Open Access Article
Open Access ArticleOn the encapsulation and assembly of anticancer drugs in a cooperative fashion†
Weikun
Wang
a,
Han
Wang
b,
Lei
Zhiquan
a,
Han
Xie
a,
Honggang
Cui
 b and
Jovica D.
Badjić
b and
Jovica D.
Badjić
 *a
*a
aDepartment of Chemistry & Biochemistry, The Ohio State University, 100 West 18th Avenue, Columbus, OH 43210, USA
bDepartment of Chemical and Biomolecular Engineering, The Johns Hopkins University, Maryland Hall 221, 3400 North Charles Street, Baltimore, MD 21218, USA
First published on 8th May 2019
Abstract
In this study, we report the remarkable recognition and assembly characteristics of D3h symmetric basket 16− containing two adjoining and nonpolar cavities with six biocompatible GABA residues at their northern and southern termini. From the results of experimental (1H NMR, fluorescence and UV-Vis spectroscopies) and computational (MM-MC/OPLS3e) investigations, we deduced that hexaanionic 16− captured two molecules of anticancer drug doxorubicin 2+ in water and accommodated them in its two deep cavities. The formation of stable 16−⊂222+ (Ka = 3 × 1012 M−2) was accompanied by the exceptional homotopic cooperativity (α = 4K2/K1 = 112) in which K1 = 3.2 ± 0.8 × 105 M−1 and K2 = 9 ± 1 × 106 M−1. Furthermore, bolaamphiphilic 16−⊂222+ assembled into spherical nanoparticles (DLS, cryo-TEM and TEM) possessing 41% drug loading. The preorganization of abiotic receptor 16− and its complementarity to 2+ have been proposed to play a part in the positive cooperativity in which ten favorable noncovalent contacts (i.e. hydrogen bonds, salt bridges, C–H⋯π and π–π contacts) are formed between doxorubicin and the dual-cavity host. In the case of topotecan 3+, however, the absence of multiple and favorable basket⊂drug interactions resulted in the predominant formation of a binary 16− ⊂ 3+ complex (K1 = 2.12 ± 0.01 × 104 M−1) and the negative homotopic allostery (α ≪ 1). To summarize, our study lays out a roadmap for creating a family of novel, accessible and multivalent hosts capable of complexing anticancer agents in a cooperative manner. As basket⊂drug complexes organize into highly loaded nanoparticles, the reported soft material is amenable to the bottom-up construction of stimuli-responsive nanomedicine capable of effective scavenging and/or delivery of drugs.
Introduction
Nanosystems carrying anticancer drugs, i.e. nanomedicine,1 hold great promise in the area of targeted chemotherapy for selective detection and extermination of cancer cells.1a,2 In particular, drug-delivery nanoparticles3 extravasate from abnormal blood vessels into the tumor microenvironment (EPR effect),4 which with their surface modification could improve the therapy.3,5,6 Besides, stimuli-responsive and biocompatible7 cyclodextrins,8 calixarenes9 and cucurbiturils10 are also capable of transporting/releasing anticancer drugs.11 As a result of the inclusion complexation,12 abiotic hosts are expected to improve the therapy by allowing spatiotemporal control13 of drug release and also assisting with (a) loading capacity,14 (b) solubility,15 (c) bioavailability16 and (d) stability of drugs.17 In addition to such virtues, allosteric and chelate cooperativity18 operating in the host–guest complexation of pharmaceuticals is expected to give rise to an amplified response so that small changes in the external signal (i.e. stimulus) trigger a large non-linear outcome.19 The on/off mode20 of delivery is, however, challenging to attain21 yet a sought-after strategy to achieve minimal release of the payload before reaching its targeted site followed by a burst in drug release.22Recently, we reported about dual-cavity baskets comprising six23 (S) alanine residues at their periphery (Fig. 1) and acting in an allosteric manner24 by trapping small molecules akin to nerve agents.25 As these complexes assembled into vesicles,25b we became increasingly curious about examining the capacity of deeper dual-cavity 16− (Fig. 1A) for including anticancer drugs doxorubicin 2+ (DOX) and topotecan 3+ in its two