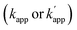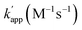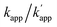Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Rational design of a DNA sequence-specific modular protein tag by tuning the alkylation kinetics†
Thang Minh
Nguyen
,
Eiji
Nakata
 ,
Zhengxiao
Zhang
,
Masayuki
Saimura
,
Huyen
Dinh
and
Takashi
Morii
,
Zhengxiao
Zhang
,
Masayuki
Saimura
,
Huyen
Dinh
and
Takashi
Morii
 *
*
Institute of Advanced Energy, Kyoto University, Uji, Kyoto 611-0011, Japan. E-mail: t-morii@iae.kyoto-u.ac.jp
First published on 20th August 2019
Abstract
Sequence-selective chemical modification of DNA by synthetic ligands has been a long-standing challenge in the field of chemistry. Even when the ligand consists of a sequence-specific DNA binding domain and reactive group, sequence-selective reactions by these ligands are often accompanied by off-target reactions. A basic principle to design DNA modifiers that react at specific sites exclusively governed by DNA sequence recognition remains to be established. We have previously reported selective DNA modification by a self-ligating protein tag conjugated with a DNA-binding domain, termed as a modular adaptor, and orthogonal application of modular adaptors by relying on the chemoselectivity of the protein tag. The sequence-specific crosslinking reaction by the modular adaptor is thought to proceed in two steps: the first step involves the formation of a DNA–protein complex, while in the second step, a proximity-driven intermolecular crosslinking occurs. According to this scheme, the specific crosslinking reaction of a modular adaptor would be driven by the DNA recognition process only when the dissociation rate of the DNA complex is much higher than the rate constant for the alkylation reaction. In this study, as a proof of principle, a set of combinations for modular adaptors and their substrates were utilized to evaluate the reactions. Three types of modular adaptors consisting of a single type of self-ligating tag and three types of DNA binding proteins fulfill the kinetic requirements for the reaction of the self-ligating tag with a substrate and the dissociation of the DNA–protein complex. These modular adaptors actually undergo sequence-specific crosslinking reactions exclusively driven by the recognition of a specific DNA sequence. The design principle of sequence-specific modular adaptors based on the kinetic aspects of complex formation and chemical modification is applicable for developing recognition-driven selective modifiers for proteins and other biological macromolecules.
Introduction
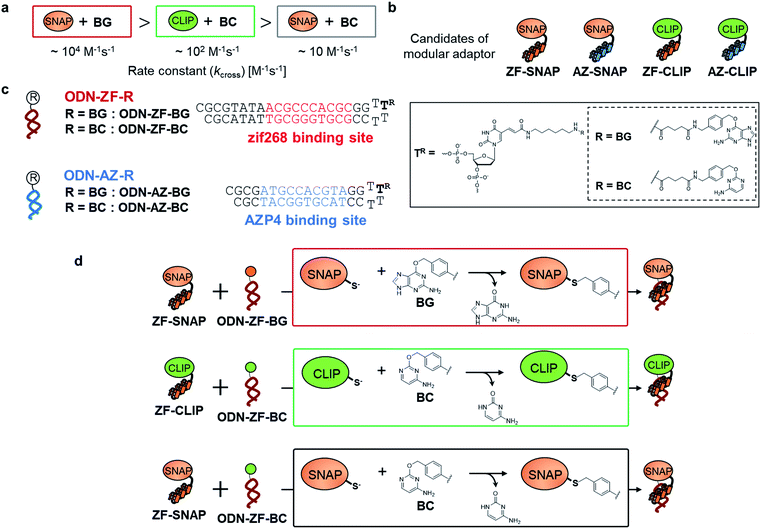
Site-specific chemical reactions that modify proteins and nucleic acids by ligands rely on specific molecular recognition of target sites and selective reactions.1 Sequence-selective chemical modification of DNA by DNA-binding molecules remains challenging in the field of chemistry.1d,e Various sequence-specific DNA binders, ranging from small molecules2 to short peptides,3 proteins,4 and synthetic oligonucleotides,5 have been coupled to a reactive group to specifically modify nucleic acid bases, sugar moieties, or phosphodiester linkages. However, coupling of a reactive group to a sequence-specific DNA binding domain is often accompanied by off-target reactions.2–5 A basic principle in designing DNA modifiers exclusively governed by DNA sequence recognition has not been established.Fully addressable nano-architectures of various shapes and geometries provide ideal scaffolds for locating molecules of interest at defined positions with a nanometer spatial precision.6 DNA scaffolds prepared by DNA origami7 can be readily incorporated with additional DNA sequences on their surface by simple extension of the staple strand. These extended DNA strands are further chemically modified to serve as binding sites for various molecules.8 Proteins are a particularly interesting class of molecules for assembly on the DNA scaffold because of their wide functional variability.9 For assembly on the DNA scaffold through hybridization, a protein of interest (POI) is chemically modified with a short DNA strand that is complementary to the attached sequence on the DNA scaffold.10 Although this method is convenient, it has some limitations; for example, the modification conditions are limited and may cause loss of activity.11,12 It would be ideal to locate the POI at a specific position on the DNA scaffold without the requirement for post-chemical modification of the POI to prepare a precisely controlled assembly suitable for further investigation.9c,13 To achieve efficient and rapid placement of a POI at a specific DNA site, we developed modular adaptors (Fig. 1a and b).14,15 A modular adaptor consists of a DNA-binding protein (zif26816) that functions as a DNA sequence-selective recognition module and self-ligating protein tag (SNAP-tag17) which forms a chemoselective covalent linkage between the protein tag and its substrate O2-benzylguanine (BG) modified at the given DNA sequence (Fig. 1a).14 A modular adaptor-fused enzyme was specifically located at the target position of the DNA scaffold with fast reaction kinetics and in high yield while fully retaining the original enzymatic activity.18 In order to assemble three types of proteins at unique positions on the DNA scaffold, a set of modular adaptors with orthogonality and which retained fast reaction kinetics under mild conditions was selected from the systematic combination of DNA-binding domains (zif268 or AZP4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19) and protein tags, SNAP-tag, CLIP-tag,20 and Halo-tag.15,21 Among them, three types of orthogonal modular adaptors simultaneously directed the three types of proteins to their respective positions on the DNA scaffold. In a previous study, orthogonal cross-linking reactions of modular adaptors were mainly conducted by taking advantage of the chemoselective characteristics of self-ligating protein tags (Fig. 1c), not exclusively by the sequence selectivity of the DNA-binding protein. In fact, zif268 conjugated with SNAP-tag (ZF-SNAP) and AZP4 conjugated with SNAP-tag (AZ-SNAP) reacted to the same extent with BG at both the zif268 and AZP4 binding sequences.15 This non-sequence-selective modification was discussed by considering the kinetics of the sequence-specific crosslinking reaction by the modular adaptor in two steps: the first step is the formation of a DNA–protein complex, and a proximity-driven intermolecular crosslinking reaction is initiated in the second step (Fig. 1b). According to this scheme, the specific crosslinking reaction of a modular adaptor would be driven by the DNA recognition process only when the dissociation rate of the DNA complex is much higher than the rate constant for the alkylation reaction.
19) and protein tags, SNAP-tag, CLIP-tag,20 and Halo-tag.15,21 Among them, three types of orthogonal modular adaptors simultaneously directed the three types of proteins to their respective positions on the DNA scaffold. In a previous study, orthogonal cross-linking reactions of modular adaptors were mainly conducted by taking advantage of the chemoselective characteristics of self-ligating protein tags (Fig. 1c), not exclusively by the sequence selectivity of the DNA-binding protein. In fact, zif268 conjugated with SNAP-tag (ZF-SNAP) and AZP4 conjugated with SNAP-tag (AZ-SNAP) reacted to the same extent with BG at both the zif268 and AZP4 binding sequences.15 This non-sequence-selective modification was discussed by considering the kinetics of the sequence-specific crosslinking reaction by the modular adaptor in two steps: the first step is the formation of a DNA–protein complex, and a proximity-driven intermolecular crosslinking reaction is initiated in the second step (Fig. 1b). According to this scheme, the specific crosslinking reaction of a modular adaptor would be driven by the DNA recognition process only when the dissociation rate of the DNA complex is much higher than the rate constant for the alkylation reaction.
