Open Access Article
Open Access ArticleCombination of gallium(III) with acetate for combating antibiotic resistant Pseudomonas aeruginosa†
Yuchuan
Wang‡
 ab,
Bingjie
Han‡
ab,
Bingjie
Han‡
 a,
Yanxuan
Xie‡
a,
Haibo
Wang‡
a,
Yanxuan
Xie‡
a,
Haibo
Wang‡
 b,
Runming
Wang
b,
Runming
Wang
 b,
Wei
Xia
b,
Wei
Xia
 a,
Hongyan
Li
a,
Hongyan
Li
 b and
Hongzhe
Sun
b and
Hongzhe
Sun
 *b
*b
aSchool of Chemistry, Sun Yat-sen University, Guangzhou, 510275, P. R. China
bDepartment of Chemistry, The University of Hong Kong, Hong Kong, P. R. China. E-mail: hsun@hku.hk
First published on 2nd May 2019
Abstract
Gallium(III) has been widely used as a diagnostic and therapeutic agent in clinics for the treatment of various diseases, in particular, Ga-based drugs have been exploited as antimicrobials to combat the crisis of antimicrobial resistance. The therapeutic properties of Ga(III) are believed to be attributable to its chemical similarity to Fe(III). However, the molecular mechanisms of action of gallium remain unclear. Herein, by integrating metalloproteomics with metabolomics and transcriptomics, we for the first time identified RpoB and RpoC, two subunits of RNA polymerase, as Ga-binding proteins in Pseudomonas aeruginosa. We show that Ga(III) targets the essential transcription enzyme RNA polymerase to suppress RNA synthesis, resulting in reduced metabolic rates and energy utilization. Significantly, we show that exogenous supplementation of acetate could enhance the antimicrobial activity of Ga(III), evidenced by the inhibited growth of persister cells and attenuated bacterial virulence. The effectiveness of co-therapy of Ga(III) and acetate was further validated in mammalian cell and murine skin infection models, which is attributable to enhanced uptake of Ga(III), and reduced TCA cycle flow and bacterial respiration. Our study provides novel insights into the mechanistic understanding of the antimicrobial activity of Ga(III) and offers a safe and practical strategy of using metabolites to enhance the efficacy of Ga(III)-based antimicrobials to fight drug resistance.
Introduction
Antimicrobial resistance (AMR) is one of the most serious global public health threats of this century. There is an urgent need to develop safe, efficacious and novel antimicrobial strategies. Owing to the lack of new antibiotics, metal-based antimicrobials have received increasing interest as promising alternatives in recent years for tackling the antimicrobial resistance crisis.1–5 Moreover, antimicrobial metals have the capability of disrupting multiple bacterial physiological processes and improving the cure rates of infections from resistant bacterial strains, thus they may also serve as antibiotic adjuvants to restore and boost antibiotic efficacy.2,3,6 This is exemplified by our recent study that an antiulcer bismuth drug (CBS, De-Nol®) could be repurposed as an antibiotic adjuvant to treat NDM1-positive bacterial infection in mouse infection models.1Pseudomonas aeruginosa (P. aeruginosa) is a classic opportunistic pathogen, which can cause a variety of hospital-acquired infections including cystic fibrosis (CF), urinary tract infections (UTIs), skin and pulmonary infections, and even sepsis in severe cases.7,8 The infections by P. aeruginosa are usually resistant to multiple antibiotics due to the bacterium's intrinsic drug resistance and additional hospital/community acquired resistance.9 Gallium compounds such as gallium nitrate, the active component of the FDA-approved formulation Ganite®, gallium-protoporphyrin IX, gallium-desferrioxamine show great potential as anti-P. aeruginosa therapeutic agents.10–12 Recently, gallium nitrate has been shown to be effective in the treatment of chronic P. aeruginosa airway infections both in a mouse infection model and in a phase I clinical trial in individuals with cystic fibrosis, and exhibits low rates in the development of resistance compared to antibiotics.5 Moreover, the synergistic effect of gallium with antibiotics was also observed. The therapeutic effects of Ga(III) are attributable to its identical chemical properties to Fe(III), being incorporated into essential Fe(III)-binding bacterial proteins/enzymes and leading to the disruption of various Fe(III)-dependent functions due to the fact that Ga(III) is unable to be reduced under physiological conditions.13–15 However, there appears to be a lack of studies at molecular levels on the mode of action of Ga(III)-based antimicrobials. Knowledge on the key molecular targets of gallium and the bacterial cellular response under the stress of gallium might allow its efficacy to be further improved.
Accumulated studies reported that the metabolic state of a bacterial cell could affect its susceptibility to antibiotics.16–18 Thus, sensitization of resistant bacteria to antibiotic treatment through metabolic stimuli represents a novel strategy to combat antimicrobial resistance.19,20 Comprehensive analysis of cellular response of a bacterium to drug treatment through diversified approaches such as proteomics and functional metabolomics provides an insight into the mechanism of action of a drug, which may lead to enhanced therapeutic efficacy.