adjoining cavities. At first, the inner space of basket 16− seemed complementary26 to 2+ and 3+ in size, shape and polarity (Fig. 1A). Moreover, structurally unique 16− possesses six biocompatible GABA (γ-aminobutyric acid) residues at its southern and northern termini for acting as “sticky” carboxylate fingers capable of grabbing functional drug molecules and holding them in two aromatic pockets.27 The results of our experimental and computational studies have revealed a fascinating way by which dual-cavity 16− encapsulates two molecules of doxorubicin 2+ with exceptional homotopic cooperativity of α = 4K2/K1 = 112!18,28 Moreover, stable ternary 16− ⊂ 222+ complexes (Ka = 1012 M−2) assembled into spherical nanoparticles possessing a high (41%) drug loading.29
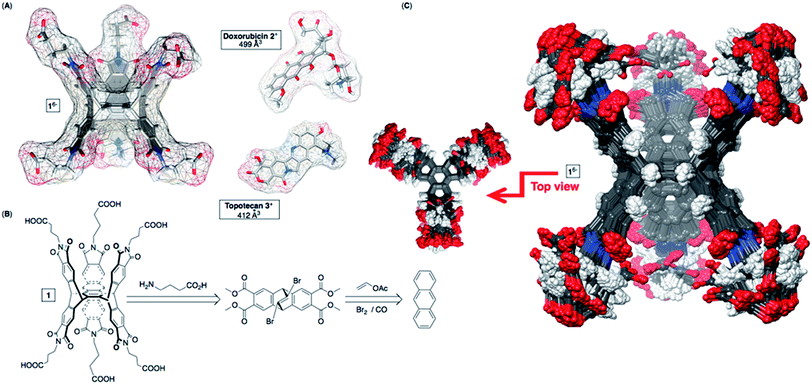 | ||
| Fig. 1 (A) van der Waals surfaces of energy-minimized basket 16−, doxorubicin 2+ and topotecan 3+ (OPLS3e, Maestro). (B) Basket 16− was prepared from anthracene, vinyl acetate and GABA (Scheme S1†). (C) Top (left) and side (right) views of the aligned conformers of 16− (<1.5 kcal mol−1) obtained from the Monte-Carlo conformational search (OPLS3e, Maestro) in implicit water. | ||
Results and discussion
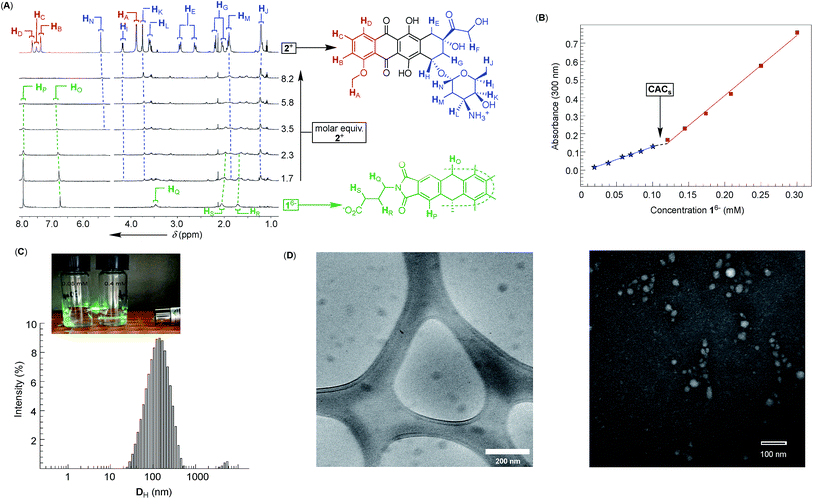
Dual-cavity 1 (Fig. 1B) was prepared from inexpensive anthracene, vinyl acetate and GABA by following a recently developed procedure (Scheme S1†).30 After basket 1 was dissolved in 10 mM phosphate buffer (PBS) at pH = 7.0, its 1H NMR spectrum showed a set of five, somewhat broad, resonances corresponding to, on average, D3h symmetric 16− (Fig. 2A); note that for six remote carboxylates, we assumed pKa < 5.31 In line with 1H NMR results, the Monte-Carlo conformational sampling of 16−, in implicit water solvent (OPLS3e, Maestro), revealed numerous conformers (Fig. 1C) populating the equilibrium (<1.5 kcal mol−1). In solution, these conformers were likely to exchange at a high rate to give the observed 1H NMR spectrum. Furthermore, six carboxylates cluster at the outer side of 16− to make the binding pockets deep, nonpolar and suitable for accommodating nonpolar segments of doxorubicin 2+ and topotecan 3+ (Fig. 1A). The phthalimide side arms are “bouncing” back and forth to alter the cavity size,32 which is central to the induced-fit mode of complexation of the first molecule of drug, as discussed in the text below.33Variously concentrated solutions of 16− exhibited a sharp change in the UV-Vis extinction coefficient (ε300, i.e. slopes in Fig. 2B) of this chromophore at ∼0.12 mM.21 The “segmental adherence to the Lambert–Beer law suggested that the basket transitioned from the monomeric to the aggregated state at 0.12 mM thereby depicting its critical aggregation concentration (CAC).25b,34 Indeed, the results of dynamic light scattering measurements of 0.76 mM 16− (DLS, Fig. 2C) were in line with the formation of nanosized particles, while the Tyndall effect was apparent for 0.4 mM but absent for the 0.05 mM solution of 16− (Fig. 2C). At last, cryo-TEM and TEM imaging (Fig. 2D) revealed the formation of circa 40–50 nm spherical objects possessing uniform coloration with no distinguishable features (i.e. double layer). Accordingly, bolaamphiphilic 16− is likely populating the interior of the spherical aggregates with a packing mode that still needs to be elucidated.34a
After an incremental addition of doxorubicin 2+ to basket 16− (Fig. 2A), there followed a broadening of 1H NMR resonances from the host while signals corresponding to the drug's nuclei became ill defined. In particular, HA–HD signals from the doxorubicin's methoxybenzene (shown in red, Fig. 2A) were difficult to resolve, as they were likely becoming magnetically shielded and broadening into the baseline. Alternatively, HE–HN signals from the doxorubicin's non-aromatic side (shown in blue, Fig. 2A) were easier to identify by comparing the spectra. From the NMR results, we hypothesized that doxorubicin 2+ formed a stable host–guest complex with 16− to result in the complexation dynamics