Open Access Article
Open Access ArticleMutation-induced alterations of intra-filament subunit organization in vimentin filaments revealed by SAXS†
Martha E.
Brennich‡
 a,
Ulla
Vainio
b,
Tatjana
Wedig
c,
Susanne
Bauch
a,
Harald
Herrmann
*cd and
Sarah
Köster
a,
Ulla
Vainio
b,
Tatjana
Wedig
c,
Susanne
Bauch
a,
Harald
Herrmann
*cd and
Sarah
Köster
 *ae
*ae
aInstitute for X-ray Physics, University of Goettingen, Friedrich-Hund-Platz 1, 37077 Göttingen, Germany. E-mail: sarah.koester@phys.uni-goettingen.de
bHASYLAB at DESY, Notkestraße 85, 22607 Hamburg, Germany
cDivision of Molecular Genetics, German Cancer Research Center, Im Neuenheimer Feld 280, 69120 Heidelberg, Germany. E-mail: h.herrmann@dkfz-heidelberg.de
dInstitute of Neuropathology, University Hospital Erlangen, Schwabachanlage 6, 91054 Erlangen, Germany
eGerman Center for Cardiovascular Research (DZHK), Göttingen, Germany
First published on 24th January 2019
Abstract
Vimentin intermediate filaments constitute a distinct filament system in mesenchymal cells that is instrumental for cellular mechanics and migration. In vitro, the rod-like monomers assemble in a multi-step, salt-dependent manner into micrometer long biopolymers. To disclose the underlying mechanisms further, we employed small angle X-ray scattering on two recombinant vimentin variants, whose assembly departs at strategic points from the normal assembly route: (i) vimentin with a tyrosine to leucine change at position 117; (ii) vimentin missing the non-α-helical carboxyl-terminal domain. Y117L vimentin assembles into unit-length filaments (ULFs) only, whereas ΔT vimentin assembles into filaments containing a higher number of tetramers per cross section than normal vimentin filaments. We show that the shape and inner structure of these mutant filaments is significantly altered. ULFs assembled from Y117L vimentin contain more, less tightly bundled vimentin tetramers, and ΔT vimentin filaments preserve the number density despite the higher number of tetramers per filament cross-section.
Introduction
Cellular shape and internal structure of eukaryotic cells are dynamically established by the cytoskeleton, a dense network of three distinct protein filament systems: actin filaments, microtubules and intermediate filaments (IFs). Their action is coordinated by a large number of associated proteins with cross-bridging function, by regulatory proteins such as protein kinases, by chaperones, and by motor proteins.1Members of the large IF protein family represent the most flexible2,3 and the most extensible4,5 component of the cytoskeleton in various cells and tissues. A prominent representative is vimentin, which is expressed in cells of mesenchymal origin such as fibroblasts, the endothelial layer of blood vessels but also in lymphocytes as well as in myoblasts during the early phase of myogenesis. Recently, new interest in the functional role of vimentin at various cellular and tissue levels arose because of its potential function in cell migration and in the epithelial-to-mesenchymal transition (EMT) during metastatic spread of cancer cells and tumorigenesis.6
IF proteins are fibrous entities without any known enzymatic function. Vimentin has a tripartite building plan: the approximately 46 nm-long α-helical domain (“rod”) lies in the center of the monomer and is flanked by a flexible, non-structured amino-terminal (“head”) and a carboxyl-terminal (“tail”) domain7 (schematic in Fig. 1a). Two such molecules are predisposed to form an α-helical in-register coiled coil. Further, two coiled-coil dimers associate in an anti-parallel, half-staggered manner to tetrameric complexes. Tetramers measure 5 nm in diameter and 60 nm in length as derived from glycerol-sprayed/low angle rotary metal shadowed EM images and determined from a structural model established at atomic resolution.8,9 These complexes stay stable in solution under low ionic strength conditions. However, they spontaneously assemble in a hierarchical fashion upon increase of the ionic strength by addition of salt to near-physiological values10,11 (shown schematically in Fig. 1b). In the first step of assembly, tetramers associate laterally to “unit-length” filaments (ULFs), which exhibit a length of ∼60 nm and a diameter of 11 to 17 nm indicating that their structural association is not complete yet at this time point and that ULFs may also not be uniform with respect to the number of tetramers.9,10,12,13 Indeed, ULF formation is nearly complete after one second of assembly.14 Lateral assembly of tetramers at this stage is based on charge periodicity along the rod and yields, depending on the ionic conditions, 8 to 12 tetramers per cross section thus exhibiting a certain degree of polymorphism.10,12,13 Quantitative data of the mass-per-length along individual ULFs have been obtained by scanning transmission electron microscopy (STEM), and from these data the number of tetramers per cross section was derived.9,15 Lateral assembly likely ends due to electrostatic forces and steric interactions. In a second step, ULFs anneal to filaments and in a third step eventually radially compact to “mature” filaments with a diameter of about 10 nm.9,16 After elongation to IFs of several μm length, these long filaments are still assembly-competent and continue to elongate by end-to-end joining to filaments that exceed 50 μm in length as visualized by electron microscopy as well as fluorescence microscopy.3,10,16,17
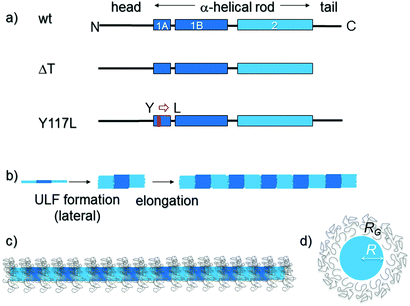 | ||