Herein, we report the identification of RNA polymerase as a key protein target that defines the bacteriostatic property of Ga(III) in P. aeruginosa. A functional metabolomics study of P. aeruginosa in response to Ga(III) treatment enabled acetate to be identified as the most effective metabolite that could enhance the bacteriostatic efficacy of Ga(III) against P. aeruginosa persisters, and the therapeutic effectiveness of co-therapy of Ga(III) and acetate was validated in mammalian cell infection and murine infection models.
Results
Gallium targets bacterial RNA polymerase and attenuates transcription
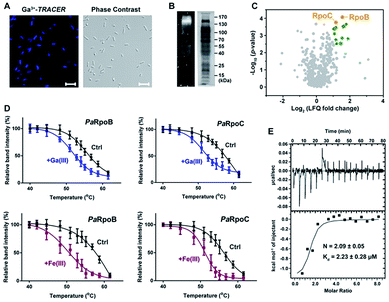
We first used the Ga-coordinated fluorescent probe Ga(III)–TRACER21,22 to track Ga-binding proteins in live P. aeruginosa. The ability of Ga(III)–TRACER to label Ga-binding proteins in vitro was demonstrated using apo-lactoferrin (hLF) as shown in Fig. S1A (ESI†). Upon incubation of P. aeruginosa cells with Ga(III)–TRACER and then irradiation with UV at 365 nm for 15 min, we observed intense blue fluorescence throughout the pathogen (Fig. 1A), suggesting that the probe entered P. aeruginosa cells and labelled intracellular Ga-binding proteins. The cells were then lysed and separated by SDS-PAGE, and intense fluorescent bands at a molecular weight of 130–170 kDa were observed (Fig. 1B). Through peptide mass fingerprinting analysis, two proteins PaRpoB and PaRpoC of ca. 150 kDa in accordance with the observed protein mass on the gel were identified as putative Ga-binding proteins (Table S1†).We also employed competitive gallium-based immobilized metal affinity chromatography (Ga-IMAC) to identify Ga-binding proteins in P. aeruginosa. Bacterial cell lysates either pre-treated or un-treated with Ga(NO3)3 were incubated with Ga-IMAC resin. Pre-treatment of Ga(NO3)3 led to pre-saturation of the actual binding sites of the target proteins and resulted in reduced abundance in the eluted protein fraction. Over 200 proteins with at least two unique peptides were identified from the Ga-IMAC experiment. Through competitive analysis, PaRpoB and PaRpoC were identified among the proteins with high enrichment factors (Fig. 1C). To validate the proteins identified by Ga-IMAC, a number of genes encoding proteins with high scores were cloned and proteins were overexpressed in E. coli cells, followed by supplementation of Ga(III)–TRACER to the cells. Only E. coli cells with PaRpoB and PaRpoC overexpressed could be fluorescent-labelled by the probe (Fig. S1B†), further corroborating the binding of Ga(III) to PaRpoB/C in cells.
The binding of Ga(III) to PaRpoB/C in cells was further examined with a cellular thermal shift assay (CETSA)23 using the PaRpoB/C polyclonal antibodies from PaRpoB/C-immunized rabbits. Addition of Ga(NO3)3 to the culture medium resulted in changes in the protein thermal stabilities, i.e., Tm from 57.2 to 53.5 °C and 58.0 to 52.3 °C for PaRpoB and PaRpoC, respectively, indicative of reduced protein thermal stabilities upon Ga(III) binding (Fig. 1D and S1C†). Such changes were also observed in the presence of Fe(NO3)3 (Fig. 1D and S1C†), demonstrating that Fe(III) also binds to PaRpoB/C in cells. We further purified PaRpoB/C proteins and investigated their Ga-binding properties in vitro. A time dependent increase in fluorescence was observed upon mixing PaRpoB with Ga(III)–TRACER, leading to ca. 2-fold fluorescence enhancement (Fig. S1D†). An isothermal titration calorimetry (ITC) experiment was then performed to monitor the titration of gallium citrate into EDTA-treated PaRpoB/C. The binding of gallium to apo-PaRpoB/C resulted in a typical S-shaped curve, while pre-saturation of PaRpoB with Fe(III) had no detectable Ga-binding, indicating that Ga(III) shares common binding sites on the protein with Fe(III) (Fig. 1E and S2†). By fitting the data with a one-set-of-sites binding model, the dissociation constants were determined to be 2.23 ± 0.28 μM and 1.68 ± 0.08 μM, with a stoichiometry of 2.09 ± 0.05 and 2.24 ± 0.23 for Ga(III) binding to PaRpoB and PaRpoC, respectively. The binding stoichiometries are consistent with those obtained from ICP-MS analysis (2.15 ± 0.18 and 2.11 ± 0.26 for Ga-PaRpoB and Ga-PaRpoC, respectively).