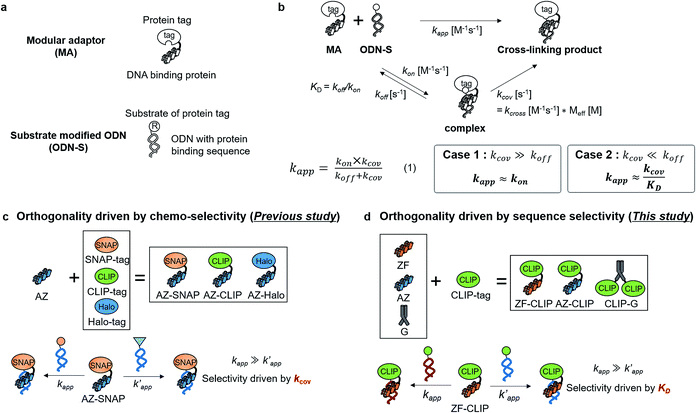 | ||
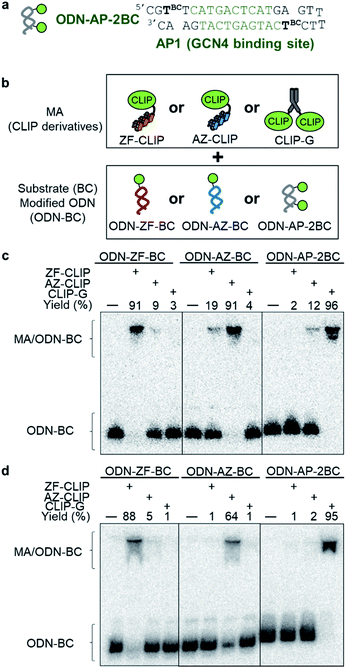
Fig. 1 (a) Structures and elements of the modular adaptor (MA) and substrate-modified oligodeoxyribonucleotide (ODN-S). (b) Scheme of the cross-linking reaction between MA and ODN-S. Complex formation between MA and ODN-S is governed by the equilibrium dissociation constant (KD) [M] as defined by KD = koff/kon. The apparent rate constant (kapp) [M−1 s−1] is expressed as kon and koff (or KD) and the rate constant kcov [s−1] for the proximity-driven intermolecular crosslinking reaction between MA and ODN-S is defined by the rate constant of intermolecular covalent formation (kcross) [M−1 s−1] and the effective concentration (Meff) [M] (eqn (1)). (c) Orthogonal cross-linking reactions driven by the chemo-selectivity of the protein tag described in our previous study.15 The apparent reaction rate constants for the matched pair and the unmatched pair are indicated as kapp [M−1 s−1] and 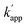 [M−1 s−1], respectively. The selective cross-linking reaction proceeds when [M−1 s−1], respectively. The selective cross-linking reaction proceeds when 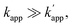 which is realized only when kcov for the matched pair of the substrate of protein tags is much larger than that for the unmatched pair of the substrate of protein tags. (d) Orthogonal cross-linking reactions driven by the sequence selectivity of DNA-binding domains and chemo-selective cross-linking reactions by the protein tag described in this study. The selective reactions of ZF-CLIP, AZ-CLIP, and CLIP-G proceed with kapp in case 2 of (b) with selectivity mainly governed by KD. which is realized only when kcov for the matched pair of the substrate of protein tags is much larger than that for the unmatched pair of the substrate of protein tags. (d) Orthogonal cross-linking reactions driven by the sequence selectivity of DNA-binding domains and chemo-selective cross-linking reactions by the protein tag described in this study. The selective reactions of ZF-CLIP, AZ-CLIP, and CLIP-G proceed with kapp in case 2 of (b) with selectivity mainly governed by KD. | ||
In this study, as a proof of principle for the above concept, we constructed sequence-selective modular adaptors by tuning the reaction kinetics between the self-ligating protein tag and its substrate (Fig. 2a and d). Conjugation of different types of DNA-binding domains to the same self-ligating protein tag provides a variety of orthogonal modular adaptors, in which the crosslinking reaction is exclusively governed by sequence-specific DNA recognition (Fig. 1d).
The reaction of the modular adaptor with its substrate at the specific DNA sequence proceeds through specific recognition of the DNA sequence and an alkylation reaction between the protein tag and its substrate (Fig. 1b). Therefore, the factors governing the specific reactions of modular adaptors are well-correlated with the design of selective DNA modifiers driven by sequence-specific DNA recognition.18,22 Our design principle of modular adaptors based on the kinetic parameters of the DNA binding and alkylation reaction would be applicable to design a wide range of site-specific modifiers driven by target recognition.
Results
Kinetic aspects of the modular adaptor provide a design strategy for sequence-specific modular adaptors for orthogonal covalent bond formation with the substrate-modified ODNs
Our previous study of the reaction of modular adaptors, consisting of a DNA-binding domain and a cross-link forming protein tag (Fig. 1a), suggested important factors that affected the kinetics and selectivity of the cross-linking reaction. Covalent bond formation between the modular adaptor and its substrate-modified oligodeoxyribonucleotide (ODN-S) (Fig. 1a) occurs in the following two steps (Fig. 1b): a reversible complex forms between the DNA-binding domain and ODN-S, and then a proximity-driven intermolecular cross-linking reaction occurs between the self-ligating protein tag domain and its substrate on ODN-S.15 The apparent rate constant of the cross-linking reaction (kapp) (M−1 s−1) between the modular adaptor and substrate-modified DNA is represented by eqn (1) shown in Fig. 1b. In eqn (1), a modular adaptor with a highly reactive protein tag to its substrate (kcov ≫ koff) falls into case 1.23,24 Because the association rates (kon) of DNA-binding proteins to their specific and non-specific sites are nearly identical,24 the modular adaptor is thought to react with ODN-S regardless of the DNA sequence (case 1 in Fig. 1b). Therefore, reducing the reactivity of the protein tag with the substrate (kcov) to fit into the condition of case 2, kcov ≪ koff (Fig. 1b), would be an effective strategy for preventing the cross-linking reaction at unmatched DNA sequences by modular adaptors. In case 2, a higher equilibrium dissociation constant (KD) for the unmatched complex of the zinc finger motif and ODNs results in fewer cross-linking reactions of unmatched complexes. The difference in the cross-linking reactions for the matched and unmatched pairs evaluated in our previous study15 and this study is described in Fig. 1c and d, respectively.Time-dependent changes in the yield of the cross-linking reaction (Fig. S1†) were simulated by varying the apparent rate constants. In addition, the apparent reaction rate constant of the matched pair (kapp) and that of the unmatched pair 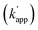 were investigated with various kcov values (Fig. S2†). From these simulations, the sequence-selective orthogonal cross-linking reaction takes place when the rate constant of the matched pair (kapp) is two orders of magnitude higher than that of the unmatched pair
were investigated with various kcov values (Fig. S2†). From these simulations, the sequence-selective orthogonal cross-linking reaction takes place when the rate constant of the matched pair (kapp) is two orders of magnitude higher than that of the unmatched pair 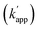 . Thus, to achieve the sequence-selective crosslinking reaction, the reactivity of the cross-linking reaction should be finely tuned to meet the condition of case 2.
. Thus, to achieve the sequence-selective crosslinking reaction, the reactivity of the cross-linking reaction should be finely tuned to meet the condition of case 2.
To test the above hypothesis, three combinations of protein tags and substrates, SNAP-tag with BG (∼104 M−1 s−1),17,20 CLIP-tag with O2-benzylcytosine (BC) (∼102 M−1 s−1),20 and SNAP-tag with BC (∼10 M−1 s−1),20 were prepared for the cross-link-forming domain of the modular adaptor (Fig. 2a). Two DNA binding proteins, zif268 (ZF) and AZP4 (AZ), were utilized as the DNA-binding domain of the modular adaptor (Fig. 2b). ZF-CLIP was constructed by fusing CLIP-tag to the C-terminus of zif268 through a GGSGGS linker (Fig. S3a†). ZF-SNAP, AZ-SNAP, and AZ-CLIP were prepared as described previously.15 ODNs containing the zif268- or AZP4-binding sequence were designed to form a loop with four T nucleotides, in which one of the T nucleotides was replaced with amino-C6-T (ODN-ZF or ODN-AZ). Amino-C6-T was modified with each of the substrates, BG or BC (ODN-ZF-BG, ODN-ZF-BC, ODN-AZ-BG or ODN-AZ-BC, respectively, as shown in Fig. 2c). Complex formations by ZF or AZ derivatives and ODN-ZF or ODN-AZ were titrated by means of fluorescence polarization measurements to determine the KD values (Fig. S4 and Table S1, S2†). The results confirmed that the KD values for the matched and unmatched complexes differed by more than two orders of magnitude. Cross-linking reactions of these modular adaptors with ODN-S containing the respective matched or unmatched sequence were analysed by denaturing polyacrylamide gel electrophoresis (PAGE) as shown in Fig. 3. In the combination of SNAP-tag and BG, kapp values for the reactions of the matched pair ZF-SNAP with ODN-ZF-BG and AZ-SNAP with ODN-AZ-BG were 3.8 × 105 and 9.1 × 105 M−1 s−1, respectively (Table 1). For the combination of CLIP-tag and BC, kapp values for the reactions of the matched pair ZF-CLIP with ODN-ZF-BC and AZ-CLIP with ODN-AZ-BC were 7.1 × 105 and 5.0 × 105 M−1 s−1, respectively (Table 1). In the combination of SNAP-tag with BC, kapp values for the reactions of the matched pair ZF-SNAP with ODN-ZF-BC and AZ-SNAP with ODN-AZ-BC were 1.7 × 103 and 3.0 × 102 M−1 s−1, respectively (Table 1). The cross-linking reactions between ZF-SNAP and ODN-ZF-BG, AZ-SNAP and ODN-AZ-BG, ZF-CLIP and ODN-ZF-BC, and AZ-CLIP and ODN-AZ-BC gave yields of 94%, 93%, 92%, and 90%, respectively (Fig. 3a and b). The reactions of ZF-SNAP with ODN-ZF-BC and AZ-SNAP with ODN-AZ-BC were very slow and showed yields of 40% and 46%, respectively, after 30 min of incubation under the described conditions (Fig. 3c).