occurring at an intermediate rate and/or rapid T2 relaxation of HA–HD protons 2+ inside the cavity(ies) of 16−.35 As the magnetic environment and dynamics of HE–HN were perturbed to a lesser degree than HA–HD protons, however, these nuclei stayed away from the concave aromatic pockets of 16−. To further quantify the inclusion complexation, we noted that incremental addition of basket 16− to 2+ (Fig. 3A) quenched the emission from doxorubicin;36 the quenching was static,37 since in the absence of the complex formation the emission from the drug remained steady (Fig. S7†). The binding isotherm (Fig. 3B; see also Fig. S8† and www.supramolecular.org) fits well to the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 complexation mode28 with a random distribution of residuals38 and K1 = 3.2 ± 0.8 × 105 M−1/K2 = 9 ± 1 × 106 M−1. The method of continuous variation (UV-Vis, Fig. 3C) was also in line with the formation of ternary 16−⊂222+ with the parabolic function peaking at [2+]/([16−] + [2+]) ∼ 0.67.38 The positive cooperativity of α = 4K2/K1 = 112 is large and quite unusual for an artificial host.21,39 We therefore suspected that the formation of ternary 16−⊂222+ could be accompanied by an aggregation of these bolaamphiphilic complexes so that the observed cooperativity is in part arising from the chelate (multivalent) cooperativity.18 In support of such logic, the observed broadening of 1H NMR signals of both 16− and 2+ in Fig. 2A could have, in part, resulted from their aggregation. First, DLS measurements of 16− ⊂ 222+ showed the presence of circa 60–240 nm particles (Fig. 3D) having electrokinetic (zeta) potential of ζ = −33.6 (Fig. S9†) and therefore moderate stability.25b Likewise, TEM (Fig. 3E) and cryo-TEM imaging (Fig. 3F) revealed the formation of nanoparticles whose size (circa 50 nm), shape and, perhaps, assembly mode were akin to those forming from 16−. As nanoparticles are composed of dual-cavity hosts (Mw = 1536) with each holding two molecules of doxorubicin (Mw = 2 × 544 = 1088), it follows that the material's loading is circa 41%, which is highly desirable13 for nanomedicine.1a
2 complexation mode28 with a random distribution of residuals38 and K1 = 3.2 ± 0.8 × 105 M−1/K2 = 9 ± 1 × 106 M−1. The method of continuous variation (UV-Vis, Fig. 3C) was also in line with the formation of ternary 16−⊂222+ with the parabolic function peaking at [2+]/([16−] + [2+]) ∼ 0.67.38 The positive cooperativity of α = 4K2/K1 = 112 is large and quite unusual for an artificial host.21,39 We therefore suspected that the formation of ternary 16−⊂222+ could be accompanied by an aggregation of these bolaamphiphilic complexes so that the observed cooperativity is in part arising from the chelate (multivalent) cooperativity.18 In support of such logic, the observed broadening of 1H NMR signals of both 16− and 2+ in Fig. 2A could have, in part, resulted from their aggregation. First, DLS measurements of 16− ⊂ 222+ showed the presence of circa 60–240 nm particles (Fig. 3D) having electrokinetic (zeta) potential of ζ = −33.6 (Fig. S9†) and therefore moderate stability.25b Likewise, TEM (Fig. 3E) and cryo-TEM imaging (Fig. 3F) revealed the formation of nanoparticles whose size (circa 50 nm), shape and, perhaps, assembly mode were akin to those forming from 16−. As nanoparticles are composed of dual-cavity hosts (Mw = 1536) with each holding two molecules of doxorubicin (Mw = 2 × 544 = 1088), it follows that the material's loading is circa 41%, which is highly desirable13 for nanomedicine.1a
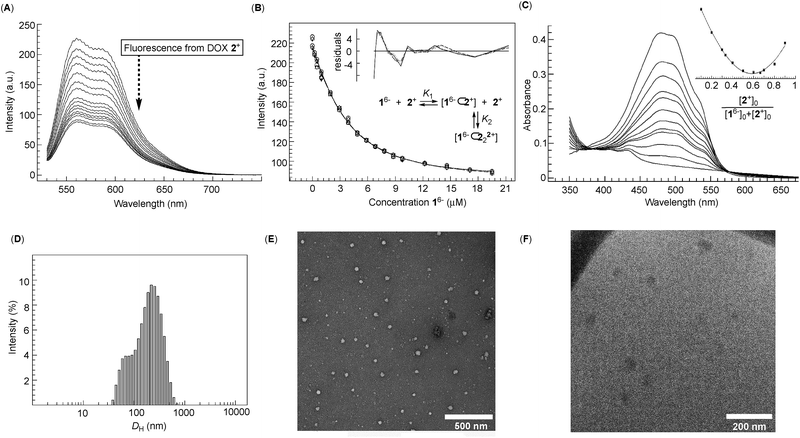 | ||