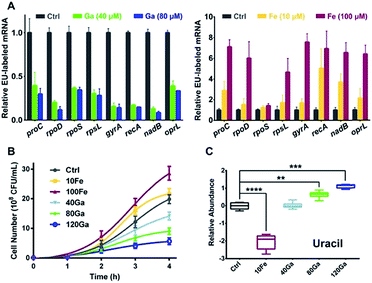
RpoB and RpoC are two subunits of DNA-dependent RNA polymerase, the key enzyme in the regulation of transcription and gene expression in living organisms.24 As Ga(III) binds to RpoB and RpoC both in vitro and in vivo, we hypothesize that the function of RNA polymerase could be interfered by Ga(III). To investigate the effect of Ga(III) on bacterial transcription, P. aeruginosa cells were pulse-labelled with 5-ethynyluridine (EU) for 4 h in the presence of Ga(NO3)3, followed by purification of the labelled nascent mRNA via click chemistry. Analysis of EU-labelled mRNA levels shows a global decrease in all analyzed P. aeruginosa house-keeping genes25 upon Ga(III) treatment (Fig. 2A), confirming the general defect in RNA polymerase-mediated transcription in the presence of Ga(III). The bacteriostatic antibiotic rifamycin, a well-known RNA polymerase inhibitor that is able to suppress nascent mRNA levels in P. aeruginosa as evaluated by the same method (Fig. S3A†), was employed as a positive control for comparing the effect of Ga(III). Interestingly, we found that bacterial growth is positively correlated with the levels of transcription regulated by Ga(III) and Fe(III). Supplementation of Ga(III) effectively inhibited the growth of P. aeruginosa in a dose-dependent manner. In contrast, Fe(III) obviously promoted bacterial growth (Fig. 2B), accompanied by enhanced bacterial transcription as revealed by the increased nascent mRNA levels (Fig. 2A). Given that uracil is an essential nucleobase for RNA synthesis, changes in its relative abundance were found to be correlated with the transcription inhibition or promotion mediated by Ga(III) or Fe(III), respectively (Fig. 2C).
Ga(III) reduces TCA cycle flow and decreases bacterial respiration
We then investigated the inhibition mechanisms of Ga(III) on P. aeruginosa at the transcriptome level. Pathway enrichment analysis of the differentially regulated genes by Ga(III) indicates that the TCA cycle and oxidative phosphorylation are the pathways significantly influenced (Fig. S4A†). We therefore evaluated the effect of Ga(III) on the bacterial central carbon metabolism. We found general dose-dependent decreases in the activities of TCA cycle enzymes upon Ga(III) treatment (Fig. S5A†), and the enhanced inhibitory effect of Ga(III) against P. aeruginosa with the supplement of malonate, an inhibitor of succinate dehydrogenase (Fig. S5B†), verifying that Ga(III) inhibits TCA cycle activity.Oxygen functions as the terminal electron acceptor for aerobic respiration in the electron transport chain (ETC), and oxygen consumption thus serves as a useful indicator for ETC activity.26 We then quantified the oxygen consumption rate (OCR) of P. aeruginosa, and observed a linear increase in the OCR over time in the bacterium without treatment (as a control). However, the increase in the OCR was progressively suppressed with the supplementation of increasing amounts of Ga(III), and reduction in the OCR was observed at 256 μM Ga (Fig. S5C†), indicating that Ga(III) inhibits respiratory activity. This was further validated by the observed enhanced inhibitory effect of Ga(III) upon supplementation of the cells with the ETC inhibitor NaN3, a cytochrome c oxidase inhibitor (Fig. S5D†).
Exogenous metabolites tune the susceptibility of P. aeruginosa to Ga(III)
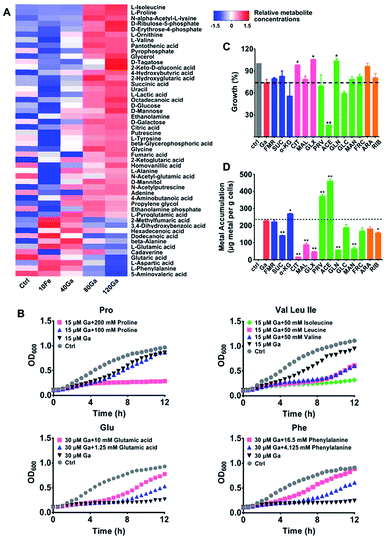
To further understand the antimicrobial mechanisms of Ga(III) against P. aeruginosa, we performed a GC-MS-based metabolomics study to analyze the changes in metabolite abundance upon Ga(III) treatment. Overall, 62 metabolites with reliable signals were detected from P. aeruginosa metabolite extracts (Fig. 3A and Table S3†), and the PCA score plots showed clear separations of the metabolic profiles of different treatment groups (Fig. S6A†). The differentially regulated metabolites were identified by one-way ANOVA analysis with significance values of p < 0.01 (Table S3†).After 4 h treatment, an increase in abundance of the majority of metabolites was observed in the group treated with a high concentration of Ga(III) (120 μM), including several essential amino acids (isoleucine, proline, valine, tyrosine and alanine) and the TCA cycle intermediates (succinic acid, citric acid, fumaric acid and α-ketoglutaric acid) (Fig. 3A, S6B and S7†), accompanied by increased cellular ATP levels (Fig. S6C†). The accumulation of various metabolites shows the reduced metabolic rates and energy utilization in P. aeruginosa, which might be ascribed to the transcription suppression mediated growth inhibition of P. aeruginosa after treatment with Ga(III). Comparing the metabolic profiles of Ga(III)-treated groups with that of Fe(III)-deficient group (control group, Fig. 3A), although Ga(III) supplementation resulted in decreased bacterial Fe(III) accumulation (Fig. S6D†), the changes in metabolite abundance upon Ga(III) treatment are mostly attributed to the addition of Ga(III) per se, and to a less extent to Ga-induced Fe(III) deficiency.