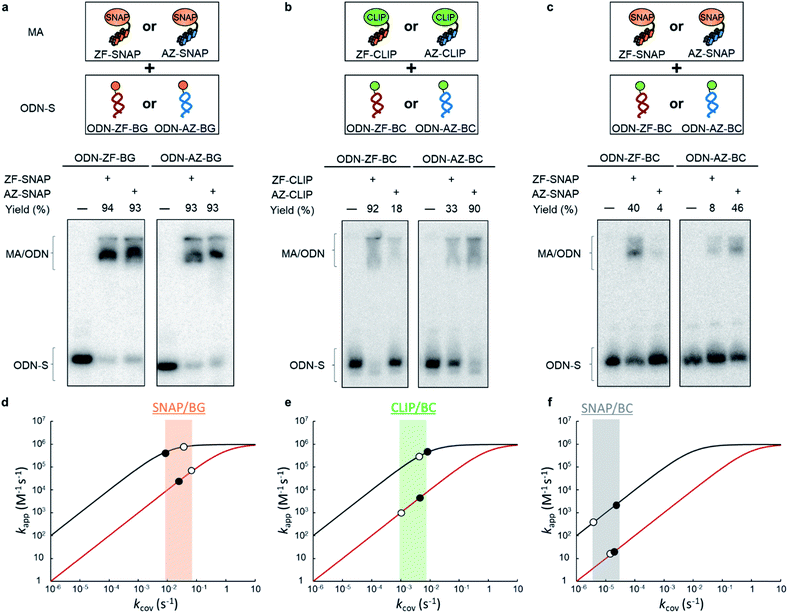 | ||
| Fig. 3 Analyses of cross-linking reactions between the modular adaptors and the substrate-modified ODNs. (a–c) Denaturing PAGE analyses of the cross-linking reaction by modular adaptors (MA: ZF-SNAP, AZ-SNAP, ZF-CLIP and AZ-CLIP) and each ODN-S (ODN-ZF-BG, ODN-AZ-BG, ODN-ZF-BC and ODN-AZ-BC). Each 5′-32P-end-labeled ODN-S was incubated with a modular adaptor (100 nM) for 30 min in a buffer (pH 8.0) containing 40 mM Tris–HCl, 20 mM acetic acid, 12.5 mM MgCl2, 1 mM DTT, 1 μM ZnCl2, 0.02% Tween 20, 200 nM BSA and 100 nM calf thymus DNA at ambient temperature. (d–f) Simulation of kapp with various kcov by following eqn (1) (Fig. 1b). The kapp values for the matched and unmatched pairs of MA and ODN-S at given kcov are shown in black and red curves, respectively. The details of simulation conditions are shown in Fig. S2.† The experimentally obtained kapp values (Table 1) are marked on the simulated lines. Filled and open circles indicate MA conjugated with ZF and AZ, respectively. (d) Reactions of SNAP-tag conjugated MAs with BG. (e) Reactions of CLIP-tag conjugated MAs with BC. (f) Reactions of SNAP-tag conjugated MAs with BC. | ||
| MAs | k app (M−1 s−1) | ||
|---|---|---|---|
| (ODN-S) | (ODN-S) | ||
| ZF-SNAP | (3.8 ± 1.2) × 105 | (2.6 ± 0.2) × 104 | 15 |
| (ODN-ZF-BG) | (ODN-AZ-BG) | ||
| AZ-SNAP | (9.1 ± 2.4) × 105 | (9.5 ± 0.6) × 104 | 10 |
| (ODN-AZ-BG) | (ODN-ZF-BG) | ||
| ZF-CLIP | (7.1 ± 0.2) × 105 | (2.8 ± 0.2) × 103 | 254 |
| (ODN-ZF-BC) | (ODN-AZ-BC) | ||
| AZ-CLIP | (5.0 ± 0.7) × 105 | (8.1 ± 2.7) × 102 | 617 |
| (ODN-AZ-BC) | (ODN-ZF-BC) | ||
| ZF-SNAP | (1.7 ± 0.2) × 103 | 22 ± 1 | 77 |
| (ODN-ZF-BC) | (ODN-AZ-BC) | ||
| AZ-SNAP | (3.0 ± 0.1) × 102 | 11 ± 3 | 27 |
| (ODN-AZ-BC) | (ODN-ZF-BC) |
Next, we evaluated the reactions of unmatched pairs for each combination of the protein tag and substrate (Table 1). The 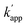 values of the unmatched pairs of ZF-SNAP with ODN-AZ-BG and AZ-SNAP with ODN-ZF-BG were 2.6 × 104 and 9.5 × 104 M−1 s−1, respectively. The
values of the unmatched pairs of ZF-SNAP with ODN-AZ-BG and AZ-SNAP with ODN-ZF-BG were 2.6 × 104 and 9.5 × 104 M−1 s−1, respectively. The 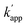 values of the unmatched pairs of ZF-CLIP with ODN-AZ-BC and AZ-CLIP with ODN-ZF-BC were 2.8 × 103 and 8.1 × 102 M−1 s−1, respectively. The
values of the unmatched pairs of ZF-CLIP with ODN-AZ-BC and AZ-CLIP with ODN-ZF-BC were 2.8 × 103 and 8.1 × 102 M−1 s−1, respectively. The 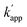 values of the unmatched pairs of ZF-SNAP with ODN-AZ-BC and AZ-SNAP with ODN-ZF-BC were 22 and 11 M−1 s−1, respectively (Table 1). The
values of the unmatched pairs of ZF-SNAP with ODN-AZ-BC and AZ-SNAP with ODN-ZF-BC were 22 and 11 M−1 s−1, respectively (Table 1). The 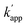 values of unmatched pairs of ZF-SNAP and AZ-SNAP were one to two orders of magnitude lower than the kapp of the matched pairs. Thus, the sequence-specific reaction only minimally occurred, as shown in Fig. S1.† In contrast, the
values of unmatched pairs of ZF-SNAP and AZ-SNAP were one to two orders of magnitude lower than the kapp of the matched pairs. Thus, the sequence-specific reaction only minimally occurred, as shown in Fig. S1.† In contrast, the 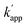 values of ZF-CLIP and AZ-CLIP were two orders of magnitude lower than kapp of the matched pairs (Fig. S5† and Table 1). In the combination of CLIP-tag and BC, the observed differences in the apparent second-order rate constants were parallel to the difference in the equilibrium dissociation constants for the matched and unmatched complexes of ZF-CLIP and AZ-CLIP, as expected from eqn (1) in case 2 (Fig. 1b). These results indicated that the cross-linking reaction of the modular adaptors consisting of CLIP-tag with the substrate BC on ODNs proceeded as shown in case 2 of eqn (1). Because the larger kapp for the matched pair was preferred as long as it falls within case 2, the pair of CLIP-tag and BC was selected as the self-ligating protein tag domain for modular adaptors that would form a covalent linkage at a specific sequence driven by the DNA recognition domain.
values of ZF-CLIP and AZ-CLIP were two orders of magnitude lower than kapp of the matched pairs (Fig. S5† and Table 1). In the combination of CLIP-tag and BC, the observed differences in the apparent second-order rate constants were parallel to the difference in the equilibrium dissociation constants for the matched and unmatched complexes of ZF-CLIP and AZ-CLIP, as expected from eqn (1) in case 2 (Fig. 1b). These results indicated that the cross-linking reaction of the modular adaptors consisting of CLIP-tag with the substrate BC on ODNs proceeded as shown in case 2 of eqn (1). Because the larger kapp for the matched pair was preferred as long as it falls within case 2, the pair of CLIP-tag and BC was selected as the self-ligating protein tag domain for modular adaptors that would form a covalent linkage at a specific sequence driven by the DNA recognition domain.
Sequence-specific reactions of modular adaptors consisting of CLIP-tag with BC modified ODNs
Three types of modular adaptors bearing the same self-ligating protein tag but different DNA-binding domains (ZF-CLIP, AZ-CLIP, and CLIP-G) were tested in a sequence-selective crosslinking reaction with ODNs modified with BC (Fig. 4). CLIP-G consisted of a basic-leucine zipper (bZIP) class of protein GCN4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 as a homo-dimeric DNA binding module and CLIP-tag fused to the N-terminus of GCN4 through a GGSGGS linker. An ODN containing the AP1 sequence as the GCN4 binding site was designed for the matched target for CLIP-G, in which two T nucleotides at either ends of the AP1 sequence were displaced by amino-C6-T (ODN-AP). ODN-AP was further modified with BC (ODN-AP-2BC) as the substrate of CLIP-G (Fig. S3b and c†). The cross-linking reactions of ZF-CLIP, AZ-CLIP, and CLIP-G with their respective matched ODNs were nearly orthogonal, as shown in Fig. 4c, with more than 90% yield for matched pairs and less than 19% covalent bond formation for unmatched pairs. The observed sequence-specific cross-linking reactions of ZF-CLIP, AZ-CLIP, and CLIP-G were driven by the specific DNA binding of each modular adaptor (Fig. S5, Table S3†). The kapp for the reaction of CLIP-G and its matched sequence ODN-AP-2BC was determined to be 2.1 × 106 M−1 s−1 and
25 as a homo-dimeric DNA binding module and CLIP-tag fused to the N-terminus of GCN4 through a GGSGGS linker. An ODN containing the AP1 sequence as the GCN4 binding site was designed for the matched target for CLIP-G, in which two T nucleotides at either ends of the AP1 sequence were displaced by amino-C6-T (ODN-AP). ODN-AP was further modified with BC (ODN-AP-2BC) as the substrate of CLIP-G (Fig. S3b and c†). The cross-linking reactions of ZF-CLIP, AZ-CLIP, and CLIP-G with their respective matched ODNs were nearly orthogonal, as shown in Fig. 4c, with more than 90% yield for matched pairs and less than 19% covalent bond formation for unmatched pairs. The observed sequence-specific cross-linking reactions of ZF-CLIP, AZ-CLIP, and CLIP-G were driven by the specific DNA binding of each modular adaptor (Fig. S5, Table S3†). The kapp for the reaction of CLIP-G and its matched sequence ODN-AP-2BC was determined to be 2.1 × 106 M−1 s−1 and 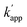 values ranged from 103 to 104 M−1 s−1. For the reactions of unmatched pairs, ZF-CLIP or AZ-CLIP with ODN-AP-2BC, the
values ranged from 103 to 104 M−1 s−1. For the reactions of unmatched pairs, ZF-CLIP or AZ-CLIP with ODN-AP-2BC, the 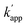 values were 102–103 M−1 s−1. Furthermore, in the presence of 200 mM NaCl, non-specific DNA binding was reduced (Fig. S6 and Table S4†)26,27 and covalent bond formation at the unmatched pairs were further reduced to less than 5% (Fig. 4d). The yields of the matched pairs were also reduced for AZ-CLIP (64%) and ZF-CLIP (88%). The kinetic constants of the matched and unmatched pairs for SNAP-tag and CLIP-tag in the presence of 200 mM NaCl are summarized in Fig. S7 and Tables S5 and S6,† respectively. Under this condition, the sequence-specificity of ZF-CLIP to the consensus sequence of zif268 (GCGTGGGCGT) and its mutated sequences was further investigated (Table S7†). ODN-ZF(G/T) and ODN-ZF(G/C) contained a single nucleotide substitution in the consensus sequence and ODN-ZF(GG/TC) and ODN-ZF(GC/CT) were mutated with two nucleotides. The KD values reported for the complexes of zif268 and these sequences were more than one order of magnitude higher than those with the consensus sequence.24 Actually, the KD values determined for the binding complexes of ZF-CLIP and these mutated ODNs by means of a fluorescence polarization assay were more than one order of magnitude higher for single nucleotide substituted ODN-ZF(G/T) and ODN-ZF(G/C) and two orders of magnitude higher for two nucleotide substituted ODN-ZF(GG/TC) and ODN-ZF(GC/CT) than that for ODN-ZF (Fig. S8 and Table S8†), being consistent with previous studies.24 The kapp for the reaction of ZF-CLIP and ODN-ZF-BC in the presence of 200 mM NaCl was determined to be 5.1 × 104 M−1 s−1. For the reaction with these mutated ODNs, the
values were 102–103 M−1 s−1. Furthermore, in the presence of 200 mM NaCl, non-specific DNA binding was reduced (Fig. S6 and Table S4†)26,27 and covalent bond formation at the unmatched pairs were further reduced to less than 5% (Fig. 4d). The yields of the matched pairs were also reduced for AZ-CLIP (64%) and ZF-CLIP (88%). The kinetic constants of the matched and unmatched pairs for SNAP-tag and CLIP-tag in the presence of 200 mM NaCl are summarized in Fig. S7 and Tables S5 and S6,† respectively. Under this condition, the sequence-specificity of ZF-CLIP to the consensus sequence of zif268 (GCGTGGGCGT) and its mutated sequences was further investigated (Table S7†). ODN-ZF(G/T) and ODN-ZF(G/C) contained a single nucleotide substitution in the consensus sequence and ODN-ZF(GG/TC) and ODN-ZF(GC/CT) were mutated with two nucleotides. The KD values reported for the complexes of zif268 and these sequences were more than one order of magnitude higher than those with the consensus sequence.24 Actually, the KD values determined for the binding complexes of ZF-CLIP and these mutated ODNs by means of a fluorescence polarization assay were more than one order of magnitude higher for single nucleotide substituted ODN-ZF(G/T) and ODN-ZF(G/C) and two orders of magnitude higher for two nucleotide substituted ODN-ZF(GG/TC) and ODN-ZF(GC/CT) than that for ODN-ZF (Fig. S8 and Table S8†), being consistent with previous studies.24 The kapp for the reaction of ZF-CLIP and ODN-ZF-BC in the presence of 200 mM NaCl was determined to be 5.1 × 104 M−1 s−1. For the reaction with these mutated ODNs, the 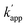 values for the single nucleotide substituted ODN-ZF(G/T) and ODN-ZF(G/C) were 1.6 × 103 and 2.3 × 103 M−1 s−1, respectively (Fig. S9 and Table S9†). The
values for the single nucleotide substituted ODN-ZF(G/T) and ODN-ZF(G/C) were 1.6 × 103 and 2.3 × 103 M−1 s−1, respectively (Fig. S9 and Table S9†). The 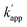 for the two nucleotide substituted ODNs ODN-ZF(GG/TC) and ODN-ZF(GC/CT) could not be obtained because of their very slow reactions. These results clearly demonstrated that the sequence selective cross-linking reaction of the modular adaptor consisting of CLIP-tag with the substrate BC on ODNs proceeded by discriminating single nucleotide substitution, and indicated that the selectivity of the crosslinking reaction was governed by the KD of the binding complex.
for the two nucleotide substituted ODNs ODN-ZF(GG/TC) and ODN-ZF(GC/CT) could not be obtained because of their very slow reactions. These results clearly demonstrated that the sequence selective cross-linking reaction of the modular adaptor consisting of CLIP-tag with the substrate BC on ODNs proceeded by discriminating single nucleotide substitution, and indicated that the selectivity of the crosslinking reaction was governed by the KD of the binding complex.
Coassembly of modular adaptors on a DNA scaffold
Three modular adaptors, ZF-CLIP, AZ-CLIP, and CLIP-G, were reacted at the designed positions on the DNA scaffold. Rectangular DNA scaffold (Fig. 5a and S12†) containing three cavities was prepared as described previously.18 Each modular adaptor (250 nM) was incubated with 5 nM of the DNA scaffold (I-1CG/II-1ZC/III-1AC) containing a single binding site for CLIP-G, ZF-CLIP, and AZ-CLIP at cavity I, II, and III, respectively, in a buffer containing 200 mM NaCl (Fig. 5 and S13†). The reaction at the matched positions achieved over 90% yield in 30 min with less than 7% modifications at unmatched positions (Fig. 5b–d). Sequential reactions in the presence of 200 mM NaCl in the order of CLIP-G, AZ-CLIP, and ZF-CLIP confirmed that modification had occurred at the unmatched positions (less than 10% in total), as shown in Fig. S14.† The one pot co-assembly reaction of the modular adaptors for their respective target positions on the DNA scaffold (Fig. 5e) resulted in 87% coassembly yield after 30 min of incubation.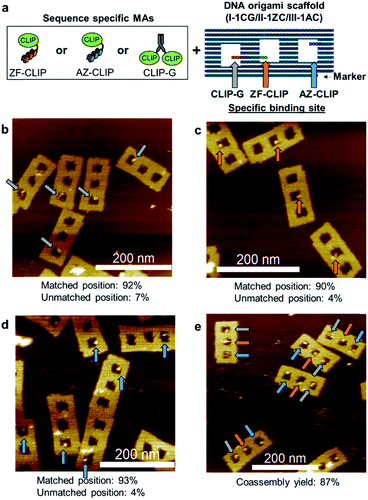 | ||
| Fig. 5 Three types of sequence-specific MAs orthogonally reacted at the predesigned positions on the DNA scaffold. (a) Three types of MAs and the DNA origami scaffold (I-1CG/II-1ZC/III-1AC). The DNA origami scaffold (I-1CG/II-1ZC/III-1AC), in which each cavity contained a single site for one MA. The stem loops in red, green, and blue denote the binding sites for CLIP-G (CG) in cavity I, ZF-CLIP (ZC) in cavity II, and AZ-CLIP (AC) in cavity III, respectively. (b–e) AFM images of the DNA scaffold reacted with MAs. (b–d) The DNA scaffold (5 nM) was incubated with MAs (250 nM) (b) CLIP-G, (c) ZF-CLIP, or (d) AZ-CLIP for 30 min at ambient temperature. (e) One-pot coassembly reaction of three MAs on the DNA scaffold (I-1CG/II-1ZC/III-1AC). The DNA scaffold (5 nM) was incubated with CLIP-G, ZF-CLIP and AZ-CLIP (250 nM each) for 30 min at ambient temperature. These reactions were carried out in a buffer (pH 8.0) containing 40 mM Tris–HCl, 20 mM acetic acid, 12.5 mM MgCl2, 200 mM NaCl, 1 mM DTT, 1 μM ZnCl2, and 0.02% Tween 20. The reaction mixture was purified by size-exclusion chromatography and then analysed by atomic force microscopy (AFM). Yields at each cavity and coassembly yields were estimated by counting the number of cavities occupied by the modular adaptors (Table S14†). | ||
Discussion
Based on eqn (1) of kapp in Fig. 1b, the factors affecting the kinetics and selectivity of the cross-linking reaction are the reactivity of the self-ligating protein tag (kcov) and the recognition process of the DNA binding domain (kon, koff, or KD). In our previous study, orthogonal cross-linking reactions of modular adaptors were mainly conducted by taking advantage of the chemoselectivity of protein tags (Fig. 1c).15 Thus, the selective reactions were driven by differences in the kcov of the reaction. Although this strategy was effective for the orthogonal cross-linking reactions, only a few variations in the orthogonal modular adaptors were available because the number of effective pairs of protein tags and substrates was limited. In contrast, DNA-binding proteins show a wide range of sequence selectivity and affinity for the recognition modules of modular adaptors.28 In case 2 of eqn (1), where kcov ≪ koff, the difference in KD values of the matched and unmatched DNA binding complexes are a critical factor for regulating the selectivity of modular adaptors with the same protein tag to a given substrate in a defined DNA sequence. At the same kcov of the protein tag module, the lower KD of the sequence specific DNA-binding complex provides a higher rate constant for the matched pair (kapp) and the higher KD for the unmatched complex results in a lower rate constant for the unmatched pair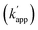 for the sequence-selective modular adaptors. Based on the simulation of the kinetic scheme in Fig. 1b (Fig. S1†), the cross-linking reaction is expected to be orthogonal when the ratio of rate constants
for the sequence-selective modular adaptors. Based on the simulation of the kinetic scheme in Fig. 1b (Fig. S1†), the cross-linking reaction is expected to be orthogonal when the ratio of rate constants 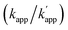 is larger than 100. The zinc finger proteins used in this study, zif268 and AZP4, showed a nearly two orders of magnitude difference in KD for complexes with matched and unmatched DNA sequences (Table S2†). In fact, modular adaptors with CLIP-tag and BC, developed in this study as the optimized protein tag and substrate pair for achieving kcov ≪ koff, showed an orthogonality of
is larger than 100. The zinc finger proteins used in this study, zif268 and AZP4, showed a nearly two orders of magnitude difference in KD for complexes with matched and unmatched DNA sequences (Table S2†). In fact, modular adaptors with CLIP-tag and BC, developed in this study as the optimized protein tag and substrate pair for achieving kcov ≪ koff, showed an orthogonality of 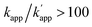 while retaining their reactivities. The GCN4 derived homodimeric modular adaptor, CLIP-G, also revealed orthogonality to ZF-CLIP and AZ-CLIP.
while retaining their reactivities. The GCN4 derived homodimeric modular adaptor, CLIP-G, also revealed orthogonality to ZF-CLIP and AZ-CLIP.
Sequence specificity of the DNA binding protein has been shown to improve in the presence of a high concentration of NaCl, as nonspecific electrostatic interactions between the protein and DNA decreases with increasing salt concentration.26,27 Although the affinity of proteins for the target DNA sequence is also reduced in high concentrations of NaCl, selectivity between the target and non-target DNA is enhanced. Indeed, the KD in the presence of 200 mM NaCl was significantly higher for both matched and unmatched complexes (Fig. S6 and Table S4†), and kapp showed nearly two-fold reduction (Fig. S7 and Table S6†), i.e., for the matched combination of ZF-CLIP and ODN-ZF-BC: kapp (no NaCl) = 7.1 × 105 M−1 s−1 and kapp (200 mM NaCl) = 3.1 × 103 M−1 s−1; for the unmatched combination of ZF-CLIP and ODN-AZ-BC: 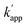 (no NaCl) = 5.3 × 103 M−1 s−1 and
(no NaCl) = 5.3 × 103 M−1 s−1 and 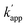 (200 mM NaCl) < 10 M−1 s−1). Notably, the reactivity of the protein tag with its substrate was not influenced by the presence of NaCl (Fig. S10 and Tables S10, S11†). The sequence specificity of the crosslinking reaction was further studied in detail by using single or two nucleotide mutated ODN-ZF-BC for ZF-CLIP in the presence of 200 mM NaCl (Table S7†). A single nucleotide mutation in the consensus sequence of ODN-ZF-BC caused more than one order of magnitude difference in the apparent reaction rate constants (Fig. S9 and Table S9†), which was consistent with the difference between the KD value for the complex of ZF-CLIP with ODN-ZF and that with the mutated sequences (Fig. S8 and Table S8†). These results strongly support our strategy in which the sequence selectivity of the cross-linking reaction by the modular adaptor was governed by KD under the condition of case 2 of eqn (1) (Fig. 1b). Interestingly, the cross-linking reaction rate of the modular adaptor was drastically accelerated on the DNA origami scaffold compared to the reaction using oligo DNA. In the presence of 200 mM NaCl, AZ-CLIP and ODN-AZ-BC reacted, showing a kapp of 3.1 × 102 M−1 s−1, which was the slowest combination among the matched pairs of the CLIP-type modular adaptors (Table S6†). AZ-CLIP rapidly reacted with BC in the matched sequence on the DNA origami scaffold while retaining selectivity and showed a kapp of 1.2 × 104 M−1 s−1, which was nearly two orders of magnitude higher than that for the reaction with ODN-AZ-BC (Fig. S13†). This increase in the stability of the complex between the DNA-binding protein and DNA may arise from the large and negatively charged surface of the DNA origami scaffold surrounding the protein–DNA complex.29
(200 mM NaCl) < 10 M−1 s−1). Notably, the reactivity of the protein tag with its substrate was not influenced by the presence of NaCl (Fig. S10 and Tables S10, S11†). The sequence specificity of the crosslinking reaction was further studied in detail by using single or two nucleotide mutated ODN-ZF-BC for ZF-CLIP in the presence of 200 mM NaCl (Table S7†). A single nucleotide mutation in the consensus sequence of ODN-ZF-BC caused more than one order of magnitude difference in the apparent reaction rate constants (Fig. S9 and Table S9†), which was consistent with the difference between the KD value for the complex of ZF-CLIP with ODN-ZF and that with the mutated sequences (Fig. S8 and Table S8†). These results strongly support our strategy in which the sequence selectivity of the cross-linking reaction by the modular adaptor was governed by KD under the condition of case 2 of eqn (1) (Fig. 1b). Interestingly, the cross-linking reaction rate of the modular adaptor was drastically accelerated on the DNA origami scaffold compared to the reaction using oligo DNA. In the presence of 200 mM NaCl, AZ-CLIP and ODN-AZ-BC reacted, showing a kapp of 3.1 × 102 M−1 s−1, which was the slowest combination among the matched pairs of the CLIP-type modular adaptors (Table S6†). AZ-CLIP rapidly reacted with BC in the matched sequence on the DNA origami scaffold while retaining selectivity and showed a kapp of 1.2 × 104 M−1 s−1, which was nearly two orders of magnitude higher than that for the reaction with ODN-AZ-BC (Fig. S13†). This increase in the stability of the complex between the DNA-binding protein and DNA may arise from the large and negatively charged surface of the DNA origami scaffold surrounding the protein–DNA complex.29
Conclusion
In summary, we have proved a design principle for sequence-specific DNA modifiers driven by specific DNA recognition based on the kinetic parameters for the DNA binding and modification reaction. The kinetic requirements for the complex formation step of DNA and the DNA binder as well as the successive proximity-driven intermolecular chemical reaction were applied to design sequence-selective modular adaptors by using appropriate combinations of a self-ligating protein tag and sequence-specific DNA-binding protein. The modular adaptors with the same self-ligating protein tag bearing different types of sequence-specific DNA-binding proteins indeed reacted with the substrate located in the respective matched DNA sequence, showing low reactivity with substrates in the unmatched sequence. Our design principle is useful for expanding the variation of sequence-specific modular adaptors by simply conjugating various types of sequence-specific DNA binding proteins28 to the same protein tag that reacts with an appropriate substrate. The resulting orthogonal set of modular adaptors can be used to add a variety of enzymes or receptors to a DNA scaffold to study the chemistry of spatially arranged biomolecules. More importantly, our design principle based on kinetic characteristics of selective modifiers can be applied for designing a wide variety of recognition-driven modifiers targeting not only DNA but also proteins and other biologically relevant molecules.Experimental procedures
Materials
Single-stranded M13mp18, restriction enzymes (NdeI and HindIII), and BC-GLA-NHS (S9237S) were purchased from New England Biolabs (Ipswich, MA, USA). The pCLIPf(T7)-2 vector was generously donated by New England Biolabs. Purified oligonucleotides as the staple strands for DNA origami, oligonucleotide primers for gene construction, and all other oligonucleotides were obtained from Sigma-Aldrich (St. Louis, MO, USA), Thermo Fisher Scientific (Waltham, MA, USA), or Gene Design (Osaka, Japan). Escherichia coli BL21 (DE3) competent cells were purchased from Invitrogen (Carlsbad, CA, USA). A Mini Elute gel extraction kit was obtained from QIAGEN (Hilden, Germany). The HiTrap SP XL cation exchange column (5 mL), HisTrap HP column (5 mL), and Sephacryl S-400 were from GE Healthcare (Little Chalfont, UK). PrimeSTAR HS DNA polymerase, T4 DNA ligase, and E. coli DH5α competent cells were obtained from TaKaRa Bio (Shiga, Japan). Ultrafree-MC-DV was obtained from Merck Millipore (Billerica, MA, USA). A Cosmosil 5C18-MS II column (4.6 i.d. × 150 mm) and HPLC-grade acetonitrile were purchased from Nacalai Tesque (Kyoto, Japan). Gel electrophoresis-grade acrylamide, bis(acrylamide), phenol, and all other chemicals and reagents were purchased from Wako Chemicals (Osaka, Japan) or Nacalai Tesque. Phosphate buffer (PB) was prepared by mixing 20 mM Na2HPO4 and 20 mM NaH2PO4.Preparation of protein-tag derivatives (ZF-SNAP, AZ-SNAP, ZF-CLIP, AZ-CLIP, and CLIP-G)
Overexpression and purification of ZF-SNAP, AZ-SNAP, and AZ-CLIP were described in a previous report.14,15 ZF-CLIP and CLIP-G were newly designed in this study. As a typical example, the preparation of ZF-CLIP is shown below. All vectors encoding protein tag derivatives (pET-30a-ZF-CLIP, pET-30a-CLIP-G) were prepared and the protein tag derivatives (ZF-CLIP and CLIP-G) were overexpressed and purified in the same manner.Construction of vectors for ZF-CLIP (pET-30a-ZF-CLIP)
CLIP-tag in the pCLIP-tag (T7)-2 vector was amplified by PCR using the following primer pairs.Forward primer (F_EcoRI_CLIP):
5′-TAATAAGAATTCGGCGGCTCCGGCGGCTCCGACAAAGATTGCGAAA-3′
Reverse primer (R_CLIP_HindIII):
5′-TTATTAAAGCTTTTAATGATGGTGATGATGATGGTGATGATGGTGGGTACCATTAACCTCGAGCCCGGGG-3′
The PCR products were separated on 1% agarose gel (TAE) and purified with a Mini Elute gel extraction kit. The PCR products and pET-30a-cys-zif268 were digested with EcoRI and HindIII and were purified in the same manner. These products were incubated with T4-DNA-ligase. The mixture was transferred into E. coli DH5α competent cells for amplification. The vector encoding ZF-CLIP (named pET-30a-ZF-CLIP) was purified, sequenced, and transferred into E. coli BL21(DE3) competent cells.
Overexpression and purification of ZF-CLIP
The transformed cells were grown at 37 °C until the OD600 reached 0.5, and protein expression was induced with 1 mM IPTG for 24 h at 25 °C. The soluble fraction of the cell lysate containing the recombinant protein was loaded onto a HisTrap HP column equilibrated with 50 mM phosphate buffer (pH 8.0) containing 200 mM NaCl and 1 mM DTT and then eluted over an imidazole gradient. The main fractions containing the target protein were collected, loaded onto a HiTrap SP HP column equilibrated with 50 mM phosphate buffer (pH 7.0) containing 1 mM DTT, and then eluted over an NaCl gradient. The purified protein was dialyzed by using 50 mM phosphate buffer (pH 8.0) containing 1 mM DTT, 50 μM ZnCl2, and 50% glycerol and stored at −20 °C. The purity of the target protein was checked by SDS-PAGE. The major band on the gel corresponded to the calculated molecular weight with an estimated purity of over 95% (Fig. S15†). The modular adaptors were characterized by MALDI-TOF mass spectrometry (AXIMA-LNR, SA matrix, Shimadzu, Kyoto, Japan). ZF-CLIP: m/z calcd 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 462, observed 32
462, observed 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 440; CLIP-G: m/z calcd 27
440; CLIP-G: m/z calcd 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 108, observed 27
108, observed 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 083. Amino acid sequences and calculated molecular weights of the recombinant proteins used in this study are shown in Table S15.†
083. Amino acid sequences and calculated molecular weights of the recombinant proteins used in this study are shown in Table S15.†
Preparation of substrate-modified ODNs and Alexa 488 modified ODNs
A coupling reaction between amino-modified ODNs (100 μM) and Alexa Fluor 488 NHS ester or succinimidyl derivative of protein tag substrates (1 mM) was carried out in 100 mM phosphate buffer (pH 8.0) for 8 h at ambient temperature. The modified ODNs were purified by reversed-phase HPLC on a Cosmosil 5C18-MS II column (4.6 × 150 mm, eluted with 100 mM triethylammonium acetate buffer, pH 7.0, with a linear gradient over 30 min from 2.5% to 30% acetonitrile at flow rate of 1.0 mL min−1) and characterized by MALDI-TOF mass spectrometry (AXIMA-LNR, Shimadzu, HPA matrix). A488-ODN-ZF: m/z calcd 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 397, observed 11
397, observed 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 395; A488-ODN-AZ: m/z calcd 11
395; A488-ODN-AZ: m/z calcd 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400, observed 11
400, observed 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399; A488-ODN-AP: m/z calcd 12
399; A488-ODN-AP: m/z calcd 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 011, observed 12
011, observed 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 014. ODN-AP-2BC: m/z calcd 12
014. ODN-AP-2BC: m/z calcd 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 290, observed 12
290, observed 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 290, ODN-8G-AP-2BC: m/z calcd 21
290, ODN-8G-AP-2BC: m/z calcd 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 577, observed 21
577, observed 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 578, ODN-11G-ZF-BC: m/z calcd 21
578, ODN-11G-ZF-BC: m/z calcd 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 160, observed 21
160, observed 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 167, ODN-ZF-BC: m/z calcd 14
167, ODN-ZF-BC: m/z calcd 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 287, observed 14
287, observed 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 287, ODN-ZF(G/T)-BC: m/z calcd 14
287, ODN-ZF(G/T)-BC: m/z calcd 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 286, observed 14
286, observed 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 289, ODN-ZF(G/C)-BC: m/z calcd 14
289, ODN-ZF(G/C)-BC: m/z calcd 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 287, observed 14
287, observed 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 282, ODN-ZF(GG/TC)-BC: m/z calcd 14
282, ODN-ZF(GG/TC)-BC: m/z calcd 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 286, observed 14
286, observed 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 288, and ODN-ZF(GG/TC)-BC: m/z calcd 14
288, and ODN-ZF(GG/TC)-BC: m/z calcd 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 286, observed 14
286, observed 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 291. The method for preparing ODN-AZ-BC, ODN-AZ-BG, and ODN-24D-AZ-BC was described in a previous report.15
291. The method for preparing ODN-AZ-BC, ODN-AZ-BG, and ODN-24D-AZ-BC was described in a previous report.15
Preparation of 32P-end-labeled ODNs and analyses of covalent-linkage formation between 32P-end-labeled ODNs and modular adaptors
The ODNs were 5′-32P-end-labeled as previously described.13b In a typical experiment for kinetic analysis, 32P-ODN-ZF-BC (less than 0.5 nM) was incubated with ZF-CLIP (10 nM) in a buffer (pH 8.0) containing 40 mM Tris–HCl, 20 mM acetic acid, 12.5 mM MgCl2, 1 mM DTT, 1 μM ZnCl2, 0.02% Tween 20, 200 nM BSA, and 100 nM calf thymus DNA, with or without 200 mM NaCl at ambient temperature. Aliquots were collected at defined reaction times and quenched by adding SDS and formamide. The aliquots were analysed by 8 M urea PAGE and the intensities of mobility-shifted bands on the gel were analysed with a Storm 860 molecular imager (Amersham plc, Amersham, UK). The kinetics data were fitted to a reaction model assuming first-order kinetics (eqn (1)), and then the second-order rate constants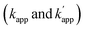 were obtained by dividing the determined first-order rate constants by the concentration of modular adaptors.
were obtained by dividing the determined first-order rate constants by the concentration of modular adaptors.| Y = 1 − e−kt | (1) |
To evaluate orthogonality, each 5′-32P-end-labeled ODN (ODN-ZF-BC, ODN-AZ-BC and ODN-AP-2BC) was incubated with a modular adaptor (10 or 100 nM) in a buffer (pH 8.0) containing 40 mM Tris–HCl, 20 mM acetic acid, 12.5 mM MgCl2, 1 mM DTT, 1 μM ZnCl2, 0.02% Tween 20, 200 nM BSA, and 100 nM calf thymus DNA, with or without 200 mM NaCl at ambient temperature.
Preparation of the DNA origami scaffold with three cavities
A solution (50 μL) containing M13mp18 single-stranded DNA (New England Biolabs, 10 nM) and staple DNA strands (5 equiv., 50 nM, nucleotide sequences of all staple DNA strands are described in our previous report18 and Table S13†) in buffer (40 mM Tris–HCl, 20 mM acetic acid, 12.5 mM MgCl2, pH 8.0) was heated at 95 °C for 1 min, annealed at 53 °C for 30 min, and held at 4 °C in a thermal cycler. The samples were purified by size-exclusion chromatography (400 μL volume of Sephacryl S-400, GE Healthcare) equilibrated with a buffer (pH 8.0) containing 40 mM Tris–HCl, 20 mM acetic acid, and 12.5 mM MgCl2 in Ultrafree-MC-DV (Millipore).Preparation of the DNA origami scaffold assembled with modular adaptors
A DNA origami scaffold was incubated with ZF-CLIP, AZ-CLIP, and/or CLIP-G under the conditions given in the captions of the figures or tables. For example, the cross-linking reaction of each of modular adaptor was carried out for 30 min at ambient temperature with 5 nM DNA scaffold and 250 nM modular adaptor (ZF-CLIP, AZ-CLIP, or CLIP-G) in buffer (pH 8.0) containing 40 mM Tris–HCl, 20 mM acetic acid, 12.5 mM MgCl2, 1 mM DTT, 1 μM ZnCl2, 200 mM NaCl, and 0.02% Tween 20. The mixture was purified by size-exclusion chromatography (400 μL volume of Sephacryl S-400 in Ultrafree-MC-DV) equilibrated with the buffer used for the reaction to remove unbound modular adaptors. The fractions containing DNA origami scaffolds were utilized for AFM analyses.AFM imaging and statistical analysis
The sample was deposited on a freshly cleaved mica (1.5 mm diameter) surface and adsorbed for 5 min at ambient temperature, followed by three washes with a buffer (pH 8.0) containing 40 mM Tris–HCl, 20 mM acetic acid, and 12.5 mM MgCl2. The sample was scanned in tapping mode using a fast-scanning AFM system (Nano Live Vision, RIBM Co., Ltd., Tsukuba, Japan) with a silicon nitride cantilever (Olympus BL-AC10DS-A2, Tokyo, Japan). At least three independent preparations of each sample were analyzed by AFM and several images were acquired from different regions of the mica surface. The total number of DNA scaffolds corresponds to the number of expected rectangular shapes possessing three cavities observed by AFM. Specific and nonspecific binding of modular adaptors was counted for only the modular adaptor bound to the perfectly folded DNA scaffold. The yield of the DNA-scaffold-assembled modular adaptor was calculated as described previously14,18 and the results are shown in Table S14.†Fluorescence polarization spectroscopy
A fluorescence polarization assay by using an Infinite 200 PRO F Plex (Tecan, Männedorf, Switzerland) was conducted to determine the equilibrium dissociation constant (KD) (Fig. S4, S6 and S8†) and kinetic parameter (kon and koff) (Fig. S11†) for the complexes of the modular adaptor with a fluorophore-modified ODN. The rate constant of covalent bond formation of the protein tag derivatives and fluorophore-modified substrates was also determined (Fig. S10†). The kinetic parameter kobs (s−1) was fitted to the reaction model assuming first-order kinetics, and then the second-order rate constants of the process were determined. These assays were carried out in a buffer (pH 8.0) containing 40 mM Tris–HCl, 20 mM acetic acid, 12.5 mM MgCl2, 1 mM DTT, 1 μM ZnCl2, 100 nM calf thymus DNA, and 0.02% Tween 20 at 25 °C with or without 200 mM NaCl.Conflicts of interest
The authors have no conflicts of interest directly relevant to the content of this article.Acknowledgements
This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan, to E. N. (Nos. 15H05492 and 17H05440) and T. M. (Nos. 15H01402 and 17H01213) and by JST CREST Grant Number JPMJCR18H5 (T.M.).Notes and references
- (a) T. A. Baillie, Angew. Chem., Int. Ed., 2016, 55, 13408 CrossRef CAS PubMed; (b) J. Singh, R. C. Petter, T. A. Baillie and A. Whitty, Nat. Rev. Drug Discovery, 2011, 10, 307 CrossRef CAS PubMed; (c) T. L. Foley and M. D. Burkart, Curr. Opin. Chem. Biol., 2007, 11, 12 CrossRef CAS PubMed; (d) M. A. Warpehoski and L. H. Hurley, Chem. Res. Toxicol., 1988, 1, 315 Search PubMed; (e) W. C. Tse and D. L. Boger, Chem. Biol., 2004, 11, 1607 CrossRef CAS PubMed.
- (a) A. Ali and S. Bhattacharya, Bioorg. Med. Chem., 2014, 22, 4506 CrossRef CAS PubMed; (b) S. Neidle, Nat. Prod. Rep., 2001, 18, 291 RSC.
- T. Bando and H. Sugiyama, Acc. Chem. Res., 2006, 39, 935 CrossRef CAS PubMed.
- R. T. Kovacic, J. T. Welch and S. J. Franklin, J. Am. Chem. Soc., 2003, 125, 6656 CrossRef CAS PubMed.
- D. D. F. Ma, T. Rede, N. A. Naqvi and P. D. Cook, Biotechnol. Annu. Rev., 2000, 5, 15 Search PubMed.
- N. C. Seeman and H. F. Sleiman, Nat. Rev. Mater., 2017, 3, 17068 CrossRef.
- P. W. K. Rothemund, Nature, 2006, 440, 297 CrossRef CAS PubMed.
- (a) F. Hong, F. Zhang, Y. Liu and H. Yan, Chem. Rev., 2017, 117, 12584 CrossRef CAS PubMed; (b) M. Madsen and K. V. Gothelf, Chem. Rev., 2019, 119, 6384 CAS.
- (a) A. R. Chandrasekaran, N. Anderson, M. Kizer, K. Halvorsen and X. Wang, ChemBioChem, 2016, 17, 1081 CrossRef CAS PubMed; (b) V. Linko, S. Nummelin, L. Aarnos, K. Tapio, J. Toppari and M. Kostiainen, Nanomaterials, 2016, 6, 139 CrossRef PubMed; (c) A. Rajendran, E. Nakata, S. Nakano and T. Morii, ChemBioChem, 2017, 18, 696 CrossRef CAS PubMed.
- (a) C. M. Niemeyer, Angew. Chem., Int. Ed., 2010, 49, 1200 CrossRef CAS PubMed; (b) B. Sacca` and C. M. Niemeyer, Chem. Soc. Rev., 2011, 40, 5910 RSC; (c) Y. R. Yang, Y. Liu and H. Yan, Bioconjugate Chem., 2015, 26, 1381 CrossRef CAS PubMed; (d) J. B. Trads, T. Tørring and K. V. Gothelf, Acc. Chem. Res., 2017, 50, 1367 CrossRef CAS PubMed.
- (a) O. I. Wilner, Y. Weizmann, R. Gill, O. Lioubashevski, R. Freeman and I. Willner, Nat. Nanotechnol., 2009, 4, 249 CrossRef CAS PubMed; (b) J. Fu, M. Liu, Y. Liu, N. W. Woodbury and H. Yan, J. Am. Chem. Soc., 2012, 134, 5516 CrossRef CAS PubMed; (c) J. Fu, Y. R. Yang, A. Johnson-Buck, M. Liu, Y. Liu, N. G. Walter, N. W. Woodbury and H. Yan, Nat. Nanotechnol., 2014, 9, 531 CrossRef CAS PubMed; (d) C. B. Rosen, A. L. B. Kodal, J. S. Nielsen, D. H. Schaffert, C. Scavenius, A. H. Okholm, N. V. Voigt, J. J. Enghild, J. Kjems, T. Tørring and K. V. Gothelf, Nat. Chem., 2014, 6, 804 CrossRef CAS PubMed.
