Open Access Article
Open Access ArticleThe destructive spontaneous ingression of tunable silica nanosheets through cancer cell membranes†
Arghya
Bandyopadhyay
a,
Priya
Yadav
b,
Keka
Sarkar
b and
Sayan
Bhattacharyya
 *a
*a
aDepartment of Chemical Sciences, Centre for Advanced Functional Materials, Indian Institute of Science Education and Research (IISER), Mohanpur – 741246, Kolkata, India. E-mail: sayanb@iiserkol.ac.in; Tel: +91-33-6136-0000 extn 1275
bDepartment of Microbiology, University of Kalyani, Nadia – 741235, India
First published on 8th May 2019
Abstract
Robust inorganic graphene analogues with atomic level sharp edges have seldom been investigated to decipher the interaction of two-dimensional materials with the cell membrane. Silica nanosheets (NSs) with four different thicknesses between 2.9 nm and 11.1 nm were synthesized by microwave irradiation and these colloidal NSs were able to spontaneously penetrate the cell membrane leaving membrane perforations at their sites of entry. The NS-ingression was most effective with the thinnest NSs, when studied in vitro. The atomistic details of the NS-membrane interaction were revealed by molecular dynamics (MD) simulations, which showed that the extraction of phospholipids was most favored when NSs were oriented vertically with respect to the membrane surface. While the folic acid modified NSs demonstrated a riveting tendency to penetrate the cancer cell membrane, co-modification with doxorubicin (DOX) unexpectedly reduced their capability. Migrating away from a conventional drug delivery approach, here we show that silica NSs can kill cancer cells primarily by mechanical scalpelling. Targeted ingress could be achieved through antibody conjugation on the NSs and thus only cancerous HeLa cells are affected by this treatment, leaving the normal HEK-293 cells intact. This destructive ingress through limited oxidative stress offers a previously unexplored route to treat fatal diseases without the necessity of transporting expensive drugs or radiation therapy, thereby bypassing deleterious side effects on healthy cells.
Introduction
The impetus received from graphene research has catapulted interest in two-dimensional (2D) inorganic graphene analogues primarily through the resurgence of materials such as MXenes1 and transition metal dichalcogenides2 among others.3–5 A parallel effort has emerged to discern the biological and environmental applications of these 2D materials,6 for example, the antibacterial activity of MXenes,7 and bio-sensing properties of MoS2.8 Within the interdisciplinary field of nanomedicine,9,10 this advent of 2D nanostructures is expected to supplant other validated nanomaterials that led towards the development of better drugs and devices to improve the sensitivity,11 early diagnosis,12 and treatment of fatal diseases like cancer.13 The limited data on the biological response to 2D-geometry compels researchers to draw parallels with earlier reports on the specific biological responses to graphene. Graphene was able to rupture the bacterial cell membrane and extract the phospholipids,14 whereby the cell uptake occurs due to spontaneous localized piercing of the membrane at the sheet asperities,15 facilitated by surface oxygen containing groups enhancing the adsorption of cellular matter.16 The physico–mechanical interactions of graphene and its inorganic analogues will largely depend on its thickness, lateral dimensions, mechanical properties and surface chemistry such as surface charge and polarity.6 The unprecedented biomolecular applications of graphene open the door for other 2D analogues such as h-BN, C3N4, silicate clays and layered double hydroxides etc. that have far reaching consequences through understanding their ability to penetrate the membrane followed by cellular uptake and extraction of cellular matter.The 2D-morphology of silica is one such unexplored nanostructure. 2D silica synthesis is challenging due to the lack of chemical/physical exfoliation routes and the spontaneous sharing of oxygen atoms between [SiO4]4− tetrahedra under ambient conditions to form single/double chain silicates finally leading to the 3D structures,17 except a few cases where ∼10 nm spherical particles were successfully synthesized.18–20 Silica with cellular toxicity levels above 100 μg mL−1,21,22 is an automatic choice for molecular delivery of drugs, nucleic acids or proteins.23–26 In this work, ultrathin silica NSs (Si-NS) were prepared by one-step microwave irradiation at 60 °C for 5 min and the NS thickness was varied by self-stacking as a function of time inside the same reaction medium under ambient temperature and pressure. These colloidal NSs can penetrate the cell membrane spontaneously creating membrane pores due to their sharp edge ingress, the extent of which depends on their dimensionality, mode of insertion and surface chemistry. Rupturing of the cancer cell membrane is observed to be more effective in the absence of a surface-conjugated drug and cell damage occurs by mechanical scalpelling. The therapeutic outcome of any drug i.e. its transport to the target site, blood circulation and tumor penetration behavior,27,28 depends on the size and composition modulations in inorganic nanocarriers,29–31 carbon,32–34 and soft-matter based drug conjugates.35–37 Nonetheless, quite often cancer cells acquire resistance through drug resistant genes leading to various side effects and toxicity to normal cells. Although drug-free, targeted mechanical formulation might be an alternative for improved therapy, mechanical treatments using nano/micro-structures are less common and involve instances such as inducing compressive stress leading to coordinated migration of cancer cells,38 dissociation of the tumour tissue into single cells by vortexing or pipetting,39 interference caused by low frequency magnetic fields,40–42 and biomechanical forces.43 2D nanomaterials with a large specific surface area are mostly explored in photothermal and photodynamic therapies,44–46 although they are especially attractive in amplifying mechanical forces for cancer therapy.47 In this regard, Si-NSs have been developed as a remarkable approach for synergistic destruction of tumors in combination with anticancer drugs and/or alone by its scalpelling excellence. Also to the best of our knowledge there is almost no data available regarding the synthesis of non-templated silica NSs in one-step microwave routes and in silico study of the interaction of Si-NSs with the cell membrane. Our innovative strategy of mechanical scalpelling the cancer cells is manifested by the destructive NS ingress through limited oxidative stress which offers an unfamiliar route to treat fatal diseases without the necessity of transporting expensive drugs or radiation therapies. This in vitro approach offers an avant-garde design to particularly treat cancer besides conventional drug delivery, photothermo-chemotherapy and photodynamic therapy.48–50
Results and discussion
Physicochemical characterization of the NSs
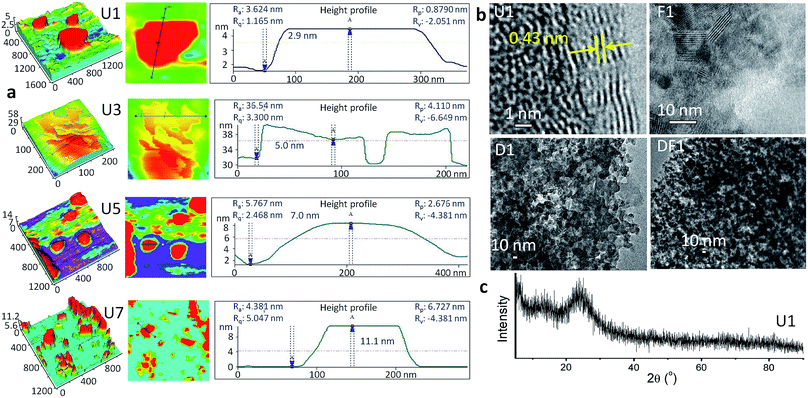
The thinnest unmodified Si-NS (U1) was prepared by microwave irradiation of an alkaline solution of methanolic tetraethyl ortho silicate (TEOS) within 5 min in the presence of a surfactant, cetyltrimethylammonium bromide, CTAB (ESI, Experimental methods, S1†). The sheet-like morphology of the NSs is evident from the atomic force microscopy (AFM) studies (Fig. 1a). The 2.9 ± 0.01 nm thick U1 NSs have a lateral span of ∼300 nm. In the high-resolution transmission electron microscope (TEM) image of U1 NSs (Fig. 1b), an interplanar spacing of 0.43 nm is observed at the NS edges, whereas the representative X-ray diffraction (XRD) pattern in Fig. 1c shows its amorphous nature. This clearly suggests the presence of crystalline domains within a global amorphous structure of the NSs. Due to the presence of >N+– groups attached to the NSs from CTAB, the U1 NSs have a positive surface charge of +12.6 mV (Fig. S1†). The positively charged surface of U1 NSs were further modified with DOX, folic acid and co-modified with DOX-folic acid to give D1, F1 and DF1 NSs, respectively. The loading concentrations of the ligands are ∼375 ng μgSi-NS−1 folic acid in F1 NSs, ∼285 ng μgSi-NS−1 DOX in D1 NSs, and ∼347 ng μgSi-NS−1 folic acid and ∼335 ng μgSi-NS−1 DOX in DF1 NSs (Table S1†). These different functionalization processes expectedly alter the topological features of the NSs (Fig. 1b) as well as increase the overall NS thickness (Fig. S2†), keeping the lateral dimensions mostly unaffected. The thickness of the U1 NSs increases from 2.9 ± 0.01 nm to 8.5 ± 0.04 nm for D1, 5.3 ± 0.5 nm for F1 and 7.2 ± 0.3 nm for DF1 NSs. The loaded drug can be clearly visualized from the roughness of the NS surface in the TEM images of D1 and DF1 (Fig. 1b).While the dimensions of U1 NSs could be stabilized by dispersing them in fresh methanol under vacuum, the colloidal NSs were allowed to stack and grow in thickness over days inside the parent reaction medium under ambient conditions to understand the impact of NS thickness on cell membrane. The NSs collected after 3, 5 and 7 days are coded as U3, U5 and U7, respectively, and similarly the DOX modified NSs are named as D3, D5 and D7, folic acid modified NSs as F3, F5 and F7 and the DOX-folic acid co-modified NSs as DF3, DF5 and DF7, respectively. The stacking-induced aggregation of the NSs causes an indiscriminate heterogeneity of the lateral dimensions, as revealed from the AFM images in Fig. 1a and the TEM image of U3 NSs (Fig. S3†). However this spontaneous growth does not alter the chemical functionalities of the NSs (Fig. S4†). In U3, the thickness increases by 73% to 5.0 ± 0.03 nm whereas those of U5 and U7 are 7.0 ± 0.02 and 11.1 ± 0.01 nm, respectively (Fig. 1a). The average lateral dimension varies as 300, 190, 270 and 150 nm for U1, U3, U5 and U7 NSs, respectively. The high-angle annular dark-field scanning TEM (HAADF-STEM) image of a representative U3 NS and the corresponding energy dispersive X-ray spectrum (EDS) mapping show the exclusive presence of Si and O within the NS area (Fig. S5†). With increasing thickness and thereby with decreasing surface area of the thicker NSs, the zeta potential decreases as ∼10, 9.9 and 8.3 mV for U3, U5 and U7 NSs, respectively (Fig. S1†).
NS ingress
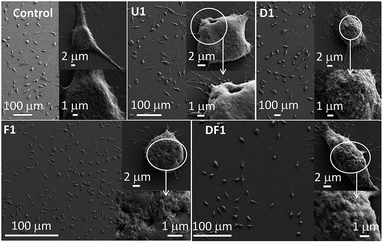
The efficacy of the improvised cellular internalization of these NSs, tested with 5 ng mL−1 NS concentration in each case, was monitored by their relative incorporation inside a HeLa cell after 24 h treatment, measured by inductively coupled mass spectrometry (ICP-MS, Fig. S6†). The temporal incorporation of U1 NSs inside the HeLa cells was manifested by the quantitative estimation of Si wt% from EDS analyses in the field emission scanning electron microscope (FESEM) instrument. While the untreated cells contain 20.6 wt% Si, the U1 NS treated cells contain 24.8, 31.1 and 37.5 wt% Si, after treatment for 2, 4 and 8 h, respectively (Fig. S7†). From ICP-MS measurements, F1 shows the highest potency to penetrate the cell membrane. Folic acid receptors are highly expressed and abundant in numerous cancers including cervical cancer cells.51 The highest uptake occurs with the thinnest F1 NSs and with increasing thickness the uptake decreases, a trend demonstrated also by other nano-constructions, U/D/DF. The lower penetration of the DF series of NSs is noteworthy. During co-modification of DOX and folic acid, both the ligands compete for the space available on the NS surface which leads to relatively lower folic acid loading. In the absence of folic acid modification, the unmodified U1–U7 and DOX modified D1–D7 NSs demonstrate moderate uptake inside the cells. Clearly, the whole mechanism of internalization depends on various environmental factors such as the pH and medium complexity, and cannot be exclusively associated with receptor-mediated uptake such as folate receptors or in the extended case of DOX.The NS treated cells were studied by FESEM as shown in Fig. 2. The membrane deformation in each treated cell suggests that the NSs have a cell killing potency, the degree of which and the type of membrane deformation varies with the thickness and surface modification of the NSs. The number of dead cells is higher with U1 (Fig. 2) over U3–U7 NSs (Fig. S8†) where with increasing NS thickness, the ratio of spindle to spherically distorted cells decreases. Since this group of NSs differs only in their relative thickness without any chemical modification or drugs, it is evident that killing of the cells occurs through membrane rupture by the sharp edge ingress of the NSs. The most striking finding is the formation of pores in the cell membrane at the sites of NS entry. The pore diameter increases steadily as 193 ± 27, 383 ± 38, 521 ± 63 and 801 ± 73 nm by treating the HeLa cells with U1, U3, U5 and U7, respectively. Distinct nuclear degranulation and formation of membrane pores are also observed by epifluorescence microscopy with U1 NS treated HeLa cells for 8 h (Fig. S9†). The unmatched membrane pore size and NS dimensions implies varying NS orientations during cell membrane internalization and the heterogeneity of NS lateral dimensions. Although not as distinct, the rarely observed externally stimulated membrane pores were previously induced by antibiotics such as polymyxins,52 and toxins belonging to the exotoxin group.53 The F1 NSs increase the membrane perforations notably in the absence of any loaded drug whereas with D1 NSs extensive membrane blebbing is observed, which is a signature marker of apoptotic cells. In the presence of DOX, significant generation of reactive oxygen species (ROS) pushes the cancerous cells to bleb and to choose the apoptotic path. Folic acid's intrinsic antioxidant property,54 reduces the production of ROS and helps to avoid membrane blebbing. Also, since folic acid is a cancer targeting molecule, it helps Si-NSs to enter the HeLa cells with higher efficiencies. Treatment with DF1 NSs has a moderate effect on the cells as is evident from the extent of pore formation and membrane blebbing. With progressively thicker NSs, the membrane perforations decrease within the respective D/F/DF groups (Fig. S8†). Here too, the extent of membrane blebbing decreases from D1 to D7.
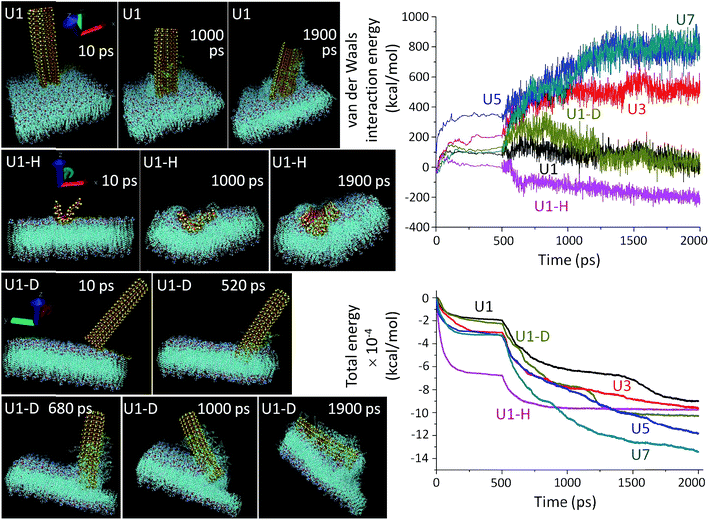
MD simulations (Fig. 3) were employed to mechanistically investigate the molecular level interactions. The all atomistic MD simulation was carried out using the Nanoscale Molecular Dynamics (NAMD) program,55 and visualized in the Visual Molecular Dynamics (VMD) software package,56 where all the unbiased atomistic simulations were performed with a model cell membrane composed of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) which is one of the important phospholipids found in eukaryotic cell membranes. Spontaneous piercing is notable when Si-NSs are placed in a vertical orientation with respect to the cell membrane.14,15 The NSs extensively damage the cell membrane and amply extract the phospholipids, which eventually damage the cells at the point of NS ingression. In reality, the cellular environment is not only composed of water, but also a mix of different ions, proteins etc., thus making the system extremely complex to replicate in the simulation. In our simulation procedure, the amount of water molecules is minimized by only stabilizing the membrane while keeping Si-NS flexible. Initial forceful insertion of the NSs within the membrane is observed and this step of penetration helps to overcome the energy barrier of penetration during which the NS-ingress breaks the weak intermolecular bonds that hold the POPC molecules together. The NSs are progressively trapped within the membrane due to strong short-range van der Waals attractions along with hydrophobic interaction between the NS and the membrane. During the course of penetration without any back and forth swinging motion of the NSs, the phospholipids of the membrane are extracted vigorously on the NS surface and are left behind in the environment or on the exposed NS surface. This scalpel effect leads to a dramatic change in the membrane bilayers and it leaves the membrane surface distorted and unstable.
The simulation results show that the van der Waals force is stabilized for an initial period of 100 ps and after this stabilization period, the NSs proceed through the insertion-cum-extraction period (Fig. 3, right top panel). The initial NS-POPC interaction profile is associated with the expected preparatory step of the NS-membrane interaction. During penetration inside the membrane, a thicker NS shows a steady increase in the van der Waals force due to a higher number of interacting atoms, whereas the U1 NS shows a significant gradual decrease providing the highest NS-membrane interaction. The diagonal orientation of U1 has a higher van der Waals force than the vertical orientation, due to a lengthened interaction though the NS corner during the first 200 ps of piercing the POPC membrane. Once the NS is fully inserted, the diagonal insertion equals the vertical orientation before the NS is engulfed within the cellular environment. It is also perceptible, that in spite of having a large number of interacting atoms in the horizontal configuration of U1, the van der Waals interaction energy remains the lowest probably due to its easier engulfment rather than piercing the membrane through a sharp edge ingress. In fact, the vertical orientation of U1 is more privileged over the other two configurations in terms of penetration capability. From the overall van dar Waals interaction energy profiles, it is clearly evident that all the U1 NSs primarily follow an attractive force towards the cell membrane, and after reaching an energy plateau, the NSs continue their penetration inside the cell. On the other hand, with the thicker NSs (especially U5 and U7), the van dar Waals interaction energy becomes repulsive from the onset of membrane interaction. The total energy consisting of molecular potential energy, including van der Waals force and kinetic energy, follows a different pattern as a function of membrane interaction time (Fig. 3, right bottom panel). The total energy profile of the thicker NSs, U3–U7, dips more as compared to U1, since kinetic energy of the thicker NSs is extremely low which also reduces their penetration capability. Also the requirement of a longer time for U3–U7 NSs to reach the energy plateau indicates their less preferential interaction. In spite of being kinetically slow, the thicker NSs have a greater chance to become entrapped within the membrane due to the higher number of individual atomic interactions and decreasing kinetic energy of the NSs which is evident from the total energy and van der Waals interaction energy profiles. The horizontal orientation of U1 involves a large number of interacting atoms, which also reduces their kinetics in comparison to the vertical or diagonal orientation of U1 NSs.
A thorough agreement between the MD simulation (Fig. 3) and the microscopic images (Fig. 2) clearly suggests that the prospects of sharp edge ingress largely depend on the orientation and thickness of the NSs, as well as their surface chemistry. As expected the piercing capability of the NSs is directly proportional to their ‘thinness’ (Fig. 3 and S10†). The point of contact between a thinner NS (U1) and the membrane is smaller and the tiny imperfections at the NS edge aids the scalpelling effect, superior to that with thicker NSs (U3–U7) where additional pressure is required for their forceful insertion. Another interesting feature of the unbiased MD simulation is the tilting of slender U1 NSs on the vertical axis during insertion. The slender NSs have a higher degree of tilting over the time period of simulation due to their flexibility which also manifests their greater tendency to interact with the membrane (Fig. S11†). The thicker NSs are tilted to a lower degree of freedom due to the rigidity of their structure and as a consequence the kinetics of penetration inside the cell membrane becomes affected. The MD simulated preferred orientation and tilting of the NSs can be further manifested by the ESI Movie files (Discussion S2†).
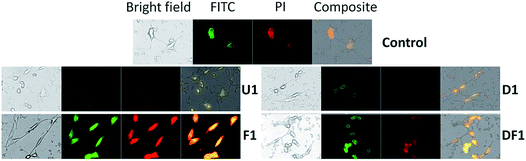
Rupturing of cancer cell membrane and cell death
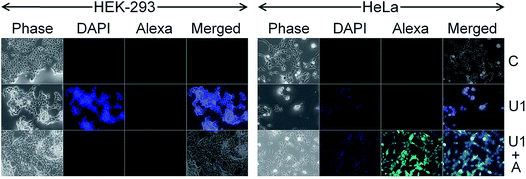
Cancer cell death by scalpelling and cell membrane perforations was confirmed by co-staining the HeLa cells with Hoechst and propidium iodide (PI) (Fig. S12†). The fluorescence detection by Hoechst–PI double stain provides an apoptosis assay whereby the blue-emitting Hoechst dye stains the apoptotic cells brighter than normal cells and red-emitting PI permeates only into dead cells, and hence the relative populations of apoptotic and dead cells can be detected against the normal cancer cells. Comparative analysis of the Hoechst and PI intensities in the treated cells suggests that the population of live cells is the least with F1–F7 NSs and the thinnest possible NSs are most proficient in cancer cell death within their respective functionalized groups of U/D/F/DF NSs. The penetration of the silica NSs leads to mechanical as well as chemical stress to the cell and causes fragmentation of DNA. The presence of folic acid on the NS surface unmistakably enhances the DNA breakage, be it only for F1 or DF1 as evidenced by the terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay followed by fluorescence microscopy (Fig. 4 and S13†) and DNA isolation followed by agarose gel electrophoresis (Fig. S14†). Since the TUNEL assay detects endogenous DNA damage and labels apoptotic cells on a single-cell level, it is considered more sensitive than the classical genomic DNA ladder test for DNA fragmentation analysis. In Fig. 4, the striking mechanical and chemical stress exerted by the thinnest NSs on the extent of DNA nicking is clearly evident. The binary influence of the sharp edge and the presence of folic acid ensures the maximum impact of F1 NSs in comparison to D1/DF1. In this context, D1 NSs act as a control to investigate the consequences of folic acid modification. With increasing NS thickness in each group, the intensity of green emission decreases, indicative of lower DNA shear through lesser invasion capability (Fig. S13†).Due to spontaneous ingress of the NSs in both healthy and cancer cells, exclusive targeting could not be achieved with either U1 or F1 NSs. A solution to this anonymous scalpelling is achieved by binding the >N+– groups on the U1 NS surface with the carboxylic acid groups of the heavy chain C-termini of a primary antibody, anti-ST6GALNAC1, which binds specifically to the ST6GALNAC1 protein (Fig. S15†). The presence of a targeting antibody on the NS surface exclusively kills the cancer cells with over-expressed ST6GALNAC1 protein (Fig. 5).44 Normal HEK-293 cells show a minimal deleterious effect in the presence of antibody conjugated NSs (U1+A) when compared to the higher fluorescence of DAPI in HeLa cells showing a better efficiency of cell damage. Low expressions of the same protein in normal HEK-293 cells exclude the antibody conjugated NSs to validate targeted killing efficacy. The entry mechanism of U1 and antibody modified NSs do not entirely follow the same pattern. Since the antibody plays the role of a guide to target the cancer cells efficiently, the receptor mediated endocytosis may be enhanced in the presence of such an active targeting molecule. However, after getting endocytosed, the antibody attached to the surface of the NS is likely to be denatured by various biochemical pathways and this ‘naked’ NS is expected to behave in the same way as that of U1 NSs.
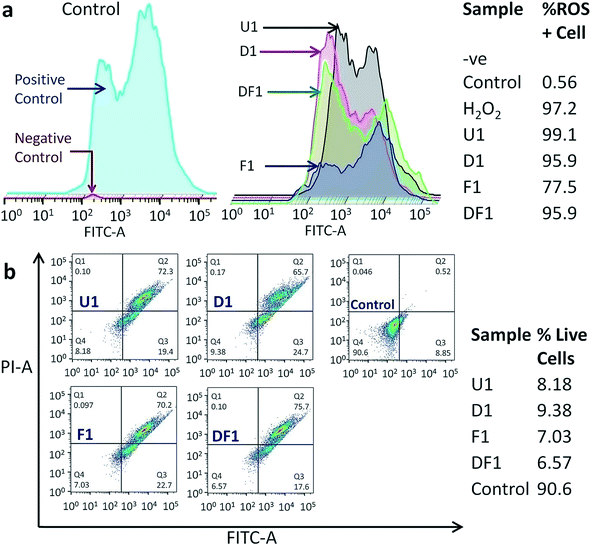
The modus operandi of in vitro cancer cell death by our nano-constructions is elucidated with the help of flow cytometric analysis, performed for three major parameters of a dying cell due to external stimuli, viz. ROS generation, amount of live and dead cell assay and cell cycle analysis. It is expected that Si-NS should kill cancer cells by high intracellular ROS generation, as ROS signaling intermediates are convoluted in the signal transduction pathway of apoptosis. Interestingly it is observed that ROS does not play a vital role in this mechanical treatment. As previously demonstrated, the highest cellular uptake occurs with folic acid modified NSs (Fig. S6†) and even though folic acid attenuates the sub-cellular ROS levels, ROS generation should have been the highest through interaction with this group due to a prominent mechanical oxidative stress. Hierarchical cluster analysis suggests that the ROS+cell percentage is the least for F1 (Fig. 6a) and F3–F7 among the thicker NSs (Fig. S16†). The most effective folic acid modification assists exclusive clustering with negative control of the experiment corresponding to the lowest ROS. On the contrary, U1 and D1 NSs show a comparatively large percentage of ROS+cells. Since U1 NSs provide a higher amount of mechanical shock due to their higher penetration capability they even create a higher amount of ROS with respect to the positive control, i.e. H2O2. As expected, in each functionalized group, the thinner the NSs the higher the mechanical stress that increases the ROS level within the treated HeLa cells (Fig. S16†). Because of this unique method of killing the cancer cells by mechanical shock, the conclusions drawn from a typical MTT assay (Fig. S17†) might be misleading, since it relies on the conventional biochemical pathway. Nevertheless, the MTT assay expectedly shows the least cell viability with F1 NSs because of their maximum incorporation inside the cells.
The treated samples were also analyzed for the post-treatment live cell percentage (Fig. 6b and S18†). It might be expected that as folic acid prevents the formation of ROS, it will leave a higher percentage of live cell population post treatment in spite of better targeting the acidic environment of cancer. However, the percentage of live cells is the lowest for F1 NS treated samples, killing ∼93% of cancer cells in 16 h (Fig. 6b), also reiterating the ICP-MS data (Fig. S6†). The least live cell count is in the samples treated with the thinnest NSs in each functionalized group (Fig. S18†). Hierarchical clustering shows that the DF NSs with surface attached DOX generating moderate ROS+cells, have the lowest impact on the cancer cell (Fig. S18†). This validates our claim that the dimensionality of the NSs plays a major role and efficient cancer cell death is possible even in the absence of an anticancer drug. The above experimental results show evidence of a vital scalpel effect of the NSs and the secondary role of DOX and ROS generation in cancer cell death. With all the NS constructions, most of the cell population shifts to Q2 (Fig. 6b and S18†), indicating the cells undergoing a late apoptosis phase. Moreover, the NSs with different thickness and surface modifications have a negligible effect on the cell cycle blocking (Fig. S19†). The cells are observed to be increasingly stalled at the sub G0–G1 phase when compared to the control and this blockage of cell cycle at sub G0–G1 is a signature of late apoptosis and significant DNA breakage,57,58 as evidenced earlier from classical DNA ladder experiment and the TUNEL assay. The least cell cycle blocking is observed with F1–F7 NSs which have the maximum effect on the sub G0–G1 phase due to reduced ROS production.
Conclusions
In summary, microwave synthesized silica NSs were employed for the mechanical treatment of cancer cells where the ingression through membrane perforation depends on the thickness of the NSs, their surface functionalization and the mode of interaction with the cell membrane. Self-stacking of the NSs reduces their efficiency at penetrating the cell membrane during their co-culturing with HeLa cells. Folic acid modified NSs had a riveting tendency to penetrate the cell membrane, whereas co-modification with DOX reduced their capability. The higher insertion capability of F1 NSs is caused by their thinness and the extent of folic acid on the NS surface that target the cancer cells. However, exclusive cancer cell targeting was achieved only by the conjugation of an antibody to the NS surface, whereafter the healthy cells were negligibly affected. MD simulations support the superior insertion of the thinnest NSs in a vertical orientation as compared to diagonal or horizontal configurations. The cellular environment engulfs the inserted NSs and cell death occurs primarily through apoptosis. The extraordinary phenomenon of cancer cell death can only be attributed to the nanoscale dimension of the 2D NSs that cut the cell membrane barrier with high competence and destroy the genetic material, as observed in DNA breakage experiments. This is the first demonstration of cancer cell death by scalpelling with ‘thin’ inorganic NSs without the need for drug molecules or photodynamic therapies. The strategy can be extended to 2D structures of other biocompatible materials and the cell rupturing will be contingent on suitable surface modification with targeting molecules. Our in vitro findings on cancer cell rupturing by Si-NSs may offer the better design of inorganic 2D structure-based scalpels for mechanical interactions with cell membranes for several biomedical applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The Department of Science and Technology (DST), Science and Engineering Board (SERB), Government of India is duly acknowledged for the financial support under grant no. EMR/2016/001703. The BD Flow cytometric facility, CRNN, University of Calcutta is duly acknowledged for providing the flow cytometric analysis platform.References
- M. Naguib, V. N. Mochalin, M. W. Barsoum and Y. Gogotsi, Adv. Mater., 2014, 26, 992–1005 CrossRef CAS PubMed.
- H. S. S. Ramakrishna Matte, A. Gomathi, A. K. Manna, D. J. Late, R. Datta, S. K. Pati and C. N. R. Rao, Angew. Chem., Int. Ed., 2010, 49, 4059–4062 CrossRef PubMed.
- S. Bhattacharyya, D. Zitoun, Y. Estrin, O. Moshe, D. H. Rich and A. Gedanken, Chem. Mater., 2009, 21, 326–335 CrossRef CAS.
- M. E. Dávila, L. Xian, S. Cahangirov, A. Rubio and G. L. Lay, New J. Phys., 2014, 16, 095002 CrossRef.
- A. Sadhu and S. Bhattacharyya, J. Phys. Chem. C, 2013, 117, 26351–26360 CrossRef CAS.
- Z. Wang, W. Zhu, Y. Qiu, X. Yi, A. von dem Bussche, A. Kane, H. Gao, K. Koski and R. Hurt, Chem. Soc. Rev., 2016, 45, 1750–1780 RSC.
- K. Rasool, M. Helal, A. Ali, C. E. Ren, Y. Gogotsi and K. A. Mahmoud, ACS Nano, 2016, 10, 3674–3684 CrossRef CAS PubMed.
- K. Kalantar-zadeh and J. Z. Ou, ACS Sens., 2016, 1, 5–16 CrossRef CAS.
- B. Pelaz, C. Alexiou, R. A. Alvarez-Puebla, F. Alves, A. M. Andrews, S. Ashraf, L. P. Balogh, L. Ballerini, A. Bestetti, C. Brendel, S. Bosi, M. Carril, W. C. W. Chan, C. Chen, X. Chen, X. Chen, Z. Cheng, D. Cui, J. Du, C. Dullin, A. Escudero, N. Feliu, M. Gao, M. George, Y. Gogotsi, A. Grünweller, Z. Gu, N. J. Halas, N. Hampp, R. K. Hartmann, M. C. Hersam, P. Hunziker, J. Jian, X. Jiang, P. Jungebluth, P. Kadhiresan, K. Kataoka, A. Khademhosseini, J. Kopeček, N. A. Kotov, H. F. Krug, D. S. Lee, C.-M. Lehr, K. W. Leong, X.-J. Liang, M. Ling Lim, L. M. Liz-Marzán, X. Ma, P. Macchiarini, H. Meng, H. Möhwald, P. Mulvaney, A. E. Nel, S. Nie, P. Nordlander, T. Okano, J. Oliveira, T. H. Park, R. M. Penner, M. Prato, V. Puntes, V. M. Rotello, A. Samarakoon, R. E. Schaak, Y. Shen, S. Sjöqvist, A. G. Skirtach, M. G. Soliman, M. M. Stevens, H.-W. Sung, B. Z. Tang, R. Tietze, B. N. Udugama, J. S. VanEpps, T. Weil, P. S. Weiss, I. Willner, Y. Wu, L. Yang, Z. Yue, Q. Zhang, Q. Zhang, X.-E. Zhang, Y. Zhao, X. Zhou and W. J. Parak, ACS Nano, 2017, 11, 2313–2381 CrossRef CAS PubMed.
- W. C. W. Chan, B. Udugama, P. Kadhiresan, J. Kim, S. Mubareka, P. S. Weiss and W. J. Parak, ACS Nano, 2016, 10, 8139–8142 CrossRef CAS PubMed.
- S. O. Kelley, C. A. Mirkin, D. R. Walt, R. F. Ismagilov, M. Toner and E. H. Sargent, Nat. Nanotechnol., 2014, 9, 969–980 CrossRef CAS PubMed.
- X. Zheng, X. Wang, H. Mao, W. Wu, B. Liu and X. Jiang, Nat. Commun., 2015, 6, 5834 CrossRef CAS PubMed.
- L. Wang, Q. Sun, X. Wang, T. Wen, J.-J. Yin, P. Wang, R. Bai, X.-Q. Zhang, L.-H. Zhang, A.-H. Lu and C. Chen, J. Am. Chem. Soc., 2015, 137, 1947–1955 CrossRef CAS PubMed.
- Y. Tu, M. Lv, P. Xiu, T. Huynh, M. Zhang, M. Castelli, Z. Liu, Q. Huang, C. Fan, H. Fang and R. Zhou, Nat. Nanotechnol., 2013, 8, 594–601 CrossRef CAS PubMed.
- Y. Li, H. Yuan, A. von dem Bussche, M. Creighton, R. H. Hurt, A. B. Kane and H. Gao, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 12295–12300 CrossRef CAS PubMed.