Metabolic response represents a strategy of cells to cope with external stimuli, which could be harnessed to regulate the metabolic status of cells and improve drug efficacy as reported in recent years.16,17,19,27 We then examined whether metabolic stimulations in P. aeruginosa would modulate the inhibitory activity of Ga(III). As revealed by the metabolomics results, Ga(III) treatment led to significant changes in the levels of several essential amino acids, i.e., the up-regulated isoleucine, proline, and valine, and the down-regulated phenylalanine, aspartic acid, and glutamic acid. We first examined the bacterial growth upon Ga(III) treatment with or without the exogenous addition of the Ga-regulated amino acids. As shown in Fig. 3B, supplementation of the up-regulated amino acids proline and the branched-chain amino acids including isoleucine, leucine and valine to the bacterial culture media suppressed bacterial growth. In contrast, addition of the down-regulated amino acids such as glutamic acid and phenylalanine promoted bacterial growth, indicating that the inhibitory activity of Ga(III) could be regulated by external metabolic stimuli.
Given that our combined study at transcriptomics and metabolomics levels showed that the central carbon metabolism of P. aeruginosa was significantly influenced by Ga(III), we therefore screened carbon source metabolites covering various central metabolic pathways, i.e., the TCA cycle, pyruvate metabolism and glycolysis. We found that these metabolites exhibited different effects on the susceptibility of P. aeruginosa to Ga(III), as revealed by the 12 h growth curves of the bacterium under different treatment conditions (Fig. 3C and S8A†). In the presence of Ga(III), addition of acetate significantly decreased bacterial growth by 80%; while addition of citrate, glyoxylate and gluconate slightly promoted bacterial growth (Fig. 3C and S8A†). Citrate and glyoxylate were also found to be able to promote the growth of P. aeruginosa under the stress of high concentration (256 μM) of Ga(III) (Fig. S8B†). Addition of metabolites alone had no significant effect on bacterial growth (Fig. S8C†). We then examined Ga(III) uptake by P. aeruginosa after co-treatment of metabolites with Ga(III), and found that the enhanced antimicrobial activity was positively correlated with the levels of Ga uptake (Fig. 3D). Acetate, which significantly inhibits bacterial growth, induced the uptake of high levels of Ga (2-fold), implying that the increased Ga uptake is responsible for the inhibited growth.
Co-treatment of Ga(III) and acetate enhances the antimicrobial efficacy of Ga(III) in cell and murine infection models and reduces P. aeruginosa virulence
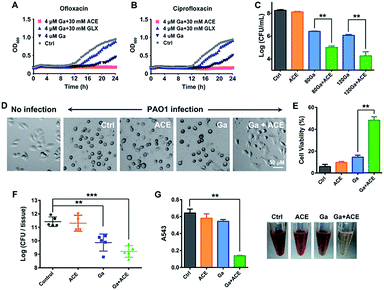
As acetate was identified as the most effective metabolite in enhancing the inhibitory activity of Ga(III) on bacterial growth, we therefore evaluated the effectiveness of the co-therapy of Ga(III) and acetate. Enhanced antimicrobial activity of Ga(III) caused by acetate was also demonstrated on a clinically isolated P. aeruginosa strain (Fig. S9†). In CF patients with chronic antibiotic treatment, P. aeruginosa frequently exists as persister cells.28 We first examined whether co-therapy of Ga(III) and acetate is effective on P. aeruginosa persister cells. The persister cells were isolated by treating stationary-phase cells with 10 μg mL−1 ciprofloxacin or 10 μg mL−1 ofloxacin, and the survival cells were then exposed to Ga(III) alone or Ga(III) in combination with metabolites. The 24 h growth curves showed that Ga(III) (4 μM) only slightly inhibited bacterial growth, while the combination of Ga(III) with acetate (30 mM) could completely inhibit the growth of persister cells. In contrast, the combined use of Ga(III) with glyoxylate (30 mM) promoted bacterial growth (Fig. 4A and B), consistent with the observations on exponential-phase P. aeruginosa.We next examined the effects of co-administration of Ga(III) with acetate in a mammalian cell infection model. A549 cells were infected with P. aeruginosa at a multiplicity of infection (MOI) of 10 for 1 h. The infected A549 cells were then exposed to Ga(III) or acetate alone or their combination for 24 h, and the cell-associated bacteria were enumerated by agar plating. Our results showed that the viable bacterial loads were at a level of 108 CFU when 30 mM acetate was used, which is comparable to those of the untreated control group. The bacterial loads dropped to a level of 106 CFU when treated with 80–120 μM Ga(III), and further dropped to a level of 104 CFU when Ga(III) (120 μM) and acetate (30 mM) were co-administered (Fig. 4C); however, the cells cultured under the stress of Fe(III) and acetate showed increased bacterial loads (Fig. S10†). Notably, we observed a marked cytoprotective effect on P. aeruginosa infected A549 cells by the combined Ga–acetate treatment. A549 cells were infected with P. aeruginosa in the presence of Ga(III) or acetate, or their combination, and the cytotoxic effect was studied after infection for 4 h. The infected cells were found to have a round morphology in the control and single therapy groups, while the cell morphology remained almost unchanged before and after bacterial infection in the group with combined Ga–acetate treatment (Fig. 4D and S11†) and cell viability was also significantly increased by 34% compared with that of Ga(III) treatment alone (Fig. 4E).
