., 2016, 19(8), 926–936 CrossRef PubMed.
- E. Barahona, A. Navazo, F. Yousef-Coronado, D. Aguirre de Cárcer, F. Martínez-Granero, M. Espinosa-Urgel, M. Martín and R. Rivilla, Efficient rhizosphere colonization by Pseudomonas fluorescens f113 mutants unable to form biofilms on abiotic surfaces, Environ. Microbiol., 2010, 12(12), 3185–3195 CrossRef CAS PubMed.
- M. F. Guerrero-Molina, B. C. Winik and R. O. Pedraza, More than rhizosphere colonization of strawberry plants by Azospirillum brasilense, Appl. Soil Ecol., 2012, 61, 205–212 CrossRef.
- M. Keiluweit, J. J. Bougoure, P. S. Nico, J. Pett-Ridge, P. K. Weber and M. Kleber, Mineral protection of soil carbon counteracted by root exudates, Nat. Clim. Change, 2015, 5(6), 588–595 CrossRef CAS.
- A. Benninghoven, Chemical-Analysis of Inorganic and Organic-Surfaces and Thin-Films by Static Time-of-Flight Secondary-Ion Mass-Spectrometry (ToF-SIMS), Angew. Chem., Int. Ed. Engl., 1994, 33(10), 1023–1043 CrossRef.
- O. Branson, E. A. Bonnin, D. E. Perea, H. J. Spero, Z. H. Zhu, M. Winters, B. Honisch, A. D. Russell, J. S. Fehrenbacher and A. C. Gagnon, Nanometer-Scale Chemistry of a Calcite Biomineralization Template: Implications for Skeletal Composition and Nucleation, Proc. Natl. Acad. Sci. U. S. A., 2016, 113(46), 12934–12939 CrossRef CAS PubMed.
- D. J. Graham and D. G. Castner, Multivariate Analysis of ToF-SIMS Data from Multicomponent Systems: The Why, When, and How, Biointerphases, 2012, 7(1–4), 49 CrossRef CAS PubMed.
- D. J. Graham, M. S. Wagner and D. G. Castner, Information from complexity: Challenges of TOF-SIMS data interpretation, Appl. Surf. Sci., 2006, 252(19), 6860–6868 CrossRef CAS.
- Y. Z. Ding, Y. F. Zhou, J. Yao, Y. J. Xiong, Z. H. Zhu and X. Y. Yu, Molecular evidence of a toxic effect on a biofilm and its matrix, Analyst, 2019, 144(8), 2498–2503 RSC.
- Y. Z. Ding, Y. F. Zhou, J. Yao, C. Szymanski, J. Fredrickson, L. Shi, B. Cao, Z. H. Zhu and X. Y. Yu, In Situ Molecular Imaging of the Biofilm and Its Matrix, Anal. Chem., 2016, 88(22), 11244–11252 CrossRef CAS PubMed.
- X. Hua, X. Y. Yu, Z. Y. Wang, L. Yang, B. W. Liu, Z. H. Zhu, A. E. Tucker, W. B. Chrisler, E. A. Hill, T. Thevuthasan, Y. H. Lin, S. Q. Liu and M. J. Marshall, In situ molecular imaging of a hydrated biofilm in a microfluidic reactor by ToF-SIMS, Analyst, 2014, 139(7), 1609–1613 RSC.
- O. Y. A. Costa, J. M. Raaijmakers and E. E. Kuramae, Microbial Extracellular Polymeric Substances: Ecological Function and Impact on Soil Aggregation, Front. Microbiol., 2018, 9, 1636 CrossRef PubMed.
- R. M. Boiteau, L. M. Markillie, H. Mitchell, D. Hu, R. Chu, D. Hoyt, L. Pasa-Tolic, J. Jansson and C. Jansson, Phytosiderophore piracy by soil bacteria, ISME J. Search PubMed , Submitted.
- P. E. Busby, C. Soman, M. R. Wagner, M. L. Friesen, J. Kremer, A. Bennett, M. Morsy, J. A. Eisen, J. E. Leach and J. L. Dangl, Research priorities for harnessing plant microbiomes in sustainable agriculture, PLos Biol., 2017, 15(3), e2001793 CrossRef PubMed.
- C. Sergaki, B. Lagunas, I. Lidbury, M. L. Gifford and P. Schafer, Challenges and Approaches in Microbiome Research: From Fundamental to Applied, Front. Plant Sci., 2018, 9, 1205 CrossRef PubMed.
- J. A. Vorholt, C. Vogel, C. I. Carlstrom and D. B. Muller, Establishing Causality: Opportunities of Synthetic Communities for Plant Microbiome Research, Cell Host Microbe, 2017, 22(2), 142–155 CrossRef CAS PubMed.
- J. A. Lau, J. T. Lennon and K. D. Heath, Trees harness the power of microbes to survive climate change, Proc. Natl. Acad. Sci. U. S. A., 2017, 114(42), 11009–11011 CrossRef CAS PubMed.
- P. Yu and F. Hochholdinger, The Role of Host Genetic Signatures on Root-Microbe Interactions in the Rhizosphere and Endosphere, Front. Plant Sci., 2018, 9, 1896 CrossRef PubMed.
Footnote |
| † These authors equal contribution. |
| This journal is © The Royal Society of Chemistry 2020 |