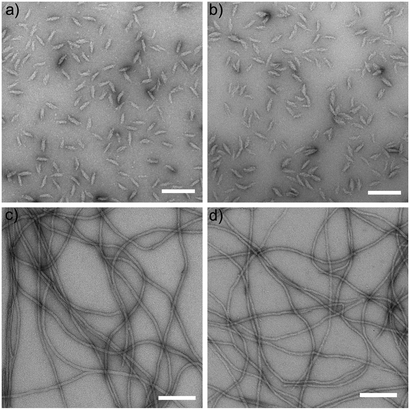
| Fig. 1 Domain organization, assembly pathway and filament appearance of vimentin and two mutant variants. (a) The WT-Vim monomer comprises a central “rod” domain of three α-helical segments (coil 1A, coil 1B and coil 2), which is flanked by intrinsically unstructured N-terminal (“head”) and C-terminal (“tail”) segments. In the Y117L variant (Y117L-Vim) the tyrosine (Y) at position 117 in coil 1A is replaced by a leucine (L). The tailless variant (ΔT-Vim) lacks the entire unstructured C-terminal segment. (b) Assembly pathway of WT-Vim: after initiating assembly by a rise in the ionic strength tetramers first laterally associate into a unit-length filament (ULF) and then ULFs longitudinally elongate into extended filaments. Subsequently a radial compaction step occurs (not shown). (c) Schematic of a fully assembled WT-Vim including the tail regions, which may form a layer of Gaussian coils around the core of the filament, thus resembling a “bottle brush”21 as originally demonstrated for neurofilaments.22,23 (d) Cross section of the filament model and parameters used for modelling the data. R is the radius of the solid core, whereas RG is the radius of gyration of the Gaussian chains surrounding it. | ||
When divalent ions instead of monovalent ions are employed to initiate assembly, vimentin also forms filaments.18,19 The principle assembly steps are apparently not affected, although the elongation reaction is considerably slowed down and the number of monomers is strongly increased up to 84 per cross-section, yielding a filament diameter of ∼25 nm instead of the regular 10 nm. Furthermore, the filaments appear much less flexible.10 Rheology experiments have revealed that the presence of divalent cations causes the formation of cross-linked networks, but the cross-linking is dependent on the presence of the non-α-helical C-terminal “tail” domain. Therefore it was concluded that the interaction of IFs is mediated by the divalent positively charged magnesium ions cross-bridging neighboring filaments via their negatively charged “tip” of the tail.9,20,21 In general, the highly negatively charged filaments are very sensitive to the ionic environment.
Here, we separate the individual hierarchical assembly steps – lateral assembly and elongation – by employing two different vimentin variants: (1) vimentin Y117L (Y117L-Vim), which stops to assemble at the ULF state, and (2) tailless vimentin (ΔT-Vim), which assembles into IFs with a higher subunit-per-cross section value compared to wildtype (wt) vimentin, i.e., authentic, unmodified vimentin (WT-Vim) (see Fig. 1a). By performing small angle X-ray scattering (SAXS) experiments in different ionic environments and at varying ion concentrations, we determine the inner structure of the assembly products and thereby can conclude upon the underlying interaction principles during assembly.
Experimental section
Protein variants
In addition to WT-Vim, two different vimentin IF variants were used. In the variant Y117L-Vim, tyrosine at position 117 is point-mutated to leucine;8,22 ΔT-Vim is lacking amino acids 412 to 466.9,20Protein purification and assembly
Human vimentin was expressed in bacteria (Escherichia coli, strain TG1) and purified from inclusion bodies.23 The protein was stored at −80 °C in 8 M urea, 5 mM Tris–HCl (pH 7.5), 1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, and 10 mM methyl ammonium chloride.24The purity of the protein was verified by SDS–polyacrylamide gel electrophoresis. For assembly, the protein was dialyzed into 2 mM sodium phosphate buffer, pH 7.5 (dialysis buffer) in a stepwise manner (8, 4, 2, 1, 0 M urea) using membranes with a 50 kDa cut-off. The protein concentration was determined by measuring the absorption at 280 nm (Nanodrop ND-1000, ThermoScientific Technologies, Inc., Wilmington, USA). Protein concentrations above 3.5 mg ml−1 were achieved by using a Nanosep 10k centrifuge concentrator (Nanosep Centrifugal Devices, PALL, Ann Arbor, Michigan, USA). Unless otherwise stated, the protein concentration after addition of the salt buffer was 2.7 mg ml−1 for WT-Vim and Y117L-Vim, and 1.0 mg ml−1 for ΔT-Vim. After about half an hour at room temperature, assembly was initiated by addition of a salt solution buffered with 2 mM sodium phosphate to the vimentin solution at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 ratio.25 Samples were filled immediately into capillaries for SAXS (mark-tubes made of quartz glass, outer diameter 1.5 mm, wall thickness 0.01 mm; Hilgenberg, Malsfeld, Germany). The capillaries were then sealed with wax and spun down briefly at room temperature to remove remaining air bubbles (1500 rpm, Eppendorf centrifuge 5810 R, Eppendorf, Hamburg, Germany). Afterwards, all samples were stored at about 4 °C for at least 24 hours before the measurements. Extended storage (up to 5 days) did not change the scattering signal (data not shown).
9 ratio.25 Samples were filled immediately into capillaries for SAXS (mark-tubes made of quartz glass, outer diameter 1.5 mm, wall thickness 0.01 mm; Hilgenberg, Malsfeld, Germany). The capillaries were then sealed with wax and spun down briefly at room temperature to remove remaining air bubbles (1500 rpm, Eppendorf centrifuge 5810 R, Eppendorf, Hamburg, Germany). Afterwards, all samples were stored at about 4 °C for at least 24 hours before the measurements. Extended storage (up to 5 days) did not change the scattering signal (data not shown).