We further evaluated the in vivo effectiveness of the Ga–acetate combination in a murine skin infection model. Female BALB/c mice were infected with 5 × 106 CFU of P. aeruginosa on an excisional skin wound. The infected mice received treatment of Ga(III) or the combination with acetate therapy twice daily, and the mice were sacrificed 48 h post-infection for colony counting. As shown in Fig. 4f, treatment with Ga(III) alone reduced the cell count by ca. 10-fold compared with that of the control group, whereas treatment with the Ga(III) and acetate combination further reduced the bacterial count by ca. 10-fold compared with that of the Ga(III) treatment group, clearly demonstrating that acetate can enhance the antimicrobial activity of Ga(III) in vivo. We then examined the effect of co-therapy of acetate and Ga(III) on the pathogenicity of P. aeruginosa. In comparison to Ga(III) or acetate treatment alone, the combination of Ga(III) and acetate at sub-inhibitory concentration significantly reduced bacterial hemolysin secretion in mammalian cells (Fig. 4G). Moreover, at higher drug concentrations (80 μM Ga(III) and 30 mM acetate), the virulence factors secreted by the pathogen including elastase, protease, pyocyanin, pyoverdine, phospholipase C, phamnolipid and exotoxin A were extensively down-regulated at transcriptome levels (Fig. S12A and Table S4†), implying reduced virulence of the bacterium.
Combined Ga(III)–acetate treatment regulates P. aeruginosa metabolome
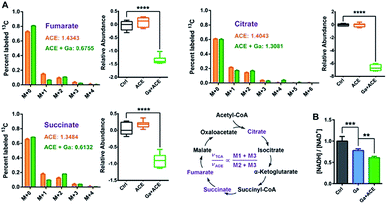
We further investigated the mechanisms of action of co-therapy of Ga(III) and acetate at transcriptome and metabolome levels. Cluster analysis and principal component analysis (PCA) of RNA-seq and GC-MS data showed certain similarity between the control and acetate treatment groups; however, the Ga(III) and combined treatment groups were clearly separated from the control group (Fig. S12B and C†), suggesting that Ga(III) is responsible for the observed antimicrobial effect. The combined Ga(III)–acetate treatment further attenuated bacterial transcription as revealed by decreased mRNA levels (Fig. S3B†).To evaluate the effect of the combined treatment on TCA cycle flow, we performed a 13C2-acetate NTFD (non-targeted tracer fate detection) experiment on P. aeruginosa in the absence and the presence of Ga(III) by GC-MS.29 Three 13C-labeled TCA cycle metabolites including citrate, succinate and fumarate were detected, suggesting that the exogenous acetate was converted by acetyl-CoA synthase to labelled acetyl-CoA and then fluxed to the TCA cycle. According to a previous study, the sum of M1 and M3 labels (νTCA) refers to the turnover of a particular metabolite in the TCA cycle, while M2 plus M3 (νACE) is the flux of the labeled acetate to the TCA cycle, thus the (M1 + M3)/(M2 + M3) ratio represents the relative flux for the certain metabolite in the TCA cycle.17 The observation of the decreases in νTCA/νACE ratios and abundance of the three metabolites in the TCA cycle under Ga(III) stress further verified the inhibition of the TCA cycle by Ga(III) (Fig. 5A). This was further corroborated by the reduced [NADH]/[NAD+] ratio upon Ga(III) and the combined treatment (Fig. 5B), representing decreased reducing power generated through TCA cycle flow.
In addition to the 13C-labeled metabolites in the TCA cycle, a number of 13C-labeled amino acids, nucleotides and fatty acids were detected upon the co-treatment of Ga(III) and 13C2-acetate (Fig. S13†), suggesting that a widespread metabolome reprogramming in P. aeruginosa occurred to deal with Ga(III) stress. Moreover, treatment of the bacterium with Ga(III) also resulted in the accumulation of glucose, and depletion of glucose 6-phosphate and ribose 5-phosphate (Fig. S14†), the key metabolites in glycolysis and the pentose phosphate pathway, indicative of suppressed glucose catabolism by Ga(III).
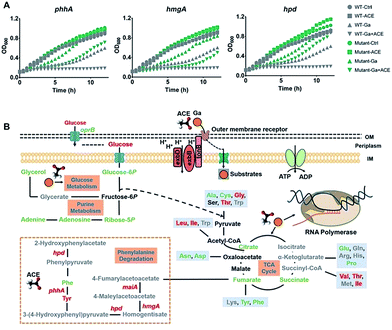
Transcriptome analysis showed that treatment of the bacterium with acetate led to marked regulation of a number of genes enriched in various amino acid metabolic pathways (Fig. S4B†), in particular, the up-regulation of the phenylalanine and tyrosine metabolic pathway enriched by the significantly up-regulated genes hpd, maiA, hmgA, phhA and phhC (Table S5†). Interestingly, proteins Hpd, PhhA and HmgA in this pathway were reported to bind ferric iron.30 To investigate the role of these proteins in the antimicrobial action of Ga(III), we generated mutant strains ΔphhA, ΔhmgA and Δhpd by transposon mutagenesis and found that all the mutant strains showed reduced susceptibility towards Ga(III) in comparison to the wild type strain (Fig. 6A), indicating the involvement of these genes in mediating Ga(III) sensitivity in P. aeruginosa.