Fig. 3 (A) An incremental addition of basket 16− to a 1.0 μM solution of doxorubicin 2+ (10 mM PBS at pH = 7.0) was monitored with fluorescence spectroscopy (λex = 500 nm; 298 K). (B) A change in the emission intensity of 2+ as a function of the increasing concentrations of 16− was subjected to global (550–590 nm) nonlinear regression analysis using a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometric model (note that doxorubicin 2+ is in this experimental setup virtually acting as a host with the stoichiometry depicted as 2 1 stoichiometric model (note that doxorubicin 2+ is in this experimental setup virtually acting as a host with the stoichiometry depicted as 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1); the reported K1 = 3.2 ± 0.8 × 105 M−1 and K2 = 9 ± 1 × 106 M−1 represent the arithmetic mean and standard deviation from two independent measurements (Fig. S8†); a random distribution of residuals is shown in the inset. (C) UV-Vis spectra of 0.04 mM doxorubicin 2+ (10 mM PBS at pH = 7.0) in the presence of different proportions of 16− with the Job plot (inset) obtained using the absorbance at 500 nm; for the y axis: (Aobs − A0)[2+]0. (D) The size distribution of particles in the solution of 16− (0.05 mM) and 2+ (0.1 mM) (10 mM PBS at pH = 7.0) was obtained from DLS measurement at 298.0 K. (E) TEM image of a 10 mM PBS solution of 16− (0.1 mM) and 2+ (0.2 mM) at pH = 7.0 deposited on a copper grid and stained with uranyl acetate. (F) Cryo-TEM image of a 0.1 mM solution of basket 16− containing 2+ (0.2 mM) in 10 mM PBS at pH = 7.0. 1); the reported K1 = 3.2 ± 0.8 × 105 M−1 and K2 = 9 ± 1 × 106 M−1 represent the arithmetic mean and standard deviation from two independent measurements (Fig. S8†); a random distribution of residuals is shown in the inset. (C) UV-Vis spectra of 0.04 mM doxorubicin 2+ (10 mM PBS at pH = 7.0) in the presence of different proportions of 16− with the Job plot (inset) obtained using the absorbance at 500 nm; for the y axis: (Aobs − A0)[2+]0. (D) The size distribution of particles in the solution of 16− (0.05 mM) and 2+ (0.1 mM) (10 mM PBS at pH = 7.0) was obtained from DLS measurement at 298.0 K. (E) TEM image of a 10 mM PBS solution of 16− (0.1 mM) and 2+ (0.2 mM) at pH = 7.0 deposited on a copper grid and stained with uranyl acetate. (F) Cryo-TEM image of a 0.1 mM solution of basket 16− containing 2+ (0.2 mM) in 10 mM PBS at pH = 7.0. | ||
Encouraged by the results with DOX 2+, we turned to examine the entrapment of another therapeutic anticancer drug, topotecan 3+ (Fig. 4). This flat molecule is in size and shape similar to doxorubicin (Fig. 1A) therefore making a good candidate for occupying the binding pockets of 16−. Upon an incremental addition of a standard solution of 3+ to 16−, there were small but steady magnetic perturbations of 1H NMR resonances from the basket (green in Fig. 4A). In particular, a greater proportion of 3+ resulted in the HL singlet from D3h symmetric 16− turning into a doublet. The observation was taken as a sign depicting the predominant formation of pseudo C3v symmetric 16−⊂3+ in which a single cavity of the host is holding the drug. Furthermore, HA/B/C and HH signals from topotecan 3+ (red in Fig. 4A) experienced a greater magnetic shielding (Δδ ∼ 0.3–0.4 ppm, red in Fig. 4A) than its other nuclei to denote the lactone but also dimethyl ammonium portions of the drug residing in the host's aromatic cavity. The method of continuous variation (Fig. 4B) indicated the formation of a binary 16−⊂3+ complex, while the fluorescence binding isotherm (Fig. 4C; see also Fig. S10†) fits well to the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometric model with a random distribution of residuals (K1 = 2.12 ± 0.01 × 104 M−1);38 note that the fluorescence of topotecan 3+ was, as in the case of doxorubicin, quenched by the basket. At last, the results from DLS (Fig. S11†), TEM (Fig. S12†) and cryo-TEM (Fig. S13†) measurements were consistent with 16− ⊂ 3+ assembling into circa 35 nm nanoparticles with the morphology akin to 16− and 16−⊂222+ nanoparticles.
1 stoichiometric model with a random distribution of residuals (K1 = 2.12 ± 0.01 × 104 M−1);38 note that the fluorescence of topotecan 3+ was, as in the case of doxorubicin, quenched by the basket. At last, the results from DLS (Fig. S11†), TEM (Fig. S12†) and cryo-TEM (Fig. S13†) measurements were consistent with 16− ⊂ 3+ assembling into circa 35 nm nanoparticles with the morphology akin to 16− and 16−⊂222+ nanoparticles.