Small angle X-ray scattering (SAXS) measurements and analysis
The SAXS measurements were performed at two sample-to-detector distances (0.885 m and 3.585 m) at the beamline B1 at DORIS III of DESY (Hamburg, Germany)26 using a Pilatus 1M detector (981 × 1043 pixels, pixel size: 172 × 172 mm2; Dectris, Baden, Switzerland). For all WT-Vim samples, as well as Y117L-Vim in the presence of KCl, the photon energy was 9 keV; for ΔT-Vim and Y117L-Vim in the presence of MgCl2 it was 12 keV. At these sample-to-detector distances, we could access real-space length scales up to about 100 nm. Exposures were split up in 5 or 10 min intervals, up to a total exposure time of 40 min at the shorter distance, and up to 60 min at the longer distance, depending on the signal-to-noise ratio and beam stability. A semi-transparent beamstop enabled measurements of the intensity of the transmitted beam directly with the Pilatus detector, and in combination with a pre-sample ionization chamber the transmission of each sample was automatically determined. On-site tools were used for the azimuthal integration of the data, averaging of sub-exposures as well as for the unification of the data for the two different sample-to-detector distances (https://github.com/uvainio/Beamline-B1-macros). This primary processing procedure includes corrections for the sample transmission and beamline background. The background scattering of capillaries filled with buffer was subtracted from the resulting SAXS curves. Finally, all curves were normalized by division by the respective protein concentration c, and the scattering intensity I was plotted against the scattering vector q using MatLab R2009b (The MathWorks, Inc., Natrick, USA).Modelling of the data
The SAXS curves were fitted to a cylindrical micelle model using the “lsqnonlin” function of MatLab18,27 R2009b. Briefly, filaments are described by a cylinder with a stiff core of constant electron density and unstructured regions at the outside with Gaussian electron density distributions.18,27–29 This kind of model results in a rough cylinder surface and a maximum electron density at the edge of the solid core, two features which have previously been reported for ULFs.12 Important fit parameters, some of which are discussed in this paper, are the radius of the core R, the radius of gyration of the Gaussian distribution RG, and the relative electron density of the Gaussian distributions in terms of the electron density of the core b, which is related to the relative mass of both regions:| F(q,R,RG) = β[FS(q,R) + λb2Fc(q,RG) + 2bSsc(q,R,RG) + b2Scc(q,R,RG)] |
Overall forward scattering intensities and cross sectional radii of gyration were determined by fits of the low-q region to the Guinier-approximation 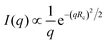 of rod-like particles using the “lsqnonlin” function of MatLab.32 The lower end of the linear range was assessed manually; the upper end was automatically determined to provide the best possible fit with (qmaxRc)2 ≤ 2.
of rod-like particles using the “lsqnonlin” function of MatLab.32 The lower end of the linear range was assessed manually; the upper end was automatically determined to provide the best possible fit with (qmaxRc)2 ≤ 2.
Electron microscopy measurements and analysis
The proteins (3.5 mg ml−1) were dialyzed into 2 mM sodium phosphate buffer (pH 7.5) and assembly was started with the corresponding buffers as described for the SAXS experiments. The assembly reaction was stopped by addition of nine volumes of assembly buffer containing 0.1% glutaraldehyde. For transmission electron microscopy (TEM), a 10 μl sample was applied to a freshly glow-discharged carbon-coated copper grid, allowed to attach for one minute followed by negative staining of protein structures with uranyl acetate. For better retention of material on the grids and observation of individual filaments, assembly was performed also at 0.2 mg ml−1. In principle, identical filaments were formed (see also ref. 33 for filaments assembled at higher protein concentration). Washed and dried samples were recorded in a transmission electron microscope, EM912 (Carl Zeiss, Oberkochen, Germany). Mass per length measurements were performed by STEM as described in ref. 9.Results
Electron microscopic analysis of assembled vimentin mutants
TEM of negatively stained specimen reveals the outer nanostructure of vimentin IFs. Comparing the assembled structures of Y117L-Vim after 10 s (Fig. 2a) and 1 h (Fig. 2b) it is clear that assembly is completed after 10 s and does not proceed beyond the level of ULFs. The prolate “cigar” shape of the ULFs is clearly visible. In contrast, although forming ULFs seemingly identical to those observed with Y117L-Vim at 10 s (see Fig. 5 in ref. 9), both ΔT-Vim and WT-Vim elongate rapidly and form extended IF by 1 h (Fig. 2c and d). These filaments, however, differ with respect to their diameter as ΔT-Vim filaments are about 1 nm thicker than WT-Vim filaments.SAXS analysis of mutant vimentin in the presence of KCl or MgCl2
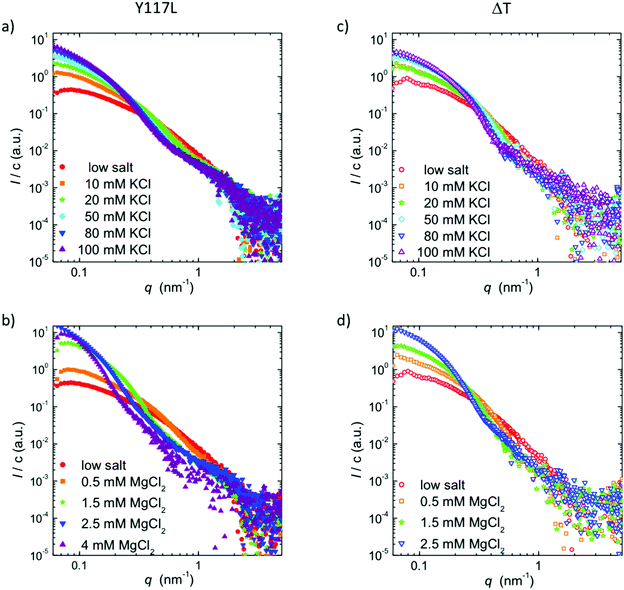
The electron micrographs discussed above nicely reveal the ability of ΔT-Vim to form extended filaments, albeit with an increased diameter. Moreover, they show for the Y117L point mutant a complete termination of assembly at the ULF state, as a consequence of which radial compaction cannot occur, because this process is connected to the elongation reaction. Nevertheless, in both cases it remains unclear whether the increased diameter and mass are accompanied by tetramer rearrangements, as electron microscopy is sensitive to the surface of imaged objects but not to the internal structure. In order to answer this question, we perform SAXS experiments on solutions of ΔT-Vim and Y117L-Vim in the presence of different KCl and MgCl2 concentrations. In Fig. 3, we plot the scattering intensity I, normalized to the protein concentration c, for the different protein mutants and ionic conditions. Corresponding Guinier plots and fits are shown in the ESI,† Fig. S1.The accessible q-range is 0.08–4.8 nm−1, and thus it is sensitive to real space length scales d = 2π/q of 1.3 to 75 nm. Note that we therefore primarily probe the lateral association step of the protein rather than the elongation reaction, which may be studied by larger wavelengths as employed in static and dynamic light scattering.14,34 In general, the increase in forward scattering, i.e., the interpolated intensity value of the cross-sectional contribution at q = 0, represents an increase in the number of subunits per cross-section, while the emergence of local maxima and minima in the SAXS curve implies the formation of a better-defined filament shape and higher monodispersity in diameter. For both vimentin variants and both types of ions, we observe that with increasing ion concentration, the forward scattering increases and the SAXS curves become steeper in the mid-q-range. Whereas the data for Y117L-Vim remain comparatively feature-less for both ion types (Fig. 3a and c) even at high ion concentrations, the SAXS curves for ΔT-Vim approach the form factor of idealized cylinders with emerging local maxima and minima in the mid-q-region upon the addition of KCl (Fig. 3b), similar to what we previously described for WT-Vim.18 This observation may be explained by the fact that the Y117L variant does not form filaments but stops assembly at the ULF state. As shown in the electron micrographs (Fig. 2a and b), the ULFs have a prolate shape and thus display different radii along the length. Thus, in the data for the Y117L mutant, features, which correspond to a defined filament radius, are smeared out and hardly visible. By contrast, the fully assembled ΔT-Vim filaments have a mostly constant diameter, with the exception of the filaments’ ends, which play a minor role for a long filament.