Discussion
Metal-based antimicrobials have received renewed interest in the era of antimicrobial resistance, which has led to the drying up of antibiotic pipelines. Recently, an FDA approved gallium drug (Ganite®) has been shown to have therapeutic effects on mice and humans with lung infections from P. aeruginosa.5 Although it has long been known that Ga(III) can disrupt bacterial iron metabolism owing to its similarity to Fe(III), a better understanding of the mode of action of Ga(III) at molecular levels allows its therapeutic potential to be fully explored. Previously, Ga(III) has been demonstrated to interact with “bacterial transferrin”, the periplasmic iron-transport protein,31 and hitA gene has been also identified to associate with Ga(III) resistance in P. aeruginosa.5,32 However, the ΔhitA mutant strain of P. aeruginosa still shows susceptibility towards Ga(III),5 strongly indicating the presence of key cellular targets of Ga(III), which have not been characterized. In the present study, by using metalloproteomics approaches, we uncovered a novel Ga(III) target, RNA polymerase (RNAP), in P. aeruginosa. We found that Ga(III) bound to two subunits RpoB and RpoC of the core RNAP enzyme with a stoichiometry of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and Fe(III) was also able to bind to the two proteins and interfered with the Ga(III) binding ability.
1, and Fe(III) was also able to bind to the two proteins and interfered with the Ga(III) binding ability.
RNAP is the basic enzyme in transcription and is well characterized to be a target of the bacteriostatic antibiotics rifamycins.24,33 Mg(II), Zn(II) and the [4Fe–4S] cluster have been reported to be the cofactors of RNAP.34 As an essential process for bacterial growth and survival, transcription is the first step of gene expression carried out by RNAP, and there exist exquisite crosstalks between transcription and metabolism to regulate cellular enzyme levels.35,36 Indeed, we observed a positive correlation between Fe(III)- and Ga(III)-mediated bacterial growth and transcriptional regulation (Fig. 2). As a bacteriostatic antimicrobial, Ga(III) markedly inhibited P. aeruginosa growth as well as transcription processes by targeting RpoB and RpoC, evidenced by the decreased nascent mRNA levels and a significant accumulation of the nucleobase uracil owing to reduced amounts incorporated into RNA chains. In contrast, the presence of Fe(III) was found to promote bacterial growth and transcription processes. However, how metal-mediated transcriptional regulation may inhibit or promote bacterial growth remains unclear. Phenotypically, Ga(III) treatment led to ATP accumulation, OCR reduction, undetectable ROS elevation and central carbon metabolism suppression featured by reduced TCA cycle flow and decreased glucose consumption, which are consistent with the phenotypic features of bacteria in response to bacteriostatic antibiotics as reported previously.26 Therefore, upon Ga(III) treatment, P. aeruginosa may enter a metabolically quiescent state as a result of drug–target interaction to withstand Ga(III) stress. We thus believe that targeting bacterial RNAP may define the bacteriostatic feature of the antimicrobial Ga(III), and such bacteriostatic agents are as effective as bactericidal agents in the treatment of bacterial infections clinically.37
The metabolome holds precise information on a cell's status, and offers a rich resource for the manipulation of the cell's function and vulnerability. For instance, regulations in bacterial metabolism could alter aminoglycoside sensitivity in bacterial persisters, stationary-phase bacteria, and antibiotic-resistant bacteria;16–18L-arginine could directly impact the metabolic fitness and enhance the anti-tumor activity of central memory T cells.38 Herein, we show that the inhibitory effect of Ga(III) could be manipulated by exogenous metabolic stimuli, and supplementation of the metabolites that are down-regulated or up-regulated by Ga(III) resulted in Ga(III) activity being suppressed or enhanced, indicating that the efficacy of a drug could be modulated according to the drug-induced metabolome changes. Our results further reveal that the central carbon metabolite acetate effectively increased Ga(III) uptake by P. aeruginosa, leading to enhanced bacteriostatic activity as demonstrated on persister cells, and A549 cell infection and murine skin infection models. Moreover, at transcriptome levels, acetate activated the Fe-demanding phenylalanine degradation pathway, and the transposon mutations of key genes in this pathway made the bacterium less sensitive to Ga(III), which may also contribute to acetate-induced enhancement of Ga(III) antimicrobial activity. Given that acetic acid has already been used clinically for the treatment of infected burn wounds,39,40 the combination of acetate with an FDA approved gallium drug may provide a rapid and cost-effective route to new therapeutic strategies to cope with the current crisis of antibiotic resistance in clinics.