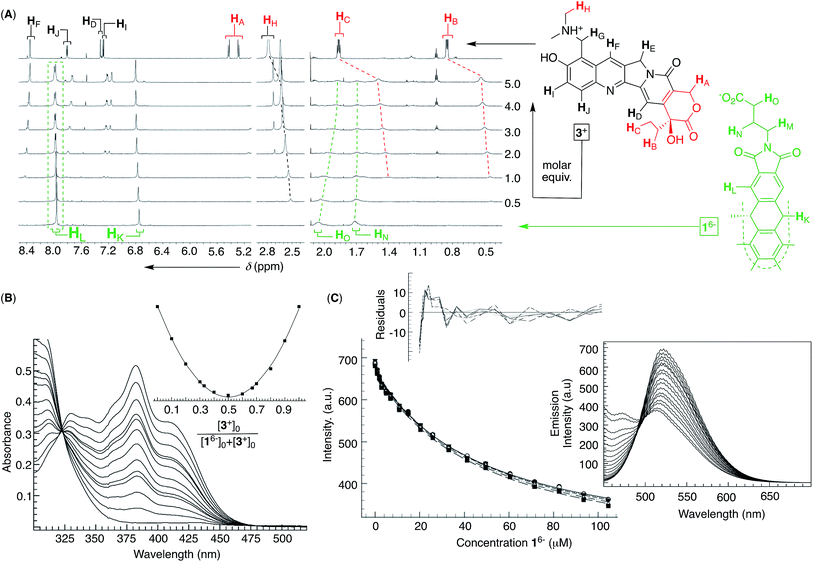 | ||
Fig. 4 (A) 1H NMR spectra (850 MHz, 298 K) depicting an incremental addition of topotecan 3+ (red, top) to 0.1 mM basket 16− (green, bottom) in 10 mM PBS at pH = 7.0. (B) UV-Vis spectra of 0.03 mM topotecan 3+ (10 mM PBS at pH = 7.0) in the presence of different proportions of 16− with the Job plot (inset) obtained using the absorbance at 382 nm; for the y axis: (Aobs − A0)[3+]0. (C) A change in the emission intensity of 3+ (2 μM in 10 mM PBS at pH = 7.0) as a function of the increasing concentrations of 16− was subjected to global (520–540 nm) nonlinear regression analysis using a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometric model to give K1 = 2.12 ± 0.01 × 104 M−1, representing the arithmetic mean and standard deviation from two independent measurements (Fig. S10†); a random distribution of residuals is shown in the inset. (Right) Fluorescence spectra of 2 μM 3+ in the presence of increasing concentrations of 16− (λex = 420 nm; 298 K) in 10 mM PBS at pH = 7.0. 1 stoichiometric model to give K1 = 2.12 ± 0.01 × 104 M−1, representing the arithmetic mean and standard deviation from two independent measurements (Fig. S10†); a random distribution of residuals is shown in the inset. (Right) Fluorescence spectra of 2 μM 3+ in the presence of increasing concentrations of 16− (λex = 420 nm; 298 K) in 10 mM PBS at pH = 7.0. | ||
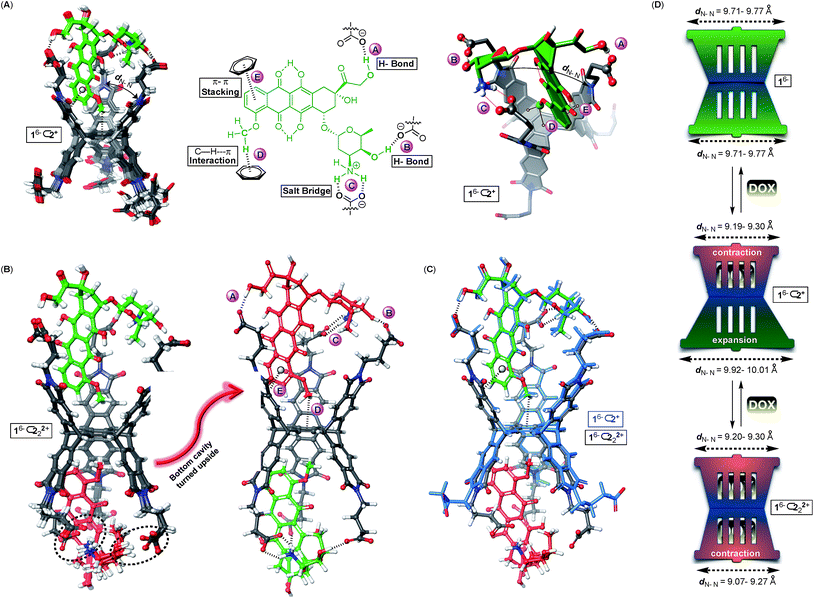
Why is the encapsulation of doxorubicin characterized with positive cooperativity (α = 112) while topotecan prefers to occupy a single pocket of dual-cavity 16− (i.e. negative cooperativity α ≪ 1)?21,24 In order to elucidate the origin of the allostery, we first subjected 16−⊂2+ and 16−⊂222+ complexes to the Monte-Carlo conformational search using the OPLS3e force field40 in implicit water solvent. The global minimum was, in all cases, assessed by running multiple calculations, each starting with the drug assuming a different pose in the basket's cavity (see the ESI†). The conformational sampling of 16−⊂2+ returned numerous yet similar conformers populating the equilibrium (<1.5 kcal mol−1, Fig. 5A): all structures encompass drug 2+ docked within the host's top cavity and the methoxybenzene ring poised to form C–H⋯π (D, Fig. 5A) and π–π stacking (E, Fig. 5A) interactions with the surrounding aromatic box. Moreover, three GABA carboxylates from basket 16− form two hydrogen bonds A/B and salt bridge C with the drug. While the occupied cavity snugs the guest by reducing the average imide dN–N distance to 9.19–9.30 Å (arithmetic mean of three dN–N distances in Fig. 5A), the unoccupied (bottom) cavity responds by flexing25a,41 its phthalimide arms to dN–N = 9.92–10.01 Å (Fig. 5D; for empty 16−, dN–N = 9.71–9.77 Å). For the doubly populated 16− ⊂ 222+, the drug molecule docked in the top cavity preserves all five noncovalent contacts A–E with the cavity 16−⊂222+ (Fig. 5B). Doxorubicin residing in the bottom cavity, though, assumes an equivalent position to that in the top one with three GABA residues forming hydrogen bonds A and B and the salt bridge C to hold it in its place. The transition of singly 16−⊂2+ into doubly populated 16−⊂222+ (Fig. 5C) is accompanied by the top cavity retaining its size dN–N = 9.20–9.30 Å and the bottom one shrinking to dN–N = 9.07–9.27 Å. The second binding event thus necessitates a reduction in the size of the bottom cavity while concurrently maintaining the shape of the top one (Fig. 5D). We reason that such increase in the steric strain (ΔH° > 0) following the second complexation is likely overcompensated by favourable noncovalent A–E contacts (ΔH° ≪ 0).