When assembly of ΔT-Vim is mediated by addition of MgCl2 instead of KCl, the SAXS curves show no identifiable local maxima or minima but develop a kink in the mid-q-region (Fig. 3d), indicating less monodisperse and well-defined filaments formed from this mutant. This observation may be explained in the light of additionally incorporated tetramers disturbing the cross-sectional order observed in full-length filaments. Of note, at 4 mM MgCl2 the SAXS curve becomes much noisier, probably due to precipitation of the protein, in agreement with light microscopy studies.35 The situation is even more severe for ΔT-Vim than for Y117L-Vim. Hence, the corresponding data are not shown in Fig. 3d.
A direct comparison on the two variants with WT-Vim is shown in Fig. 4, for two representative ionic conditions. For both 100 mM KCl and 2.5 mM MgCl2, we observe an increased forward scattering for either mutant, indicative of an increased number of tetramers per cross-section. For measurements in the presence of MgCl2 we do not observe a difference between ΔT-Vim and Y117L-Vim. By contrast, for the data recorded in the presence of KCl, ΔT-Vim shows similar minima and maxima as the full-length protein, albeit moved towards smaller q-values, which correspond to larger real space length scales. This finding agrees with our EM data that reveal thicker filaments for the tailless than for the wt protein. As the SAXS curves show a difference between Y117L-Vim and ΔT-Vim, at least in the presence of KCl, we conclude that ΔT-Vim filaments do compact to a fairly homogeneous diameter, albeit thicker than WT-Vim filaments. The larger diameter most likely stems directly from the packing into filaments of more vimentin tetramers per cross-section, as indicated by the increased forward scattering intensity at equal KCl concentration and supported by the EM data.10
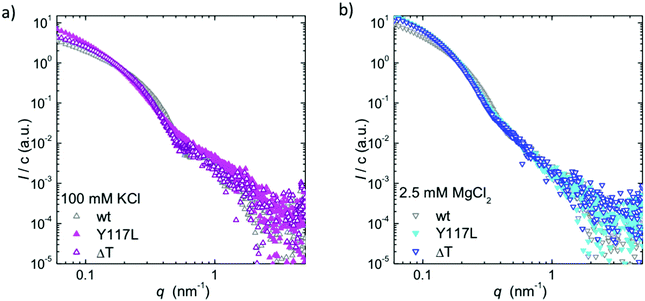 | ||
| Fig. 4 Direct comparison of the SAXS data of the vimentin variants and WT-Vim, (a) in the presence of 100 mM KCl and (b) in the presence of 2.5 mM MgCl2. | ||
To summarize our observations from the SAXS data for the mutants, Y117L-Vim responds more strongly to the salt concentration than WT-Vim. We interpret this behaviour as a consequence of the intermediate assembly state exhibited by ULFs and a higher polydispersity due to the looser, topologically less defined packing of the tetramers. As previously reported,10 ΔT-Vim forms thicker filaments upon the addition of KCl than WT-Vim, as these filaments are less densely packed. Furthermore, our data show that upon increasing the salt concentration, the filaments become more well-defined. For the assembly with MgCl2, we do not observe the same degree of differences between the variants.
Quantification of the mass-per-length and the mass-per-volume of the filaments
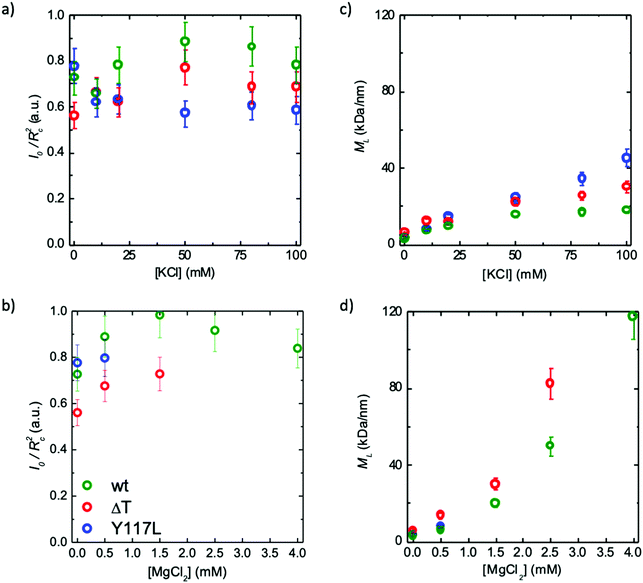
In order to link our SAXS data to previous quantifications obtained by STEM, we analyze both the mass density (see Fig. 5a and b) and the mass-per-length (see Fig. 5c and d). Note that the Guinier analysis to determine I0 failed for the Y117L mutant beyond 0.5 mM MgCl2, and thus no corresponding data are presented in Fig. 5. For both mutants, as seen in Fig. 3b, at 4 mM MgCl2 the protein precipitates and the data are omitted here. The mass density is directly estimated32 from the forward scattering intensities I0 and the cross-sectional radii of gyration Rcvia MV ∝ I0/Rc2. The mass-per-length is given by ML = I0d2/π(Δρ)2c where d is the mass density of the protein, c the protein concentration and Δρ the difference in scattering length densities between solvent and protein.36 Here, we assume that d2/(Δρ)2 is constant and thus use the equation ML = MtetramerL·I0/Itetramer0 for our computation. The tetramer data are obtained from the “low salt” measurements (see Fig. 3). It has been shown previously by analytical ultracentrifugation that at these conditions the protein is present in tetrameric form.12,37,38 The mass-per-length for a tetramer is 3.6 kDa nm−1. This value is calculated from the molecular weight of a tetramer (215 kDa) and its length (60 nm). For a mature filament assembled from WT-Vim with 8 tetramers per cross section we expect 40 kDa nm−1, because the tetramers partially overlap longitudinally.44,45 For all protein variants, ML increases with increasing ion concentration, and the effect is more pronounced for MgCl2 than for KCl. However, MV remains fairly constant over the whole ion concentration range. This indicates that the mass density of the filaments is not dependent on the ionic environment, whereas the number of tetramers per cross section is.For both types of ions, WT-Vim filaments have the highest mass density MV but the lowest mass per length ML, thus their radius is below the value for both mutants. The removal of the tail segment reduces the mass of a ΔT-Vim tetramer by about 12%, hence the same packing density of tetramers would result in a reduction of ML by 12%. Together with the EM data, this confirms an increase in the number of tetramers per cross section in comparison to WT-Vim filaments but suggests a similar arrangement of these tetramers within the cross-section. The data also explicitly show that the ULFs of Y117L-Vim, as well, do not only have a larger diameter than the WT-Vim filaments but also a looser packing, i.e., they are not compacted yet. Hence, one may assume that the compaction is a result of the elongation reaction between individual octameric protofibrils of successive ULF segments, in that it provides the opportunity to undergo an intra-filamentous stabilization event by reorganization of “protofibrils”, i.e., the presumed continuous octameric strands within a mature filament.39
Comparison of structural parameters
In order to further quantify the structural packing of the emerging filaments and understand how it changes with increasing cation concentration, we fit the data using a “bottle brush” model,18,27,40 which includes a filament core with radius R decorated with Gaussian chains with radius of gyration RG (Fig. 1c).Previously, we had studied WT-Vim at different KCl and MgCl2 concentrations employing the same model.18 In order to interpret the mutant results in the context of those data, we first provide a short summary of the findings: for WT-Vim, for both KCl and MgCl2 the filament radius increases with increasing salt concentration. This effect is more pronounced for MgCl2 than for KCl. Furthermore, magnesium, but not potassium ions accumulate in the unstructured tail segments, due to the high number of negatively charged amino acids in the tail.
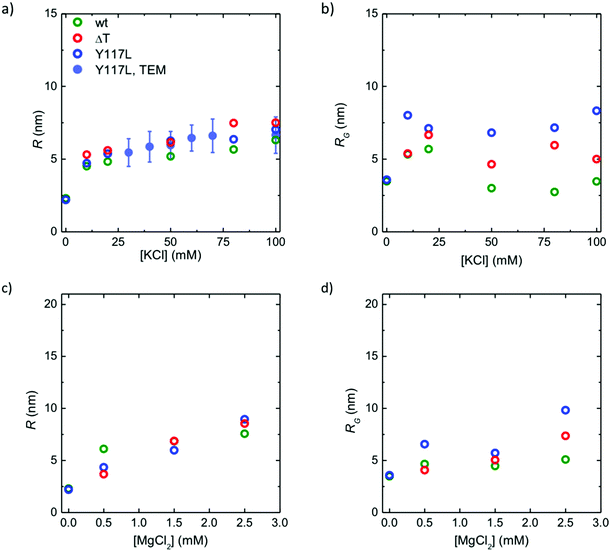
In Fig. 6, we compare the data for the two vimentin variants to the WT-Vim data: it shows the radius of the core R in dependence of the ion concentration, and it reflects exactly what we have learned from the analysis of the mass-per-length and mass density above. R increases in a similar manner for all three investigated protein variants with increasing ion concentration and eventually saturates. The ΔT-Vim filaments are thickest, assumedly because more subunits are incorporated per cross sections, followed by ULFs of Y117L-Vim, which are not compacted, and finally WT-Vim filaments (Fig. 6a). The effect is similar, albeit less pronounced, for MgCl2 (Fig. 6c).
The Gaussian “corona” with a radius of gyration RG stems from the intrinsically disordered head and tail domains of the filaments. We assume that the tail domains are presented to a large part on the surface of the filament. Although it has not been shown directly, there is strong evidence that a certain part of the vimentin heads (102 amino acids), stick out as well, and only the first part of the head is essential for the assembly reaction.23 GFAP (glial fibrillary acidic protein), another cytoplasmic IF protein with high amino acid sequence similarity to vimentin, has a short head of only 68 amino acids but assembles in an analogous manner. We can thus safely assume that part of the heads and the tails together form a “hydrogel-like”41 structure on the surface of the filaments.
When comparing the radii of gyration for the three protein variants (Fig. 6b and d), the values for the Y117L mutant are the highest and lie above the corresponding values for the core R. This implies that the ULF does not possess a solid core but instead is a rather loose structure across the entire diameter. Interestingly, the Gaussian chains decorating the WT-Vim filaments collapse at increasing KCl concentration, but remain loosely packed (large values for RG) for the ULFs formed by Y117L-Vim (Fig. 6b). Furthermore, whereas the corona size RG, stays roughly constant between 10 mM and 100 mM KCl, the radius of the inner core R matches the radius determined by TEM (Fig. 6a, filled circles). The values corresponding to ΔT-Vim lie in-between: on the one hand, they combine a higher number of contributing monomers per cross section than WT-Vim, on the other hand, the truncated tails cannot contribute to the corona.