Conclusions
The rapid emergence and spread of antibiotic resistant bacterial strains and the diminished antibiotic pipelines have led to renewed interest in using metal-based antimicrobials for fighting against infectious diseases in clinics. Knowledge of the mechanisms of action of these antimicrobials allows us to further enhance their effectiveness or to rationally design more potent antimicrobial agents. Ga(III) has broad-spectrum antimicrobial activity against a range of infectious pathogens and shows evidence to be a safe and effective antimicrobial in the treatment of human lung infections in clinical trials. By integration of transcriptomics, proteomics and metabolomics, we show for the first time that Ga(III) targets bacterial transcription to inhibit cell growth, which defines the bacteriostatic feature of Ga(III) against P. aeruginosa (Fig. 6B). Moreover, we identified that the central carbon metabolite acetate could effectively enhance the bacteriostatic activity of Ga(III) through increased uptake and activated the Fe-demanding genes in the phenylalanine degradation pathway (Fig. 6B). The combination of acetate and Ga(III) enabled eradication of P. aeruginosa persister cells, and the increased antimicrobial activity of the co-therapy was further demonstrated in mammalian cell and murine infection models. Our results provide novel insights into the modes of action of Ga(III), as well as strategies for the development of efficient Ga-based therapies.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21601209 and 21671203), the Natural Science Foundation of Guangdong Province (2017A030313063), the Research Grants Council of Hong Kong (17333616P and 17307017P), the Fundamental Research Funds for the Central Universities, a starting fund from Sun Yat-sen University, and the University of Hong Kong. We thank Dr Zifeng Wang for the suggestions on transcription assay design; Dr Minji Wang for the information on transposon mutant P. aeruginosa strains; Dr Hui Zhang and Wenhua Lu for the assistance in the Seahorse Experiment.Notes and references
- R. Wang, T.-P. Lai, P. Gao, H. Zhang, P.-L. Ho, P. C.-Y. Woo, G. Ma, R. Y.-T. Kao, H. Li and H. Sun, Nat. Commun., 2018, 9, 439, DOI:10.1038/s41467-018-02828-6.
- Y. Wang, L. Hu, F. Xu, Q. Quan, Y.-T. Lai, W. Xia, Y. Yang, Y.-Y. Chang, X. Yang, Z. Chai, J. Wang, I. K. Chu, H. Li and H. Sun, Chem. Sci., 2017, 8, 4626–4633 RSC.
- J. R. Morones-Ramirez, J. A. Winkler, C. S. Spina and J. J. Collins, Sci. Transl. Med., 2013, 5, 190ra181, DOI:10.1126/scitranslmed.3006276.
- M. B. Harbut, C. Vilcheze, X. Luo, M. E. Hensler, H. Guo, B. Yang, A. K. Chatterjee, V. Nizet, W. R. Jacobs Jr, P. G. Schultz and F. Wang, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 4453–4458 CrossRef CAS PubMed.
- C. H. Goss, Y. Kaneko, L. Khuu, G. D. Anderson, S. Ravishankar, M. L. Aitken, N. Lechtzin, G. Zhou, D. M. Czyz, K. McLean, O. Olakanmi, H. A. Shuman, M. Teresi, E. Wilhelm, E. Caldwell, S. J. Salipante, D. B. Hornick, R. J. Siehnel, L. Becker, B. E. Britigan and P. K. Singh, Sci. Transl. Med., 2018, 10, eaat7520, DOI:10.1126/scitranslmed.aat7520.
- J. A. Lemire, J. J. Harrison and R. J. Turner, Nat. Rev. Microbiol., 2013, 11, 371–384 CrossRef CAS PubMed.
- T. S. Cohen and A. Prince, Nat. Med., 2012, 18, 509–519 CrossRef CAS PubMed.
- M. W. Azam and A. U. Khan, Drug Discovery Today, 2019, 24, 350–359 CrossRef CAS PubMed.
- K. Poole, Front. Microbiol., 2011, 2, 65, DOI:10.3389/fmicb.2011.00065.
- C. Bonchi, F. Imperi, F. Minandri, P. Visca and E. Frangipani, BioFactors, 2014, 40, 303–312 CrossRef CAS PubMed.
- S. Hijazi, P. Visca and E. Frangipani, Front. Cell. Infect. Microbiol., 2017, 7, 12, DOI:10.3389/fcimb.2017.00012.
- E. Banin, A. Lozinski, K. M. Brady, E. Berenshtein, P. W. Butterfield, M. Moshe, M. Chevion, E. P. Greenberg and E. Banin, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 16761–16766 CrossRef CAS PubMed.
- C. R. Chitambar, Biochim. Biophys. Acta, 2016, 1863, 2044–2053 CrossRef CAS PubMed.
- F. Minandri, C. Bonchi, E. Frangipani, F. Imperi and P. Visca, Future Microbiol., 2014, 9, 379–397 CrossRef CAS PubMed.
- Y. Kaneko, M. Thoendel, O. Olakanmi, B. E. Britigan and P. K. Singh, J. Clin. Invest., 2007, 117, 877–888 CrossRef CAS PubMed.
- K. R. Allison, M. P. Brynildsen and J. J. Collins, Nature, 2011, 473, 216–220 CrossRef CAS PubMed.