- J. Fu, Y. R. Yang, S. Dhakal, Z. Zhao, M. Liu, T. Zhang, N. G. Walter and H. Yan, Nat. Protoc., 2016, 11, 2243 CrossRef CAS PubMed.
- (a) B. Saccà, R. Meyer, M. Erkelenz, K. Kiko, A. Arndt, H. Schroeder, K. S. Rabe and C. M. Niemeyer, Angew. Chem., Int. Ed., 2010, 49, 9378 CrossRef PubMed; (b) E. Nakata, F. F. Liew, C. Uwatoko, S. Kiyonaka, Y. Mori, Y. Katsuda, M. Endo, H. Sugiyama and T. Morii, Angew. Chem., Int. Ed., 2012, 51, 2421 CrossRef CAS PubMed; (c) T. A. Ngo, E. Nakata, M. Saimura, T. Kodaki and T. Morii, Methods, 2014, 67, 142 CrossRef CAS PubMed; (d) K. J. Koßmann, C. Ziegler, A. Angelin, R. Meyer, M. Skoupi, K. S. Rabe and C. M. Niemeyer, ChemBioChem, 2016, 17, 1102 CrossRef PubMed; (e) S. Sagredo, T. Pirzer, R. A. Aghebat, M. A. Goetzfried, G. Moncalian, F. C. Simmel and F. De La Cruz, Angew. Chem., Int. Ed., 2016, 55, 4348 CrossRef CAS PubMed; (f) K. N. Lovendahl, A. N. Hayward and W. R. Gordon, J. Am. Chem. Soc., 2017, 139, 7030 CrossRef CAS PubMed; (g) T. Kurokawa, S. Kiyonaka, E. Nakata, M. Endo, S. Koyama, E. Mori, N. H. Tran, H. Dinh, Y. Suzuki, K. Hidaka, M. Kawata, C. Sato, H. Sugiyama, T. Morii and Y. Mori, Angew. Chem., Int. Ed., 2018, 57, 2586 CrossRef CAS PubMed.
- E. Nakata, H. Dinh, T. A. Ngo, M. Saimura and T. Morii, Chem. Commun., 2015, 51, 1016 RSC.
- T. M. Nguyen, E. Nakata, M. Saimura, H. Dinh and T. Morii, J. Am. Chem. Soc., 2017, 139, 8487 CrossRef CAS PubMed.
- N. P. Pavletich and C. O. Pabo, Science, 1991, 252, 809 CrossRef CAS PubMed.
- A. Keppler, S. Gendreizig, T. Gronemeyer, H. Pick, H. Vogel and K. Johnsson, Nat. Biotechnol., 2003, 21, 86 CrossRef CAS PubMed.
- T. A. Ngo, E. Nakata, M. Saimura and T. Morii, J. Am. Chem. Soc., 2016, 138, 3012 CrossRef CAS PubMed.
- T. Sera and C. Uranga, Biochemistry, 2002, 41, 7074 CrossRef CAS PubMed.
- A. Gautier, A. Juillerat, C. Heinis, I. R. Corrêa Jr, M. Kinder mann, F. Beaufils and K. Johnsson, Chem. Biol., 2008, 15, 128 CrossRef CAS PubMed.
- (a) G. V. Los, L. P. Encell, M. G. McDougall, D. D. Hartzell, N. Karassina, C. Zimprich, M. G. Wood, R. Learish, R. F. Ohana, M. Urh, D. Simpson, J. Mendez, K. Zimmerman, P. Otto, G. Vidugiris, J. Zhu, A. Darzins, D. H. Klaubert, R. F. Bulleit and K. V. Wood, ACS Chem. Biol., 2008, 3, 373 CrossRef CAS PubMed; (b) C. G. England, H. Luo and W. Cai, Bioconjugate Chem., 2015, 26, 975 CrossRef CAS PubMed.
- G. Schreiber, G. Haran and H.-X. Zhou, Chem. Rev., 2009, 109, 839 CrossRef CAS PubMed.
- (a) F. C. Collins and G. E. Kimball, J. Colloid Sci., 1949, 4, 425 CrossRef CAS; (b) P. A. Maslak, J. Am. Chem. Soc., 1989, 111, 8201 CrossRef CAS; (c) O. G. Berg and P. H. von Hippel, Annu. Rev. Biophys. Biophys. Chem., 1985, 14, 131 CrossRef CAS PubMed; (d) C. Berger, L. Piubelli, U. Haditsch and H. Rudolf Bosshard, FEBS Lett., 1998, 425, 14 CrossRef CAS PubMed.
- (a) P. H. Von Hippel and O. G. Berg, J. Biol. Chem., 1989, 264, 675 CAS; (b) J.-S. Kim and C. O. Pabo, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 2812 CrossRef CAS PubMed; (c) M. Geertz, D. Shore and S. J. Maerkl, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 16540 CrossRef CAS PubMed.
- (a) I. A. Hope, S. Mahadevan and K. Struhl, Nature, 1988, 333, 635 CrossRef CAS PubMed; (b) E. K. O'Shea, R. Rutkowski and P. S. Kim, Science, 1989, 243, 538 CrossRef PubMed; (c) T. E. Ellenberger, C. J. Brandl, K. Struhl and S. C. Harrison, Cell, 1992, 71, 1223 CrossRef CAS PubMed; (d) P. König and T. J. Richmond, J. Mol. Biol., 1993, 233, 139 CrossRef PubMed; (e) W. Keller, P. Konig and T. J. Richmond, J. Mol. Biol., 1995, 254, 657 CrossRef CAS PubMed.
- (a) M. T. Record, C. F. Anderson and T. M. Lohman, Q. Rev. Biophys., 1978, 11, 103 CrossRef CAS PubMed; (b) D. Jantz and J. M. Berg, Biophys. J., 2010, 98, 852 CrossRef CAS PubMed; (c) B. L. Jutras, A. Verma and B. Stevenson, Curr. Protoc. Microbiol., 2012, 24, 1F.1.1 Search PubMed; (d) A. Esadze and J. Iwahara, J. Mol. Biol., 2014, 426, 230 CrossRef CAS PubMed.
- (a) P. L. Privalov, A. I. Dragan and C. Crane-robinson, Nucleic Acids Res., 2011, 39, 2483 CrossRef CAS PubMed; (b) A. Atzori, S. Liggi, A. Laaksonen, M. Porcu, A. P. Lyubartsev, G. Saba and F. Mocci, Can. J. Chem., 2016, 94, 1181 CrossRef CAS; (c) A. Esadze, C. A. Kemme, A. B. Kolomeisky and J. Iwahara, Nucleic Acids Res., 2014, 42, 7039 CrossRef CAS PubMed.
- (a) C. O. Pabo and R. T. Sauer, Annu. Rev. Biochem., 1992, 61, 1053 CrossRef CAS PubMed; (b) P. Prabakaran, J. G. Siebers, S. Ahmad, M. M. Gromiha, M. G. Singarayan and A. Sarai, Structure, 2006, 14, 1355 CrossRef CAS PubMed; (c) R. Gamsjaeger, C. K. Liew, F. E. Loughlin, M. Crossley and J. P. Mackay, Trends Biochem. Sci., 2007, 32, 63 CrossRef CAS PubMed; (d) W. H. Hudson and E. A. Ortlund, Nat. Rev. Mol. Cell Biol., 2014, 15, 749 CrossRef CAS PubMed.
- (a) A. G. Cherstvy, Phys. Chem. Chem. Phys., 2011, 13, 9942 RSC; (b) R. C. Harris, T. MacKoy, A. C. D. MacHado, D. Xu, R. Rohs and M. O. Fenley, in RSC Biomolecular Sciences No. 24 Innovations in Biomolecular Modeling and Simulations, ed. T. Schlick, Royal Society of Chemistry, Cambridge, 2012, vol. 2, pp. 53–80 Search PubMed; (c) M. Barbi and F. Paillusson, in Advances in Protein Chemistry and Structural Biology, ed. T. Karabencheva-Christova, Elsevier, Netherlands, 2013, vol. 92, pp. 253–297 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Rate simulation of the cross-linking reaction (Fig. S1, S2), construction of the modular adaptor (Fig. S3, S15, and Table S15), evaluation of activity and orthogonality (Table S1–S12, Fig. S4–S11), structures of DNA origami scaffolds (Fig. S12), sequence of staple strand DNAs used to assemble the DNA origami scaffold (Table S13), and statistical analyses of AFM images for determining the occupancies of DNA scaffolds by modular adaptors (Fig. S13, S14, and Table S14). See DOI: 10.1039/c9sc02990g |
| This journal is © The Royal Society of Chemistry 2019 |