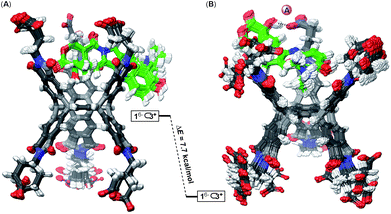
The formation of binary 16−⊂2+ is a favourable process (K1 = 3.2 ± 0.8 × 105 M−1) supported with the notion that the basket's top grips onto the drug by using all three of its phthalimide arms and engaging in five favourable non-covalent interactions (Fig. 5A): the top cavity shrinks to hold the drug, while the bottom one expands; the hydrophobic effect is expected to contribute to this complexation as well.42 With all three of the top cavity's sides being “tied”, the bottom three phthalimides become more rigid25a and thus more preorganized to, via favourable entropy, increase the affinity for the second drug molecule (K2 = 9 ± 1 × 106 M−1). That is to say, the first complexation preorganizes the host for the second one to occur with a minimal loss in entropy, and gain in enthalpy and the overall positive homotopic cooperativity. The role of the assembly of 16− ⊂ 222+ has not been taken into consideration although it is more than likely that the process has an effect on the cooperativity and the mechanism by which the recognition takes place.
A validation for our computational findings comes from the results of 1H NMR spectroscopic experiments (Fig. 2A). The methoxybenzene nuclei from doxorubicin 2+ sustained the greatest degree of magnetic perturbation to implicate it occupying the host's cavities as the theory suggests (Fig. 5).
The Monte-Carlo conformational sampling of 16−⊂3+ (Fig. 6) revealed topotecan docking in the basket's cavity by anchoring its lactone ring (Fig. 6A) or dimethyl ammonium group in the aromatic box (Fig. 6B). The apparent absence of additional noncovalent contacts between 3+ and 16− (except one hydrogen bond A in Fig. 6B) makes this host–guest pair less complementary than the prior one. The basket is thus loosely held by topotecan to make the 16−⊂3+ complex less preorganized for binding another molecule of the drug. Moreover, a weaker complexation could also be alleged to provide insufficient driving force for overcoming the strain required in the second binding event. In support of topotecan 3+ being less complementary to dual-cavity 16− than doxorubicin 2+, we note that binary 16−⊂3+ is an order of magnitude less stable (K1 = 2.12 ± 0.01 × 104 M−1) than 16−⊂2+ (K1 = 3.2 ± 0.8 × 105 M−1). On another note, the results of the NMR titration (Fig. 4A) showed that protons at both termini of topotecan became magnetically shielded to, perhaps, indicate the concurrent existence of both computed poses shown in Fig. 6.
Conclusions
We discovered an abiotic host capable of encapsulating two molecules of anticancer drug doxorubicin in a positive cooperative fashion with ternary drug–host complexes assembling into nanoparticles. The origin of the cooperativity can, in part, be traced back to extraordinary complementarity of the drug to the abiotic receptor in addition to its preorganization. That is to say, at least ten favorable noncovalent contacts are established between doxorubicin and the dual-cavity basket, resembling those found for receptors in the natural world. The basket acts as a “claw grabber” capturing drug molecules with its six “sticky carboxylate fingers” and snugging them within the adjoining aromatic cavities in an induced-fit manner. In the case of topotecan, however, we posited that the paucity of favorable noncovalent contacts between the basket and the drug, i.e. lack of complementarity, played the key role in the observed complexation stoichiometry. In general, our discovery sets the stage for (a) improving our fundamental understanding of cooperativity in artificial systems and (b) creating a family of novel hosts capable of complexing plane-shaped anticancer agents with high affinity and selectivity.Holding a high drug payload (41%),13 nanoparticles release doxorubicin at a lower pH43 (Fig. S14/S15†) or in the presence of spermine44 (Fig. S16†), as signaled by the increase in fluorescence.36b,45 Accordingly, our aims shift to the examination of this soft material for delivery46 as well as scavenging47 of anticancer drugs in biological and other competitive media.22 In fact, with dual-cavity baskets amenable to screening protocols,30 one should be able to optimize their capacity for selective removal48 as well as amplified delivery22 of targeted pharmaceuticals.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported with funds obtained from the Army Research Office (W911NF-17-1-0140). We would like to thank Prof. C. M. Hadad from the Ohio State University, Department of Chemistry and Biochemistry, for useful suggestions and inspiring discussions.Notes and references
- (a) W. C. W. Chan, Acc. Chem. Res., 2017, 50, 627 CrossRef CAS PubMed; (b) F. Caruso, T. Hyeon and V. M. Rotello, Chem. Soc. Rev., 2012, 41, 2537 RSC.
- (a) S. Aftab, A. Shah, A. Nadhman, S. Kurbanoglu, S. Aysil Ozkan, D. D. Dionysiou, S. S. Shukla and T. M. Aminabhavi, Int. J. Pharm., 2018, 540, 132 CrossRef CAS PubMed; (b) R. Awasthi, A. Roseblade, P. M. Hansbro, M. J. Rathbone, K. Dua and M. Bebawy, Curr. Drug Targets, 2018, 19, 1696 CrossRef CAS PubMed; (c) T. J. Anchordoquy, Y. Barenholz, D. Boraschi, M. Chorny, P. Decuzzi, M. A. Dobrovolskaia, Z. S. Farhangrazi, D. Farrell, A. Gabizon, H. Ghandehari, B. Godin, N. M. La-Beck, J. Ljubimova, S. M. Moghimi, L. Pagliaro, J.-H. Park, D. Peer, E. Ruoslahti, N. J. Serkova and D. Simberg, ACS Nano, 2017, 11, 12 CrossRef CAS PubMed; (d) L. Isaacs, Adv. Drug Delivery Rev., 2012, 64, 763 CrossRef CAS; (e) C. M. Hartshorn, M. S. Bradbury, G. M. Lanza, A. E. Nel, J. Rao, A. Z. Wang, U. B. Wiesner, L. Yang and P. Grodzinski, ACS Nano, 2018, 12, 24 CrossRef CAS PubMed.
- S. Wilhelm, A. J. Tavares, Q. Dai, S. Ohta, J. Audet, H. F. Dvorak and W. C. W. Chan, Nat. Rev. Mater., 2016, 1, 16014 CrossRef CAS.
- C. Kinnear, T. L. Moore, L. Rodriguez-Lorenzo, B. Rothen-Rutishauser and A. Petri Fink, Chem. Rev., 2017, 117, 11476 CrossRef CAS PubMed.
- V. P. Torchilin, Handb. Exp. Pharmacol., 2010, 197, 3 CAS.
- R. Mout, F. Moyano Daniel, S. Rana and M. Rotello Vincent, Chem. Soc. Rev., 2012, 41, 2539 RSC.
- Y.-R. Zheng, K. Suntharalingam, T. C. Johnstone and S. J. Lippard, Chem. Sci., 2015, 6, 1189 RSC.
- K. Uekama, F. Hirayama and T. Irie, Chem. Rev., 1998, 98, 2045 CrossRef CAS PubMed.
- M. Giuliani, I. Morbioli, F. Sansone and A. Casnati, Chem. Commun., 2015, 51, 14140 RSC.
- L. Cao, G. Hettiarachchi, V. Briken and L. Isaacs, Angew. Chem., Int. Ed., 2013, 52, 12033 CrossRef CAS PubMed.
- X. Ma and Y. Zhao, Chem. Rev., 2015, 115, 7794 CrossRef CAS PubMed.
- L. Wang, L.-l. Li, Y.-s. Fan and H. Wang, Adv. Mater., 2013, 25, 3888 CrossRef CAS PubMed.
- K. Park, ACS Nano, 2013, 7, 7442 CrossRef CAS PubMed.
- C. Weeden, K. J. Hartlieb and L. Y. Lim, J. Pharm. Pharmacol., 2012, 64, 1403 CrossRef CAS PubMed.
- D. Ma, G. Hettiarachchi, D. Nguyen, B. Zhang, J. B. Wittenberg, P. Y. Zavalij, V. Briken and L. Isaacs, Nat. Chem., 2012, 4, 503 CrossRef CAS PubMed.
- W. Shao, X. Liu, G. Sun, X.-Y. Hu, J.-J. Zhu and L. Wang, Chem. Commun., 2018, 54, 9462 RSC.
- A. Abdul Karim, Q. Dou, Z. Li and X. J. Loh, Chem.–Asian J., 2016, 11, 1300 CrossRef CAS PubMed.
- C. A. Hunter and H. L. Anderson, Angew. Chem., Int. Ed., 2009, 48, 7488 CrossRef CAS PubMed.
- Y. Li, T. Zhao, C. Wang, Z. Lin, G. Huang, J. Gao, C. Wang and D. Sumer Baran, Nat. Commun., 2016, 7, 13214 CrossRef PubMed.
- J. A. Riddle, X. Jiang and D. Lee, Analyst, 2008, 133, 417 RSC.
- S. Shinkai, M. Ikeda, A. Sugasaki and M. Takeuchi, Acc. Chem. Res., 2001, 34, 494 CrossRef CAS PubMed.
- Y. Li, Y. Wang, G. Huang and J. Gao, Chem. Rev., 2018, 118, 5359 CrossRef CAS PubMed.
- K. C. Tjandra and P. Thordarson, Bioconjugate Chem., 2019, 30, 503 CrossRef CAS PubMed.
- J. D. Badjic, A. Nelson, S. J. Cantrill, W. B. Turnbull and J. F. Stoddart, Acc. Chem. Res., 2005, 38, 723 CrossRef CAS PubMed.
- (a) S. Chen, M. Yamasaki, S. Polen, J. Gallucci, C. M. Hadad and J. D. Badjic, J. Am. Chem. Soc., 2015, 137, 12276 CrossRef CAS PubMed; (b) S. Chen, L. Wang, S. M. Polen and J. D. Badjic, Chem. Mater., 2016, 28, 8128 CrossRef CAS.
- J. B. Wittenberg and L. Isaacs, Supramol. Chem.: Mol. Nanomater., 2012, 1, 25 CAS.
- K. Hermann, M. Nakhla, J. Gallucci, E. Dalkilic, A. Dastan and J. D. Badjic, Angew. Chem., Int. Ed., 2013, 52, 11313 CrossRef CAS PubMed.
- P. Thordarson, Chem. Soc. Rev., 2011, 40, 1305 RSC.
- S. Mitragotri and P. Stayton, MRS Bull., 2014, 39, 219 CrossRef CAS.
- T. Neal, W. Wang, L. Zhiquan, R. Peng, P. Soni, H. Xie and J. D. Badjic, Chem.–Eur. J., 2019, 25, 1115 CrossRef CAS.
- L. Zhiquan, S. Polen, C. M. Hadad, T. V. RajanBabu and J. D. Badjic, J. Am. Chem. Soc., 2016, 138, 8253 CrossRef CAS PubMed.
- Y. Ruan, H. A. Taha, R. J. Yoder, V. Maslak, C. M. Hadad and J. D. Badjic, J. Phys. Chem. B, 2013, 117, 3240 CrossRef CAS PubMed.
- L. Zhiquan, H. Xie, S. E. Border, J. Gallucci, R. Z. Pavlovic and J. D. Badjic, J. Am. Chem. Soc., 2018, 140, 11091 CrossRef CAS PubMed.
- (a) S. E. Border, R. Z. Pavlovic, L. Zhiquan, M. J. Gunther, H. Wang, H. Cui and J. D. Badjic, Chem. Commun., 2019 10.1039/c8cc08938h; (b) L. Wang, T. Neal, S. Chen and J. D. Badjic, Chem.–Eur. J., 2017, 23, 8829 CrossRef CAS PubMed.
- S. Chen, Y. Ruan, J. D. Brown, J. Gallucci, V. Maslak, C. M. Hadad and J. D. Badjic, J. Am. Chem. Soc., 2013, 135, 14964 CrossRef CAS PubMed.
- (a) S. Chen, S. M. Polen, L. Wang, M. Yamasaki, C. M. Hadad and J. D. Badjic, J. Am. Chem. Soc., 2016, 138, 11312 CrossRef CAS PubMed; (b) W.-C. Geng, S. Jia, Z. Zheng, Z. Li, D. Ding and D.-S. Guo, Angew. Chem., Int. Ed., 2019 DOI:10.1002/anie.201813397.
- D.-S. Guo, V. D. Uzunova, X. Su, Y. Liu and W. M. Nau, Chem. Sci., 2011, 2, 1722 RSC.
- F. Ulatowski, K. Dabrowa, T. Balakier and J. Jurczak, J. Org. Chem., 2016, 81, 1746 CrossRef CAS PubMed.
- I. Saha, J. H. Lee, H. Hwang, T. S. Kim and C.-H. Lee, Chem. Commun., 2015, 51, 5679 RSC.
- E. Harder, W. Damm, J. Maple, C. Wu, M. Reboul, J. Y. Xiang, L. Wang, D. Lupyan, M. K. Dahlgren, J. L. Knight, J. W. Kaus, D. S. Cerutti, G. Krilov, W. L. Jorgensen, R. Abel and R. A. Friesner, J. Chem. Theory Comput., 2016, 12, 281 CrossRef CAS PubMed.
- P. Thordarson, E. J. A. Bijsterveld, J. A. A. W. Elemans, P. Kasak, R. J. M. Nolte and A. E. Rowan, J. Am. Chem. Soc., 2003, 125, 1186 CrossRef CAS PubMed.
- L. Zhiquan, S. M. Polen, C. M. Hadad, T. V. RajanBabu and J. D. Badjic, Org. Lett., 2017, 19, 4932 CrossRef CAS PubMed.
- (a) J.-Z. Du, H.-J. Li and J. Wang, Acc. Chem. Res., 2018, 51, 2848 CrossRef CAS PubMed; (b) B. Li, Z. Meng, Q. Li, X. Huang, Z. Kang, H. Dong, J. Chen, J. Sun, Y. Dong, J. Li, X. Jia, J. L. Sessler, Q. Meng and C. Li, Chem. Sci., 2017, 8, 4458 RSC.
- Y. Chen, Z. Huang, H. Zhao, J.-F. Xu, Z. Sun and X. Zhang, ACS Appl. Mater. Interfaces, 2017, 9, 8602 CrossRef CAS PubMed.
- Y. Yuan and B. Liu, Chem. Sci., 2017, 8, 2537 RSC.
- D. Bousmail, L. Amrein, J. J. Fakhoury, H. H. Fakih, J. C. C. Hsu, L. Panasci and H. F. Sleiman, Chem. Sci., 2017, 8, 6218 RSC.
- H. J. Oh, M. S. Aboian, M. Y. J. Yi, J. A. Maslyn, W. S. Loo, X. Jiang, D. Y. Parkinson, M. W. Wilson, T. Moore, C. R. Yee, G. R. Robbins, F. M. Barth, J. M. DeSimone, S. W. Hetts and N. P. Balsara, ACS Cent. Sci., 2019 DOI:10.1021/acscentsci.8b00700.
- J.-C. Leroux, Nat. Nanotechnol., 2007, 2, 679 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: The preparation and characterization of 1 with additional experimental and computational details. See DOI: 10.1039/c9sc01380f |
| This journal is © The Royal Society of Chemistry 2019 |