Discussion
Both ULFs formed by Y117L-Vim and filaments formed by ΔT-Vim are thicker than filaments formed by WT-Vim. In this study we now show that the underlying mechanisms for the increase differ with both mutants. For ΔT-Vim, the diameter increases because additional tetramers are added at constant packing density. One possible explanation is that the removal of the tail results in more possible configurations and “space” to fit in additional sub-units. However, in this case, we would expect an increase in the packing density and compactness of the filaments, i.e., the parameter I0/Rc2 in Fig. 5 should be larger. An alternative explanation for the increased diameter of ΔT-Vim filaments is the reduced excess charge of ΔT-Vim tetramers, which carry a negative charge of only about 60 e as opposed to about 75 e for WT-Vim (based on the amino acid sequence and pKa values at pH 7.5). This reduction of charge results in a overall decrease of electrostatic repulsion forces for the ΔT-Vim filaments. We assume that the attractive forces, which are due to specific charge patterns in the amino acid sequence and therefore more localized, are to a lesser extent influenced by the removal of the tail, i.e. that the attraction is mostly due to interaction of the head and rod domains. Therefore, thicker filaments, with more subunits per cross-section can form, as previously determined by STEM, due to the removal of the tail. This assumption makes sense in the light that rod and head domains are necessary for filament formation,7,9,42 whereas the tails are not: the similar form of the scattering curves and the parallels of the structural parameters indicate a very limited specific influence of the tail region on the actual structure of mature filaments.This agrees with the more general observation that the rod domain is to a large extent conserved between different IF protein species whereas the tail domain can vary considerably both in amino acid composition and length. We assume that specific interactions of the human vimentin tail affect filament–filament interactions within the network but not the intramolecular filament ordering, i.e., the tail region is less important for the assembly process though it does have an effect on the number of subunits in the cross-section. In this context, it may be interesting to note that ΔT-Vim filaments when stably transfected into vimentin-free cells exhibit larger diameters than WT-Vim filaments transfected in parallel, i.e., 13.4 versus 10.2 nm.9 Hence, the, in principal, similar organization of the tail-deleted vimentin into filaments occurs also after in vivo synthesis and assembly.
As evident from EM data (Fig. 2), the point mutation tyrosine (Y) to leucine (L) at position 117 turns WT-Vim into a non-elongating variant and the length and shape of the resulting assemblies corresponds to that of ULFs obtained when the assembly of WT-Vim is “frozen” after one second by chemical fixation. The complementary SAXS measurements reveal a very different inner structure than for the fully assembled wt filament. The diameter of these ULFs is similar to ΔT-Vim filaments, but in this case the inner structure is different. This observation implies a re-arrangement of the tetramers during the elongation and/or the compaction process. This implication leads to the question to what extent the structure of the ULF determines the structure of the mature filament. Strong rearrangements have indeed been revealed by hydrogen–deuterium exchange experiments combined with mass spectrometry (HDex-MS). It has been demonstrated that different segments within the rod of WT-Vim exhibit a high mobility in different stages of assembly.38 Moreover, the HDex-MS patterns of ULFs assembled from Y117L-Vim differ significantly from those of WT-Vim filaments as in the Y117L-Vim ULFs the first one third of coil 2 is protected against exchange of hydrogen by deuterium, whereas the ends of the rod, mediating the filament elongation reaction, is protected in WT-Vim but not in Y117L-Vim. The reason is that the C-terminal ends, which stick out on either side of the ULF, are not able to connect to another ULF and are thus still open. Hence, both parts of coil 2 have different properties in the course of assembly. Similar to the case of the ΔT-Vim filaments, also Y117L-Vim is processed in vimentin-free cells consistent with its in vitro assembly characteristics. Here, it manifests in transfected cells as very fine dot-like structures consistent with them being ULFs.22
Therefore, we entertain the possibility that the observed structural feature of the Y117L-Vim ULF is a specific signature of the arrested state and that WT-Vim ULFs, as a transition structure on the way to fully assembled filaments, display a similarly open structure and salt dependence like Y117L-Vim ULFs in this first phase of assembly. Previous EM data show that WT-Vim ULFs are indeed very open with a diameter up to 20 nm.9,16 Hence, we expect that more insight will be obtained by combining the SAXS measurements with a microfluidic environment employing continuous flow11,43 as well as by studying the temperature-sensitive mutant K139C-vimentin, whose assembly at 21 °C is arrested at an ULF-like state but for which elongation can be triggered by an increase of the temperature to 37 °C.25
Conclusions
We have analyzed three different variants of the IF protein vimentin by a combination of EM and SAXS, (i) the wildtype protein, (ii) the point-mutated version Y117L, which forms ULFs by lateral assembly but does not elongate and (iii) the tail-truncated ΔT protein, which assembles into filaments, but is known to not form networks that are cross-linked by ions.20 We can now draw a consistent picture of the inner (SAXS) and outer (EM) nanostructure of these protein assemblies.The ΔT-Vim filaments, devoid of tails, are thicker than the WT-Vim filaments and have a larger mass-per-length ML, as well as a lower mass density MV. This is possibly due to the absence of the tail, causing a reduced charge of each monomer. Furthermore, the Gaussian corona surrounding the ULF core is thicker for the ΔT-Vim filaments. In our opinion, this is due to the higher number of monomers per cross-section.
For the Y117L variant, which to some extent represents the intermediate ULF state in the assembly hierarchy, we find a much more heterogeneous diameter, which makes sense in the light of the prolate “cigar” shape of the ULF with smaller cross-sectional area at the ends than at the center. Their mass density MV is lower than for the ΔT variant and the mass-per-length ML higher, meaning that the average thickness of the uncompacted ULFs is higher than the WT-Vim filaments. Finally, the corona of these comparatively unstructured assemblies exceeds that of both filament forming variants.
Our findings show that the progression from ULFs to mature filaments includes both a reduction in the number of tetramers in the cross section and a compaction in their arrangement. While the tail domains do not seem to affect the arrangement of tetramers within the filament, they influence the final diameter by decreasing the number of tetramers allowed per cross-section.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank Bernd Nöding, Christian Dammann, Britta Weinhausen, Sarah Schwarz, Oliva Saldanha and Rabea Sandmann for assistance during beam times. We thank Simon Castorph, Sajal Ghosh and Tim Salditt for providing experimental expertise and for discussions. We thank Norbert Mücke for providing data on the thickness distribution of vimentin filaments and Dorothee Möller for help with the electron micrographs. Part of this research was carried out at the light source DORIS III at DESY, a member of the Helmholtz Association (HGF). This work was supported by the Helmholtz Gemeinschaft in the framework of Virtual Institute VH-VI-403 “In Situ Nano-Imaging of Biological and Chemical Processes” and by the Deutsche Forschungsgemeinschaft (DFG) in the framework of SFB 755 “Nanoscale Photonic Imaging” (project B07), projects KO 3752/5-1, HE 1853/11-1 and HE 1853/4-3, and the Excellence Initiative.Notes and references
- H. Herrmann, H. Bär, L. Kreplak, S. V. Strelkov and U. Aebi, Nat. Rev. Mol. Cell Biol., 2007, 8, 562–573 CrossRef CAS PubMed.
- N. Mücke, L. Kreplak, R. Kirmse, T. Wedig, H. Herrmann, U. Aebi and J. Langowski, J. Mol. Biol., 2004, 335, 1241–1250 CrossRef.
- B. Nöding and S. Köster, Phys. Rev. Lett., 2012, 108, 88101 CrossRef PubMed.
- L. Kreplak, H. Bär, J. F. Leterrier, H. Herrmann and U. Aebi, J. Mol. Biol., 2005, 354, 569–577 CrossRef CAS PubMed.
- (a) J. Block, H. Witt, A. Candelli, J. C. Danes, E. J. G. Peterman, G. J. L. Wuite, A. Janshoff and S. Köster, Sci. Adv., 2018, 4, eaat1161 CrossRef PubMed; (b) J. Block, H. Witt, A. Candelli, E. J. G. Peterman, G. J. L. Wuite, A. Janshoff and S. Köster, Phys. Rev. Lett., 2017, 118, 48101 CrossRef PubMed.
- (a) E. D. Hay, Am. J. Med. Genet., 1989, 34, 14–29 CrossRef CAS PubMed; (b) K. Vuoriluoto, H. Haugen, S. Kiviluoto, J.-P. Mpindi, J. Nevo, C. Gjerdrum, C. Tiron, J. B. Lorens and J. Ivaska, Oncogene, 2010, 30, 1436 CrossRef PubMed; (c) F. Danielsson, M. K. Peterson, H. Caldeira Araújo, F. Lautenschläger and A. K. B. Gad, Cells, 2018, 7, 147 CrossRef PubMed.
- D. A. D. Parry, S. V. Strelkov, P. Burkhard, U. Aebi and H. Herrmann, Exp. Cell Res., 2007, 313, 2204–2216 CrossRef CAS PubMed.
- A. A. Chernyatina, S. Nicolet, U. Aebi, H. Herrmann and S. V. Strelkov, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 13620–13625 CrossRef CAS PubMed.
- H. Herrmann, M. Haner, M. Brettel, S. A. Müller, K. N. Goldie, B. Fedtke, A. Lustig, W. W. Franke and U. Aebi, J. Mol. Biol., 1996, 264, 933–953 CrossRef CAS PubMed.
- H. Herrmann, M. Häner, M. Brettel, N. O. Ku and U. Aebi, J. Mol. Biol., 1999, 286, 1403–1420 CrossRef CAS PubMed.
- M. E. Brennich, J. F. Nolting, C. Dammann, B. Nöding, S. Bauch, H. Herrmann, T. Pfohl and S. Köster, Lab Chip, 2011, 11, 708–716 RSC.
- A. V. Sokolova, L. Kreplak, T. Wedig, N. Mücke, D. I. Svergun, H. Herrmann, U. Aebi and S. V. Strelkov, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 16206–16211 CrossRef CAS PubMed.
- N. Mücke, R. Kirmse, T. Wedig, J. F. Leterrier and L. Kreplak, J. Struct. Biol., 2005, 150, 268–276 CrossRef PubMed.
- N. Mücke, L. Kämmerer, S. Winheim, R. Kirmse, J. Krieger, M. Mildenberger, J. Baßler, E. Hurt, W. H. Goldmann, U. Aebi, K. Toth, J. Langowski and H. Herrmann, Biophys. J., 2018, 114, 2408–2418 CrossRef PubMed.
- A. C. Steven, J. Wall, J. Hainfeld and P. M. Steinert, Proc. Natl. Acad. Sci. U. S. A., 1982, 79, 3101–3105 CrossRef CAS.
- S. Georgakopoulou, D. Möller, N. Sachs, H. Herrmann and U. Aebi, J. Mol. Biol., 2009, 386, 544–553 CrossRef CAS PubMed.
- S. Winheim, A. R. Hieb, M. Silbermann, E.-M. Surmann, T. Wedig, H. Herrmann, J. Langowski and N. Mücke, PLoS One, 2011, 6, e19202 CrossRef CAS PubMed.
- M. E. Brennich, S. Bauch, U. Vainio, T. Wedig, H. Herrmann and S. Köster, Soft Matter, 2014, 10, 2059–2068 RSC.
- M. Kooijman, M. Bloemendal, P. Traub, R. van Grondelle and H. van Amerongen, J. Biol. Chem., 1997, 272, 22548–22555 CrossRef CAS PubMed.
- Y. C. Lin, C. P. Broedersz, A. C. Rowat, T. Wedig, H. Herrmann, F. C. MacKintosh and D. A. Weitz, J. Mol. Biol., 2010, 399, 637–644 CrossRef CAS PubMed.
- (a) Y. C. Lin, N. Y. Yao, C. P. Broedersz, H. Herrmann, F. C. MacKintosh and D. A. Weitz, Phys. Rev. Lett., 2010, 104, 4 Search PubMed; (b) S. Köster, Y. C. Lin, H. Herrmann and D. A. Weitz, Soft Matter, 2010, 6, 1910–1914 RSC.
- M. Meier, G. P. Padilla, H. Herrmann, T. Wedig, M. Hergt, T. R. Patel, J. Stetefeld, U. Aebi and P. Burkhard, J. Mol. Biol., 2009, 390, 245–261 CrossRef CAS PubMed.
- H. Herrmann and U. Aebi, Annu. Rev. Biochem., 2004, 73, 749–789 CrossRef CAS PubMed.
- P. Traub, Methods Enzymol., 1971, 306–316 Search PubMed.
- N. Mücke, T. Wedig, A. Burer, L. N. Marekov, P. M. Steinert, J. Langowski, U. Aebi and H. Herrmann, J. Mol. Biol., 2004, 340, 97–114 CrossRef PubMed.
- H. G. Haubold, K. Gruenhagen, M. Wagener, H. Jungbluth, H. Heer, A. Pfeil, H. Rongen, G. Brandenberg, R. Moeller, J. Matzerath, P. Hiller and H. Halling, Rev. Sci. Instrum., 1989, 60, 1943–1946 CrossRef CAS.
- C. Y. J. Hemonnot, M. Mauermann, H. Herrmann and S. Köster, Biomacromolecules, 2015, 16, 3313–3321 CrossRef CAS PubMed.
- (a) J. S. Pedersen, J. Appl. Crystallogr., 2000, 33, 637–640 CrossRef CAS; (b) C. L. P. Oliveira, M. A. Behrens, J. S. Pedersen, K. Erlacher, D. Otzen and J. S. Pedersen, J. Mol. Biol., 2009, 387, 147–161 CrossRef CAS PubMed; (c) J. S. Pedersen, Adv. Colloid Interface Sci., 1997, 70, 171–210 CrossRef CAS.
- G. Oster and D. P. Riley, Acta Crystallogr., 1952, 5, 272–276 CrossRef CAS.
- P. Debye, J. Phys. Colloid Chem., 1947, 51, 18–32 CrossRef CAS PubMed.
- T. Lichtenstern, N. Mücke, U. Aebi, M. Mauermann and H. Herrmann, J. Struct. Biol., 2012, 177, 54–62 CrossRef CAS PubMed.
- O. Glatter and O. Kratky, Small Angle X-ray Scattering, Academic Press, London, 1982 Search PubMed.
- M. Schopferer, B. H. H. Bär, S. Sharma, N. Mücke, H. Herrmann and N. Willenbacher, J. Mol. Biol., 2009, 388, 133–143 CrossRef CAS PubMed.
- (a) C. G. Lopez, O. Saldanha, K. Huber and S. Köster, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 11152–11157 CrossRef CAS PubMed; (b) C. G. Lopez, O. Saldanha, A. Aufderhorst-Roberts, C. Martinez-Torres, M. Kuijs, G. H. Koenderink, S. Köster and K. Huber, Soft Matter, 2018, 8445–8454 RSC.
- (a) C. Dammann, B. Nöding and S. Köster, Biomicrofluidics, 2012, 6, 22009 CrossRef PubMed; (b) C. Dammann and S. Köster, Lab Chip, 2014, 14, 2681–2687 RSC; (c) C. Dammann, H. Herrmann and S. Köster, Isr. J. Chem., 2016, 56, 614–621 CrossRef CAS.
- (a) P. Thiyagarajan, T. S. Burkoth, V. Urban, S. Seifert, T. L. S. Benzinger, D. M. Morgan, D. Gordon, S. C. Meredith and D. G. Lynn, J. Appl. Crystallogr., 2000, 33, 535–539 CrossRef CAS; (b) L. A. Feigin and D. I. Svergun, Structure Analysis by Small-Angle X-Ray and Neutron Scattering, Plenum Press, New York, 1987 CrossRef.
- N. Mücke, T. Wedig, A. Bürer, L. N. Marekov, P. M. Steinert, J. Langowski, U. Aebi and H. Herrmann, J. Mol. Biol., 2004, 340, 97–114 CrossRef PubMed.
- A. Premchandar, N. Mücke, J. Poznański, T. Wedig, M. Kaus-Drobek, H. Herrmann and M. Dadlez, J. Biol. Chem., 2016, 291, 24931–24950 CrossRef CAS PubMed.
- U. Aebi, J. Cell Biol., 1983, 97, 1131–1143 CrossRef CAS PubMed.
- R. Beck, J. Deek and C. R. Safinya, Biochem. Soc. Trans., 2012, 40, 1027–1031 CrossRef CAS PubMed.
- S. Frey, R. P. Richter and D. Görlich, Science, 2006, 314, 815–817 CrossRef CAS PubMed.
- A. Eckelt, H. Herrmann and W. W. Franke, Eur. J. Cell Biol., 1992, 58, 319–330 CAS.
- (a) M. Denz, G. Brehm, C. Y. J. Hémonnot, H. Spears, A. Wittmeier, C. Cassini, O. Saldanha, E. Perego, A. Diaz, M. Burghammer and S. Köster, Lab Chip, 2017, 18, 171–178 RSC; (b) O. Saldanha, M. E. Brennich, M. Burghammer, H. Herrmann and S. Köster, Biomicrofluidics, 2016, 10, 24108 CrossRef PubMed.
- D. Henderson, N. Geisler and K. Weber, J. Mol. Biol., 1982, 155, 173–176 CrossRef CAS PubMed.
- H. Herrmann and U. Aebi, Cold Spring Harbor Perspect. Biol., 2016, 8, a018242 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Guinier plots for all ionic conditions presented in the main text. See DOI: 10.1039/c8sm02281j |
| ‡ Present address: European Molecular Biology Laboratory, Grenoble Outstation, 71 Avenue des Martyrs, 38042 Grenoble, France. |
| This journal is © The Royal Society of Chemistry 2019 |