- B. Peng, Y. B. Su, H. Li, Y. Han, C. Guo, Y. M. Tian and X. X. Peng, Cell Metab., 2015, 21, 249–261 CrossRef CAS PubMed.
- S. Meylan, C. B. Porter, J. H. Yang, P. Belenky, A. Gutierrez, M. A. Lobritz, J. Park, S. H. Kim, S. M. Moskowitz and J. J. Collins, Cell Chem. Biol., 2017, 24, 195–206 CrossRef CAS PubMed.
- B. Peng, H. Li and X. X. Peng, Protein Cell, 2015, 6, 628–637 CrossRef CAS PubMed.
- S. Meylan, I. W. Andrews and J. J. Collins, Cell, 2018, 172, 1228–1238 CrossRef CAS PubMed.
- Y.-T. Lai, Y.-Y. Chang, L. Hu, Y. Yang, A. Chao, Z.-Y. Du, J. A. Tanner, M.-L. Chye, C. Qian, K.-M. Ng, H. Li and H. Sun, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 2948–2953 CrossRef CAS PubMed.
- Y. T. Lai, Y. Yang, L. Hu, T. Cheng, Y. Y. Chang, M. Koohi-Moghadam, Y. Wang, J. Xia, J. Wang, H. Li and H. Sun, Metallomics, 2017, 9, 38–47 RSC.
- D. M. Molina, R. Jafari, M. Ignatushchenko, T. Seki, E. A. Larsson, C. Dan, L. Sreekumar, Y. Cao and P. Nordlund, Science, 2013, 341, 84–87 CrossRef CAS PubMed.
- S. Borukhov and E. Nudler, Trends Microbiol., 2008, 16, 126–134 CrossRef CAS PubMed.
- B. Alqarni, B. Colley, J. Klebensberger, D. McDougald and S. A. Rice, J. Microbiol. Methods, 2016, 127, 182–187 CrossRef CAS PubMed.
- M. A. Lobritz, P. Belenky, C. B. Porter, A. Gutierrez, J. H. Yang, E. G. Schwarz, D. J. Dwyer, A. S. Khalil and J. J. Collins, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 8173–8180 CrossRef CAS PubMed.
- Y. B. Su, B. Peng, H. Li, Z. X. Cheng, T. T. Zhang, J. X. Zhu, D. Li, M. Y. Li, J. Z. Ye, C. C. Du, S. Zhang, X. L. Zhao, M. J. Yang and X. X. Peng, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E1578–E1587 CrossRef CAS PubMed.
- R. A. Fisher, B. Gollan and S. Helaine, Nat. Rev. Microbiol., 2017, 15, 453–464 CrossRef CAS PubMed.
- K. Hiller, C. M. Metallo, J. K. Kelleher and G. Stephanopoulos, Anal. Chem., 2010, 82, 6621–6628 CrossRef CAS PubMed.
- C. The UniProt, Nucleic Acids Res., 2017, 45, D158–D169 CrossRef PubMed.
- M. Guo, I. Harvey, W. Yang, L. Coghill, D. J. Campopiano, J. A. Parkinson, R. T. MacGillivray, W. R. Harris and P. J. Sadler, J. Biol. Chem., 2003, 278, 2490–2502 CrossRef CAS PubMed.
- R. Garcia-Contreras, E. Lira-Silva, R. Jasso-Chavez, I. L. Hernandez-Gonzalez, T. Maeda, T. Hashimoto, F. C. Boogerd, L. Sheng, T. K. Wood and R. Moreno-Sanchez, Int. J. Med. Microbiol., 2013, 303, 574–582 CrossRef CAS PubMed.
- H. G. Floss and T.-W. Yu, Chem. Rev., 2005, 105, 621–632 CrossRef CAS PubMed.
- M. N. Wojtas, M. Mogni, O. Millet, S. D. Bell and N. G. A. Abrescia, Nucleic Acids Res., 2012, 40, 9941–9952 CrossRef CAS PubMed.
- C. Ma, X. Yang and P. J. Lewis, Microbiol. Mol. Biol. Rev., 2016, 80, 139–160 CrossRef CAS PubMed.
- S. Donati, T. Sander and H. Link, Wiley Interdiscip. Rev.: Syst. Biol. Med., 2018, 10, e1396, DOI:10.1002/wsbm.1396.
- N. Wald-Dickler, P. Holtom and B. Spellberg, Clin. Infect. Dis., 2018, 66, 1470–1474 CrossRef CAS PubMed.
- R. Geiger, J. C. Rieckmann, T. Wolf, C. Basso, Y. Feng, T. Fuhrer, M. Kogadeeva, P. Picotti, F. Meissner, M. Mann, N. Zamboni, F. Sallusto and A. Lanzavecchia, Cell, 2016, 167, 829–842 CrossRef CAS PubMed.
- B. S. Nagoba, S. P. Selkar, B. J. Wadher and R. C. Gandhi, J. Infect. Public Health, 2013, 6, 410–415 CrossRef CAS PubMed.
- V. L. Madhusudhan, Int. Wound J., 2016, 13, 1129–1136 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental procedures, supplementary figures and omics data. See DOI: 10.1039/c9sc01480b |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |