Open Access Article
Open Access ArticleA H-bond strategy to develop acid-resistant photoswitchable rhodamine spirolactams for super-resolution single-molecule localization microscopy†
Qingkai
Qi
a,
Weijie
Chi
b,
Yuanyuan
Li
c,
Qinglong
Qiao
a,
Jie
Chen
a,
Lu
Miao
a,
Yi
Zhang
a,
Jin
Li
a,
Wei
Ji
c,
Tao
Xu
*c,
Xiaogang
Liu
*b,
Juyoung
Yoon
 *d and
Zhaochao
Xu
*d and
Zhaochao
Xu
 *a
*a
aCAS Key Laboratory of Separation Science for Analytical Chemistry, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China. E-mail: zcxu@dicp.ac.cn
bSingapore University of Technology and Design, 8 Somapah Road, Singapore 487372, Singapore. E-mail: xiaogang_liu@sutd.edu.sg
cNational Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China. E-mail: xutao@ibp.ac.cn
dDepartment of Chemistry and Nano Science, Ewha Womans University, Seoul 120-750, Korea. E-mail: jyoon@ewha.ac.kr
First published on 4th April 2019
Abstract
Rhodamine spirolactam based photoswitches have been extensively applied in super-resolution single-molecule localization microscopy (SMLM). However, the ring-opening reactions of spirolactams are cross-sensitive to acid, limiting their photoswitch use to neutral pH conditions. In addition, the ring-closing reactions of spirolactams are environment-sensitive and slow (up to hours), virtually making rhodamine spirolactams caged fluorescent dyes instead of reversible photoswitches in SMLM. Herein, by introducing hydrogen bonds to stabilize spirolactams, we report a series of acid-resistant rhodamine spirolactams with accelerated ring-closing reactions from fluorescent xanthyliums to non-fluorescent spirolactams, endowing them with good photoswitchable properties even in acidic environments. By further substitution of 6-phenylethynyl naphthalimide on the spirolactam, we shifted the photoactivation wavelength into the visible region (>400 nm). Subsequently, we have successfully applied these dyes in labeling and imaging the cell surface of Bacillus subtilis at pH 4.5 using SMLM.
Introduction
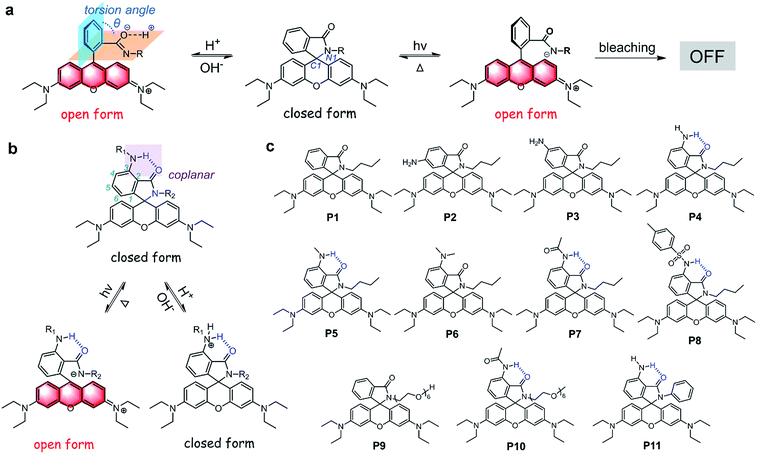
Super-resolution fluorescence microscopy breaks the diffraction limit and achieves nanometre-scale resolution.1 The fundamental principle underlying this technique relies on the “dark” and “fluorescent” pair states of fluorescent markers, enabling temporal separation of adjacent molecules in a subdiffraction-sized region. The requirements for both dark/emitting state-switching and outstanding emission properties (including high brightness and photostability), particularly for super-resolution single-molecule localization microscopy (SMLM),2 have driven the creation of improved photoswitchable fluorescent probes via two means: combining a photochromic moiety with excellent fluorophores3 and incorporating fluorescent fragments in photochromic compounds.4Among various photoswitchable fluorophores, rhodamine spirolactams have attracted considerable research interest, owing to their excellent photophysical properties and bio-compatibility. These dyes were widely applied in constructing various chemosensors for pH, metal ions and bioactive molecules, based on ring-opening reactions from colorless and non-fluorescent spirolactams to colored and strongly fluorescent xanthyliums.5 Since their photoswitching properties were first reported in 1977, rhodamine spirolactams have been used as photochromic materials.6 Recently, owing to dark/emitting state switching, rhodamine spirolactams have also been successfully deployed in SMLM.7 While many exciting super-resolution imaging applications have been demonstrated,8 two significant challenges limit the application of rhodamine spirolactams. First, rhodamine spirolactams are strictly limited to neutral pH environments, due to their considerable acid-activated switching to fluorescent xanthyliums (the open form) across a wide acidic pH range (usually 2.0 < pH < 6.5) (Fig. 1a).5 As these dyes enter an acidic environment (such as in lysosomes (pH 4.5–6.0), endosomes (pH 5.0–6.5), or locations next to proton-donating groups in proteins), acid-activation becomes substantial, leading to the disablement of photoactivation. Yet, low pH or acidic environments are ubiquitous in biological systems. Intracellular acidification is associated with many critical diseases (such as cancer).9 Second, environmental changes greatly alter the lifetime of the open form, which ranges from milliseconds to hours.6,7a,c With a long “open form” lifetime in aqueous solutions, photobleaching reactions could considerably compete with thermal fading ring-closing reactions, compromising the photostability of rhodamine spirolactams.6c,10 Moreover, on the timescale of seconds or longer, the photoswitching reversibility of rhodamine spirolactams is virtually negligible for image acquisition purposes; these dyes effectively behave as typical caged fluorescent compounds: they are activated, imaged, and then bleached (Fig. 1a).7a By introducing a lag-time in between frames to allow relaxation, these dyes could be used as localized markers several times. In a landmark paper, a spontaneously blinking fluorophore based on an intramolecular spirocyclization reaction was reported to increase reversible photoswitching cycles and allow long-term dynamic studies in live cells.10 In a recent study, the photoswitch of rhodamine spirolactam was combined with fluxionality to enable long time-lapse SMLM imaging in live cells.7i However, these fluorophores were still acid sensitive or pH-dependent, limiting their application to neutral pH environments. Consequently, dedicated photoswitchable rhodamine spirolactams with acid immunity and short “open form” lifetime are required for SMLM.
Herein, we report a series of acid-resistant rhodamine spirolactams with a dedicated photoswitchable property and short “open form” lifetime. These characteristics are attributed to the substitution of 3-amino groups in the carboxyphenyl ring and associated intramolecular hydrogen bonding in spirolactams (Fig. 1b). Inspired by Moerner's work,7c we further extended the photoactivation wavelength of our dyes into the visible region (>400 nm), via conjugation with 6-phenylethynyl naphthalimide. The excellent acid-resistance and visible-light photoswitching make our rhodamine spirolactams suitable for SMLM imaging, and more importantly, generate reliable fluorescence signals even in acidic environments.
Results and discussion
Molecular design strategy
Our strategy of introducing intramolecular hydrogen bonding to increase the acid-resistance of spirolactams and shorten the “open form” lifetime originated from three facts: (1) the acid-activated ring-opening reaction is initiated with the protonation of the carbonyl oxygen (Fig. 1a);1 (2) the resulting ring opening reaction is associated with a rotation of the amide group (–CONR) with respect to the phenyl ring (Fig. 1a, S1; Tables S1 and S2†); (3) introducing an electron-donating group into spirolactams reduces the driving force of ring-opening reactions.11 We anticipated that the intramolecular hydrogen bonding would reduce the reactivity of carbonyl oxygen with exterior acid and lock the spirolactam group. In addition, both the suppressed rotation of the amide group and the incorporation of an electron-donating group contributed to the stabilization of the closed form. Accordingly, we synthesized compounds P1–P6 to verify our hypothesis (Fig. 1c and Schemes S1–S3†). Compounds P4 and P5 possessed 3-substituted NHR groups which were expected to form intramolecular hydrogen bonds with the lactam moiety, while the rest served as control compounds without intramolecular hydrogen bonds.Characterization of the acid-resistance of rhodamine spirolactams
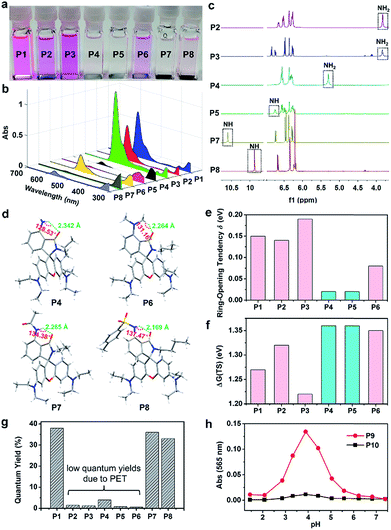
We first examined the acid resistance of P1–P6. With the addition of CF3COOH, a new emission band centered at 560 nm in the absorption spectra of P1–P3 and P6 was observed (Fig. 2a, b, S2 and S3†). Accordingly, the solutions of P1–P3 and P6 displayed noticeable color changes from colorless to pink, indicating the acid-controlled xanthylium formation. In stark contrast, the UV-vis absorption spectra of P4 and P5 did not show obvious changes in the visible region, and the solutions almost remained colorless (Fig. 2a, b and S2†). These results indicated that the fluorescence switching of P4 and P5 was far less acid-sensitive, while that of P1–P3 and P6 was acid-sensitive.Verification of the H-bond mechanism
We next conducted NMR and crystallographic experiments to examine the existence of intramolecular hydrogen bonding (Fig. 2c, d and Table S3†). 1H NMR spectra show large downfield shifts of the resonance of NH protons in P4 (5.30) and P5 (6.75), compared with those in P2 (3.83) and P3 (3.83) (Fig. 2c). The crystal structure of P4 directly revealed strong intramolecular hydrogen bonding between NH and carbonyl oxygen (distance: 2.347 Å; angle: 128.61°) (Fig. 2d), which explained the downfield shift of NH in P4. Though we failed to obtain the crystal structure of P5, we believed that the much larger downfield shift of NH in P5 indicated even stronger intramolecular hydrogen bonding in P5. According to the absorption and emission spectral evolution of P4 and P5 (after the addition of CF3COOH), P5 was more acid-resistant than P4, which should be ascribed to the stronger intramolecular hydrogen bonding (Fig. S2–S4†). In addition, we also noticed weak intramolecular CH⋯O interactions in P6 (distance: 2.249 Å; angle: 130.80°). The above results clearly demonstrate that intramolecular hydrogen bonding plays a key role in enhancing the acid-resistance of rhodamine spirolactams.We also performed density functional theory (DFT) calculations on P1–P6 to rationalize the hydrogen-bond induced acid-resistance. As expected, optimized molecular geometries show that intramolecular hydrogen bonds are present only in P4 and P5 (Fig. S5†). Moreover, by modelling eight representative protonation configurations for each compound to fully simulate their responses in an acidic aqueous environment (Fig. S6†), our results show that among the control compounds, fluorescent open-ring structures possess significantly lower Gibbs free energy than non-emissive closed-ring structures by up to 0.19 eV (Table S4 and Fig. S7–S12†). Therefore, DFT calculations suggest that the emissions of these control compounds could easily turn on in an acidic environment. In contrast, the closed-ring structures of P4 and P5 are relatively more stable than those of P1–P3 and P6 by ∼0.12 eV, suggesting that these structures are resistant to acid-activation. We further calculated the difference in Gibbs free energy between the most stable closed-ring and open-ring protonated structures (δ) to quantify the tendency of ring-opening in P1–P6. Herein, a large δ corresponds to a strong thermodynamic driving force for ring-opening reactions to proceed. Indeed, our results showed that P4 and P5 exhibited a significantly lower ring-opening tendency than P1–P3 and P6 did (Fig. 2e). We also computed the energy barriers during the ring-opening reactions of P1–P6 (Table S5 and Fig. S13–S19†). Our results show that the barriers in P4 and P5 are higher than those of the control compounds, due to the presence of intramolecular hydrogen bonds (Fig. 2f). All these computational results rationalize that P4 and P5 possess significantly enhanced acid-resistance towards fluorescence activation. At the same time, we expected that P4 and P5 remained photoswitchable, given the high energy of photoactivation light.
Improving quantum yields and water solubility of rhodamine spirolactams
Unfortunately, the quantum yields of P2–P6 are low (Fig. 2g), possibly quenched by photo-induced electron transfer (PET) from the 3-aminophenyl moiety to the xanthene scaffold. To enhance brightness and preserve acid-resistance, we synthesized P7 and P8 (Scheme S4†) with 3-acetamido and 3-toluenesulfonamide substituents to inhibit PET, respectively. As a result, the quantum yields of P7 and P8 were significantly improved (Fig. 2g). At the same time, P7 and P8 remained acid-resistant (Fig. 2a and b). 1H NMR analysis and the single crystal structures of P7 and P8 clearly showed the existence of intramolecular hydrogen bonds (Fig. 2c and d). Notably, the stronger intramolecular hydrogen bonding in P7 than that in P8 resulted in stronger acid-resistance of P7 than that of P8 (Fig. S2–S4†). Owing to its high brightness and excellent acid-resistance, we chose P7 for further derivations and applications.Subsequently, we synthesized water-soluble compounds P9 and P10 with polyethylene glycol (PEG) groups to verify the applicability of our strategy in water (Scheme S5†). As shown in Fig. 2h and S20,† the UV-vis absorption and emission intensities of the open form of P9 substantially increased when the pH was decreased from 7.40 to 3.89, and then decreased when the pH was decreased from 3.89 to 1.44. In contrast, P10 did not show any noticeable absorption and fluorescence bands in the visible region in the pH range from 1.44 to 7.40 (Fig. 2h and S20†). These results indicated that our strategy to increase the acid-resistance of rhodamine spirolactams worked well in aqueous solutions and was suitable for biological systems.
Evaluation of open form lifetime
We next evaluated the lifetime of the open forms via laser flash photolysis (Table S6 and Fig. S21†). It's reported that the lifetime of the open form was greatly affected both by the environment and molecular structures.6,7a,c The open form has a lifetime of a few milliseconds in polar solvents.6 In polyvinylalcohol, the lifetime of the open form was reported to lengthen to hours.7a By introducing aromatic substituents on the lactam nitrogen, the open isomer with the charge at the amide nitrogen atom can be stabilized and then it has a longer lifetime.6b,7a,c In our experiments, the lifetime of P1vs.P7 in CH2Cl2 was too short to be examined (Table S6†). But in dioxane, the lifetime was detected to be in the range of 60 ns. Importantly, we found that the intramolecular hydrogen bonding could shorten the lifetime of the open forms (62.5 ns for P1 and 59.7 ns for P7). The effect of intramolecular hydrogen bonding on lifetime becomes more significant when an aromatic substituent is introduced on the lactam nitrogen. Compared with the 200 ms lifetime of N-phenyl-rhodaminelactame in dioxane,6a,b the lifetime of the reference compound P11 was much shorter, measured to be ∼31.5 ns in the same solvent. These results indicated that the thermal fading processes in our dyes may compete with photobleaching.In vitro bioimaging experiments
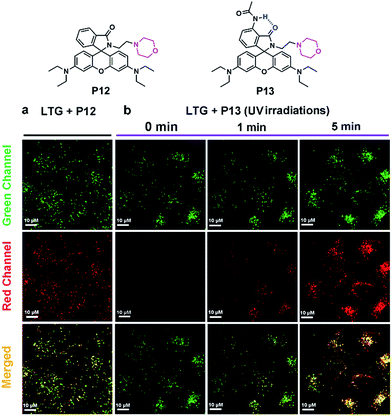
With this progress, we continued to synthesize P12 and P13 to validate our design strategy with in vitro bioimaging experiments (Fig. 3 and Scheme S7†). The “control” probe P12 and the “experimental” probe P13 are coupled with a morpholine moiety, which enables these probes to become protonated and accumulated in lysosomes. The pH value of lysosomes is in the range of 4.0–6.0, which provides a good platform to test the acid-resistance of rhodamine spirolactams. After incubation for 30 min, P12 emitted strong fluorescence in MCF-7 cells (Fig. S22†). Subsequently, we performed co-localization experiments of P12 (the red channel) with LysoTracker Green DND-26 (LTG; the green channel), a commercially available lysosome probe (Fig. 3a). The good overlap of fluorescence signals in the red and green channels demonstrates that P12 was distributed in the lysosomes of MCF-7 cells. Overall, these results show that the acidic environment in lysosomes has activated intensive fluorescence in the control probe P12.In contrast, after incubating MCF-7 cells with P13 for 2 hours, we did not observe any fluorescence signals (Fig. S22†). To ensure the presence of P13 in lysosomes, continuous UV light of 375 nm (∼1 mW cm−2) was applied to activate the fluorescence of P13. By increasing the illumination time, we noticed a gradual increase of fluorescence signals in the red channel (Fig. 3b). These signals overlapped with the fluorescence of the lysosome probe LTG in the green channel. It is thus clear that P13 maintains acid-resistance in lysosomes and possesses a dedicated photoactivation property for cellular imaging. It's worth mentioning that the fluorescent form of P13 in lysosomes did not thermally revert back to the closed form in a short time. Considering the environmental sensitivity of the open form, this is probably because a large number of P13 accumulated in lysosomes, and many xanthyliums formed under continuous strong UV irradiation tended to stabilize each other, thus slowing down the ring-close reactions. The photoactivation experiments of P1 and P7 in CH2Cl2 may support our hypothesis. The open form lifetimes of P1 and P7 in CH2Cl2 were too short to observe a large amount of open isomers. But if we excited these compounds in CH2Cl2 solution with continuous UV irradiation, simultaneously photoactivated xanthyliums may accumulate and then significantly slow down the decay dynamics of P1 and P7, where the lifetime of the open isomers was on the order of min (Fig. S23†).
Photoactivation of rhodamine spirolactams using visible light
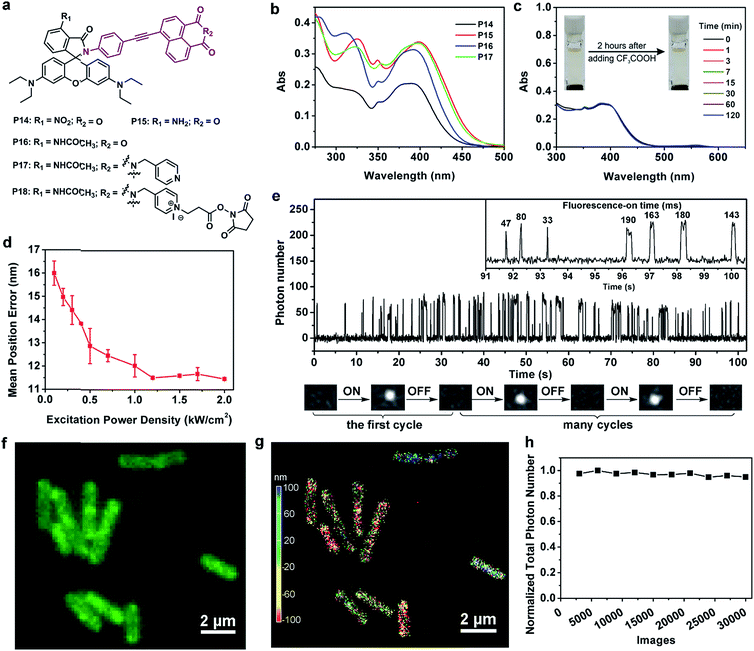
Encouraged by the success of P13 in bioimaging applications, we moved on to extend the photoactivation wavelength of rhodamine spirolactams into the visible region for cell imaging. We thus synthesized P14–P17 by conjugating a 6-phenylethynyl naphthalimide moiety to the spirolactam (Fig. 4a and Scheme S8†). The enlarged π-conjugation extends the UV-vis absorption spectra of P14–P17 into the visible region, with a peak absorption wavelength at ∼400 nm (Fig. 4b). It is thus possible to photoactivate these dyes with a 405 nm laser and read out the fluorescence signals with an excitation laser of 561 nm. The visible photoactivation wavelength is critical for minimizing phototoxicity in live-cell imaging.Among P14–P17, we selected P17 for further experiments, foreseeing its excellent acid-resistance and good quantum yield. Indeed, P17 remained colorless in acidic medium (Fig. 4c). We then doped P17 into a PMMA film at a low concentration to characterize its single-molecule fluorescence properties. We applied a continuous 405 nm laser light (60 W cm−2) to photoactivate P17 (Fig. S24†), and a 561 nm laser light to generate fluorescence from activated P17. Our experiments showed that a laser excitation power of ∼1.2 kW cm−2 at 561 nm led to a localization precision of the single molecule of ∼12 nm in PMMA (Fig. 4d and S25†). Subsequent experiments showed that good imaging quality could be achieved by reducing the photoactivation light intensity at 405 nm down to 1.8 W cm−2 in a cellular environment.
Super-resolution imaging of Bacillus subtilis in acidic environments
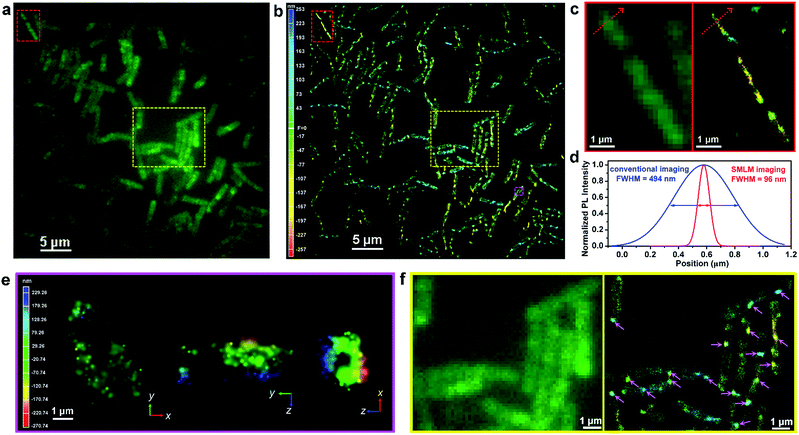
To fully demonstrate the imaging applications of our photoswitchable dyes, we linked N-hydroxysuccinimide ester to P17, resulting in the formation of P18 (Fig. 4a and Scheme S9†). Laser flash photolysis showed that the open form lifetime of P18 was 105 ns in aqueous solutions with 5% DMSO (Table S6 and Fig. S21†). In a single molecule fluorescence imaging experiment with a 561 nm laser (0.36 kW cm−2) as an excitation source, P18 labeled on a glass coverslip showed reversible fluorescence blinking ∼100 times (Fig. 4e). Moreover, in this experiment the fluorescence-on time of P18 increased to ∼100 ms-scale, probably owing to the environmental sensitivity of the open form lifetime. For example, this lifetime of a rhodamine spirolactam increased from milliseconds in polar solvents to hours in polyvinyl alcohol.7a The reversible blinking and good fluorescence signals inspired us to label P18 on the membrane surface of Gram-positive Bacillus subtilis cells in PBS solution (pH 4.5), since these bacteria can maintain satisfactory activity in an acidic pH environment. We expected that P18 could specifically label bacterial membranes for the following reasons: (1) the positively charged P18 could bind with the anionic bacterial membranes through electrostatic interactions, and these interactions could further inhibit the diffusion of P18 through cell membranes; (2) the NHS ester of P18 could rapidly label the amino groups in the membrane proteins; and (3) the hydrophobic 1,8-naphthalimide group may intercalate into the membrane bilayer. We then imaged acid-resistant Bacillus subtilis cells, using a continuous 405 nm laser (1.8 W cm−2) as photoactivation light and a 561 nm laser (1.2 kW cm−2) as an excitation source. The fluorescence of P18 exhibited an excellent photo-blinking property, allowing the reconstruction of the super-resolution image via single-molecule localization. In comparison to a conventional diffraction-limited fluorescence image (Fig. 4f), the 3D-SMLM image of Bacillus subtilis displayed a greatly improved spatial resolution (Fig. 4g). Besides, owing to the low intensity of the 405 nm excitation laser, and slow photobleaching rates of the 561 nm laser (as a result of fast ring-close reactions and a short open-form lifetime), P18 exhibited excellent photostability. The number of collected photons per 3000 frames (20 ms per frame) remained almost constant throughout our experiments of >10 min (Fig. 4h), demonstrating the potential of P18 to perform long time-lapse nanoscopy imaging in future studies.We continued to investigate the staining properties and labeling specificity of P18. As shown in Fig. 5a, b and Movie 1,†P18 stained bacteria evenly with high density, and exhibited excellent photoblinking properties. Interestingly, the divisions of three Bacillus subtilis cells were clearly visible in a 3D-SMLM image (Fig. 5b and c), while they were barely resolved in a diffraction-limited fluorescence image (Fig. 5a and c). The full-width at half-maximum (FWHM) of a single Bacillus subtilis is ∼96 nm in the super-resolution images, cf. ∼494 nm in the conventional fluorescence images (Fig. 5d). Cross-sections of a single Bacillus subtilis highlighted in the pink box show a hollow structure from the xz view, indicating that our probes localized on the cell membranes of bacteria (Fig. 5e). The flipped 3D-SMLM image of bacterial cell membranes is further shown in Movie 2.† We thus demonstrated that P18 enabled SMLM studies of the membranes of bacteria cells at nanometric resolution (without using additional targeting strategies). Moreover, we found that the membranes of most Bacillus subtilis bacteria did not show uniform fluorescence intensities in the 3D-SMLM image shown in Fig. 5b. The slight irregularity of the membrane staining likely indicates the presence of many fluid microdomains (as highlighted by pink arrows in the right image of Fig. 5f). These fluid microdomains had been well studied by several research groups.12 However, these fluid microdomains were hardly resolved in conventional diffraction-limited fluorescence images (in the left image of Fig. 5f). Besides, we also discovered that the majority of these fluid microdomains appeared in certain areas, especially around cell poles.
Conclusions
In summary, this study presents an effective molecular engineering approach for developing acid-resistant rhodamine spirolactams with a dedicated photoswitchable property and short open-form lifetime. These unique characteristics resulted from intramolecular hydrogen bonding between the 3-amino substituents in the carboxyphenyl ring and the lactam moiety. Moreover, we have extended the photoactivation wavelengths of rhodamine spirolactams into the visible region (>400 nm), by conjugating a 6-phenylethynyl naphthalimide moiety to the spirolactams. The visible photoactivation and dedicated photoswitchable properties of our dyes make them highly reliable fluorescent probes for SMLM, as demonstrated by the successful labeling and imaging of Bacillus subtilis at pH 4.5 using 3D-SMLM. We believe that our work sheds light on rational engineering of fluorescent properties of rhodamine dyes to facilitate the advancement of super-resolution imaging techniques.Experimental section
Single-crystal structures
Single crystals of P4, P6, P7 and P8 that were suitable for X-ray structural analysis were obtained by slow evaporation from a chloroform/n-hexane solution at room temperature. Single crystal X-ray diffraction data were collected on an Xcalibur, Atlas, Gemini ultra-diffractometer using the ω-scan mode with a graphite-monochromator and Mo Kα radiation. The crystal structures were solved with the Superflip13 structure solution program and refined with the SHELXL14 refinement package, as implemented in OLEX2.15 All H atoms were placed geometrically and treated with riding constraints, and displacement parameters were derived from the C atoms to which they were attached. All CH2 groups had Uiso(H) values fixed at 1.2 times the Ueq value of the attached C atom. CH3 groups were idealized as freely rotating groups, with Uiso(H) values fixed at 1.5 times the Ueq value of the attached C atom. H atoms on N atoms were located via Fourier difference map inspection and positionally refined isotropically with thermal parameters based upon the N atom to which they are bonded (Uiso(H) = 1.2Ueq(N)). These data can be obtained free of charge from the Cambridge Crystallographic Data Centre (CCDC reference numbers: 1582847, 1901370–1901372†).Measurement of single molecule optical properties
P17 was immobilized in polymer films to investigate the effects of the excitation laser power density on (1) the photon output from dyes, (2) background noise, and (3) localization precision. Polymer films were prepared using 1% (by mass) solution of polymethyl methacrylate (PMMA) in toluene. The polymer films were doped with P17 (10 nM) and then spin-cast onto argon-plasma-etched coverslips. After that, the coverslips were placed in the sample holder of an SMLM instrument for imaging. The intensity of the 405 nm photoactivation laser was adjusted to activate only a few molecules in each frame. The fluorescence images of the polymer film were acquired using an electron-multiplying charge-coupled device (EMCCD) camera at room temperature.Sample preparation and data analysis for single-molecule fluorescence imaging experiment
Coverslips were cleaned by sonication for 10 min in each of the following: Milli-Q water, absolute ethyl alcohol, and acetone. The above coverslips were further boiled in a solution mixture of concentrated sulfuric acid and hydrogen peroxide (v/v, 7/3) for 1 h and then washed with Milli-Q water and dried using nitrogen. 20 μL of very dilute P18 (10 nM) in DMSO solution was dripped on the dry coverslips. Then, P18 with the N-hydroxysuccinimide ester adsorbed and reacted with the hydroxyl groups on the surface of the cleaned coverslip at a low density such that individual dye molecules could be clearly resolved. Then the surface of the cleaned coverslip was rinsed with PBS (pH 7.4) to remove unbound P18 before imaging. Finally, the coverslip was placed on the sample holder of the SMLM instrument and illuminated with 561 nm laser light (0.36 kW cm−2). 5000–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 frames were recorded at a frame rate of 20 ms per frame. Single-molecule fluorescence time trace was calculated for a single molecule by analyzing the total integrated signal in a 2 × 2 pixel region centered on the single molecule during each frame of the movie. The number of photons was calculated as the total intensity count minus the background intensity count, which was converted to photon number using the calibrated parameter for electron multiplication and analog-to-digital conversion gain settings used during acquisition.
000 frames were recorded at a frame rate of 20 ms per frame. Single-molecule fluorescence time trace was calculated for a single molecule by analyzing the total integrated signal in a 2 × 2 pixel region centered on the single molecule during each frame of the movie. The number of photons was calculated as the total intensity count minus the background intensity count, which was converted to photon number using the calibrated parameter for electron multiplication and analog-to-digital conversion gain settings used during acquisition.
Cell culture and CLSM imaging
MCF-7 cells were grown on DMEM (Gibco) supplemented with 10% FBS (Gibco). In CLSM imaging, the MCF-7 cells were incubated with 1 mL of DMEM containing 10 μM P11 (experimental group) or P12 (control group) and the images were collected at 0, 30, 60 and 120 min by using an ANDORTM live cell imaging system. Then, the adherent cells were further incubated with 1 mL of fresh DMEM containing LysoTracker Green DND-26 (LTG) (0.1 μM) for another 30 min. The adherent cells were washed 3 times with PBS (pH 7.4) and utilized for CLSM imaging. In the experimental group, the CLSM images of MCF-7 cells were taken under 0–5 min continuous UV (375 nm) irradiation at 37 °C.Live bacteria sample preparation and labeling
A single colony of Bacillus subtilis was inoculated in 5 mL of Luria–Bertani (LB) medium with shaking at 37 °C. Subsequently, 1 mL of the Bacillus subtilis suspension was inoculated in 10 mL of fresh LB media, which was incubated for 5 h with shaking (250 rpm) at 37 °C to achieve mid-log phase growth. After the Bacillus subtilis had grown to the mid-log phase, 1 mL of the suspension was washed at least 3 times by centrifuging for 3 minutes at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 RPM and resuspending the pellet in 1 mL of clean PBS (pH 4.5). To label the surface of Bacillus subtilis, 50 μL of P17 in DMSO solution (∼10 nM) was added slowly to the suspension of the Bacillus subtilis cells and left to incubate for 30 minutes. The unreacted P17 was rinsed 6 times with PBS (pH 4.5) before imaging. Then, the cleaned Bacillus subtilis cells were resuspended in a small amount of PBS (pH 4.5) to produce a concentrated cell suspension. Finally, 2 μL of this cell suspension was deposited onto an agarose pad (1.5% (by mass) low melting point agarose in PBS) and mounted onto an Ar plasma etched glass slide (size: 24 × 50 mm, thickness: 0.13–0.16 mm) and immediately used for the SMLM imaging.
000 RPM and resuspending the pellet in 1 mL of clean PBS (pH 4.5). To label the surface of Bacillus subtilis, 50 μL of P17 in DMSO solution (∼10 nM) was added slowly to the suspension of the Bacillus subtilis cells and left to incubate for 30 minutes. The unreacted P17 was rinsed 6 times with PBS (pH 4.5) before imaging. Then, the cleaned Bacillus subtilis cells were resuspended in a small amount of PBS (pH 4.5) to produce a concentrated cell suspension. Finally, 2 μL of this cell suspension was deposited onto an agarose pad (1.5% (by mass) low melting point agarose in PBS) and mounted onto an Ar plasma etched glass slide (size: 24 × 50 mm, thickness: 0.13–0.16 mm) and immediately used for the SMLM imaging.
3D-SMLM imaging and data processing
The super-resolution imaging was carried out using a Nikon N-STORM 5.0 super-resolution microscope system with a motorized inverted microscope ECLIPSE Ti2-E, a 100×/NA 1.49 oil immersion TIRF objective lens (CFI HP) and an ORCA-Flash 4.0 SCMOS camera (Hamamatsu Photonics K.K.). During super-resolution imaging, a 405 nm laser (1.8 W cm−2) was employed to photoactivate rhodamine spirolactams from ring-closed isomers to ring-opened isomers. At the same time, a 561 nm laser (1.2 kW cm−2) was used as the excitation source to produce sparse emissions from ring-opened rhodamines. Repeating this cycle many times (i.e., to collect ∼30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 images with the SCMOS camera with an integration time of 20 ms per frame) allowed us to reconstruct a super-resolution image. Astigmatism was used in the 3D-imaging experiments. The software NIS-Elements Ar and N-STORM Analysis were used to analyze the collected images and computationally reconstruct the 3D-SMLM image.
000 images with the SCMOS camera with an integration time of 20 ms per frame) allowed us to reconstruct a super-resolution image. Astigmatism was used in the 3D-imaging experiments. The software NIS-Elements Ar and N-STORM Analysis were used to analyze the collected images and computationally reconstruct the 3D-SMLM image.
Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21708039, 21502189, 21878286), CPSF (2017M611278, 2018T110234), and DICP (DMTO201603, TMSR201601, 2015YB07). X. L. and W. C. are indebted to the financial support from SUTD (T1SRCI17126, IDD21700101, IDG31800104). J. Y. acknowledges the support from the National Research Foundation of Korea (NRF) (No. 2012R1A3A2048814).Notes and references
- (a) S. W. Hell, Science, 2007, 316, 1153 CrossRef CAS PubMed; (b) A. von Diezman, Y. Shechtman and W. E. Moerner, Chem. Rev., 2017, 117, 7244 CrossRef PubMed; (c) E. Betzig, Angew. Chem., Int. Ed., 2015, 54, 8034 CrossRef CAS PubMed.
- G. Patterson, M. Davidson, S. Manley and J. Lippincott-Schwartz, Annu. Rev. Phys. Chem., 2010, 61, 345 CrossRef CAS PubMed.
- (a) M. Heilemann, S. van de Linde, M. Schuttpelz, R. Kasper, B. Seefeldt, A. Mukherjee, P. Tinnefeld and M. Sauer, Angew. Chem., Int. Ed., 2008, 47, 6172 CrossRef CAS PubMed; (b) J. B. Grimm, T. Klein, B. G. Kopek, G. Shtengel, H. F. Hess, M. Sauer and L. D. Lavis, Angew. Chem., Int. Ed., 2016, 55, 1723 CrossRef CAS PubMed.
- (a) O. Nevskyi, D. Sysoiev, A. Oppermann, T. Huhn and D. Wöll, Angew. Chem., Int. Ed., 2016, 55, 12698 CrossRef CAS PubMed; (b) B. Roubinet, M. L. Bossi, P. Alt, M. Leutenegger, H. Shojaei, S. Schnorrenberg, S. Nizamov, M. Irie, V. N. Belov and S. W. Hell, Angew. Chem., Int. Ed., 2016, 55, 15429 CrossRef CAS PubMed; (c) B. Roubinet, M. Weber, H. Shojaei, M. Bates, M. L. Bossi, V. N. Belov, M. Irie and S. W. Hell, J. Am. Chem. Soc., 2017, 139, 6611 CrossRef CAS PubMed.
- (a) H. N. Kim, M. H. Lee, H. J. Kim, J. S. Kim and J. Yoon, Chem. Soc. Rev., 2008, 37, 1465 RSC; (b) X. Chen, T. Pradhan, F. Wang, J. S. Kim and J. Yoon, Chem. Rev., 2012, 112, 1910 CrossRef CAS PubMed.
- (a) K. H. Knauer and R. Gleiter, Angew. Chem., Int. Ed., 1977, 16, 113 CrossRef; (b) H. Willwohl, J. Wolfrum and R. Gleiter, Laser Chem., 1989, 10, 63 CrossRef CAS; (c) H. Montenegro, M. D. Paolo, D. Capdevila, P. F. Aramendia and M. L. Bossi, Photochem. Photobiol. Sci., 2012, 11, 1081 RSC; (d) K. Li, Y. Xiang, X. Wang, J. Li, R. Hu, A. Tong and B. Z. Tang, J. Am. Chem. Soc., 2014, 136, 1643 CrossRef CAS PubMed.
- (a) J. Folling, V. Belov, R. Kunetsky, R. Medda, A. Schonle, A. Egner, C. Eggeling, M. Bossi and S. W. Hell, Angew. Chem., Int. Ed., 2007, 46, 6266 CrossRef CAS PubMed; (b) V. N. Belov, M. L. Bossi, J. Fölling, V. P. Boyarskiy and S. W. Hell, Chem.–Eur. J., 2009, 15, 10762 CrossRef CAS PubMed; (c) M. K. Lee, P. Rai, J. Williams, R. J. Twieg and W. E. Moerner, J. Am. Chem. Soc., 2014, 136, 14003 CrossRef CAS PubMed; (d) B. Roubinet, M. Bischoff, S. Nizamov, S. Yan, C. Geisler, S. Stoldt, G. Y. Mitronova, V. N. Belov, M. L. Bossi and S. W. Hell, J. Org. Chem., 2018, 83, 6466 CrossRef CAS PubMed; (e) M. Bossi, J. Folling, V. N. Belov, V. P. Boyarskiy, R. Medda, A. Egner, C. Eggeling, A. Schonle and S. W. Hell, Nano Lett., 2008, 8, 2463 CrossRef CAS PubMed; (f) V. N. Belov, C. A. Wurm, V. P. Boyarskiy, S. Jakobs and S. W. Hell, Angew. Chem., Int. Ed., 2010, 49, 3520 CrossRef CAS PubMed; (g) J. Folling, V. Belov, D. Riedel, A. Schonle, A. Egner, C. Eggeling, M. Bossi and S. W. Hell, ChemPhysChem, 2008, 9, 321 CrossRef PubMed; (h) H. Aoki, K. Mori and S. Ito, Soft Matter, 2012, 8, 4390 RSC; (i) E. A. Halabi, D. Pinotsi and P. Rivera-Fuentes, Nat. Commun., 2019, 10, 1232 CrossRef PubMed; (j) E. A. Halabi, Z. Thiel, N. Trapp, D. Pinotsi and P. Rivera-Fuentes, J. Am. Chem. Soc., 2017, 139, 13200 CrossRef CAS PubMed; (k) E. A. Halabi, S. Püntener and P. Rivera-Fuentes, Helv. Chim. Acta, 2018, 101, e1800165 CrossRef.
- V. N. Belov and M. L. Bossi, Isr. J. Chem., 2013, 53, 1 CrossRef.
- (a) R. A. Gottlieb, H. A. Giesing, J. Y. Zhu, R. L. Engler and B. M. Babior, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 5965 CrossRef CAS PubMed; (b) R. A. Gottlieb, J. Nordberg, E. Skowronski and B. M. Babior, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 654 CrossRef CAS PubMed; (c) M. A. Barry, J. E. Reynolds and A. Eastman, Cancer Res., 1993, 53, 2349 CAS.
- (a) S. N. Uno, M. Kamiya, T. Yoshihara, K. Sugawara, K. Okabe, M. C. Tarhan, H. Fujita, T. Funatsu, Y. Okada, S. Tobita and Y. Urano, Nat. Chem., 2014, 6, 681 CrossRef CAS PubMed; (b) H. Takakura, Y. Zhang, R. S. Erdmann, A. D. Thompson, Y. Lin, B. McNellis, F. Rivera-Molina, S. N. Uno, M. Kamiya, Y. Urano, J. E. Rothman, J. Bewersdorf, A. Schepartz and D. Toomre, Nat. Biotechnol., 2017, 35, 773 CrossRef CAS PubMed.
- G. Xi, L. Sheng, I. Zhang, J. Du, T. Zhang, Q. Chen, G. Li, Y. Zhang, Y. Song, J. Li, Y. Zhang and S. X. Zhang, ACS Appl. Mater. Interfaces, 2017, 9, 38032 CrossRef CAS PubMed.
- (a) K. Matsumoto, J. Kusaka, A. Nishibori and H. Hara, Mol. Microbiol., 2006, 61, 1110 CrossRef CAS PubMed; (b) A. S. Johnson, S. van Horck and P. J. Lewis, Microbiology, 2004, 150, 2815 CrossRef CAS PubMed; (c) F. Kawai, M. Shoda, R. Harashima, Y. Sadaie, H. Hara and K. Matsumoto, J. Bacteriol., 2004, 186, 1475 CrossRef CAS PubMed; (d) I. Fishov and C. L. Woldringh, Mol. Microbiol., 1999, 32, 1166 CrossRef CAS PubMed; (e) A. Nishibori, J. Kusaka, H. Hara, M. Umeda and K. Matsumoto, J. Bacteriol., 2005, 187, 2163 CrossRef CAS PubMed.
- O. V. Dolomanov, L. J. Bourhis, R. J. Gildea, J. A. K. Howard and H. Puschmann, J. Appl. Crystallogr., 2009, 42, 339 CrossRef CAS.
- L. Palatinus and G. Chapuis, J. Appl. Crystallogr., 2007, 40, 786 CrossRef CAS.
- G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Crystallogr., 2008, 64, 112 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Data-mining of 3D molecular structures of rhodamine spiroamides, computational results, general information on synthesis and characterization, Fig. S1–S26 and Tables S1–S7 (PDF), and Movies 1 and 2 showing the 3D-SMLM imaging of Bacillus subtilis cells. CCDC 1582847, 1901370–1901372. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c9sc01284b |
| This journal is © The Royal Society of Chemistry 2019 |