- J. Chen, G. Zhou, L. Chen, Y. Wang, X. Wang and S. Zeng, J. Phys. Chem. C, 2016, 120, 6225–6231 CrossRef CAS.
- L. V. Azaroff, Introduction to Solids, Tata McGraw Hill Education Pvt Ltd, New Delhi, 2010, ch. 15, pp. 425–428 Search PubMed.
- D. R. Hristov, E. Mahon and K. A. Dawson, Chem. Commun., 2015, 51, 17420–17423 RSC.
- K. Ma, C. Mendoza, M. Hanson, U. Werner-Zwanziger, J. Zwanziger and U. Wiesner, Chem. Mater., 2015, 27, 4119–4133 CrossRef CAS.
- M. A. Snyder, J. A. Lee, T. M. Davis, L. E. Scriven and M. Tsapatsis, Langmuir, 2007, 23, 9924–9928 CrossRef CAS PubMed.
- H. Meng, W. X. Mai, H. Zhang, M. Xue, T. Xia, S. Lin, X. Wang, Y. Zhao, Z. Ji, J. I. Zink and A. E. Nel, ACS Nano, 2013, 7, 994–1005 CrossRef CAS PubMed.
- H. L. Herd, A. Malugin and H. Ghandehari, J. Controlled Release, 2011, 153, 40–48 CrossRef CAS PubMed.
- X. Wang, J. Xu, Q. Wang, A. Xu, Y. Zhai, J. Luo, Y. Jiang, N. He and Z. Wang, Small, 2017, 13, 1603369 CrossRef PubMed.
- P. Y. Huang, S. Kurasch, J. S. Alden, A. Shekhawat, A. A. Alemi, P. L. McEuen, J. P. Sethna, U. Kaiser and D. A. Muller, Science, 2013, 342, 224 CrossRef CAS PubMed.
- B. Yang, E. Emmez, W. E. Kaden, X. Yu, J. A. Boscoboinik, M. Sterrer, S. Shaikhutdinov and H. J. Freund, J. Phys. Chem. C, 2013, 117, 8336–8344 CrossRef CAS.
- C. Fang, A. Van Blaaderen and M. A. Van Huis, J. Phys. Chem. C, 2015, 119, 14343–14350 CAS.
- Y. Geng, P. Dalhaimer, S. Cai, R. Tsai, M. Tewari, T. Minko and D. E. Discher, Nat. Nanotechnol., 2007, 2, 249–255 CrossRef CAS PubMed.
- V. P. Chauhan, Z. Popović, O. Chen, J. Cui, D. Fukumura, M. G. Bawendi and R. K. Jain, Angew. Chem., Int. Ed., 2011, 50, 11417–11420 CrossRef CAS PubMed.
- G. B. Braun, T. Friman, H.-B. Pang, A. Pallaoro, T. H. de Mendoza, A.-M. A. Willmore, V. R. Kotamraju, A. P. Mann, Z.-G. She, K. N. Sugahara, N. O. Reich, T. Teesalu and E. Ruoslahti, Nat. Mater., 2014, 13, 904–911 CrossRef CAS PubMed.
- E. Huynh, Y. C. LeungBen, B. L. Helfield, M. Shakiba, J.-A. Gandier, C. S. Jin, E. R. Master, B. C. Wilson, D. E. Goertz and G. Zheng, Nat. Nanotechnol., 2015, 10, 325–332 CrossRef CAS PubMed.
- T. Yu, K. Greish, L. D. McGill, A. Ray and H. Ghandehari, ACS Nano, 2012, 6, 2289–2301 CrossRef CAS PubMed.
- S. Kapri, S. Maiti and S. Bhattacharyya, Carbon, 2016, 100, 223–235 CrossRef CAS.
- H. J. Yoon, T. H. Kim, Z. Zhang, E. Azizi, T. M. Pham, C. Paoletti, J. Lin, N. Ramnath, M. S. Wicha, D. F. Hayes, D. M. Simeone and S. Nagrath, Nat. Nanotechnol., 2013, 8, 735–741 CrossRef CAS PubMed.
- R. Singhal, Z. Orynbayeva, R. V. Kalyana Sundaram, J. J. Niu, S. Bhattacharyya, E. A. Vitol, M. G. Schrlau, E. S. Papazoglou, G. Friedman and Y. Gogotsi, Nat. Nanotechnol., 2011, 6, 57–64 CrossRef CAS PubMed.
- J. Kopeček, Adv. Drug Delivery Rev., 2013, 65, 49–59 CrossRef PubMed.
- C. Shi, D. Guo, K. Xiao, X. Wang, L. Wang and J. Luo, Nat. Commun., 2015, 6, 7449 CrossRef PubMed.
- D. Peer, J. M. Karp, S. Hong, O. C. Farokhzad, R. Margalit and R. Langer, Nat. Nanotechnol., 2007, 2, 751–760 CrossRef CAS PubMed.
- J. M. Tse, G. Cheng, J. A. Tyrrell, S. A. Wilcox-Adelman, Y. Boucher, R. K. Jain and L. L. Munn, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 911–916 CrossRef CAS PubMed.
- X. Qiu, J. De Jesus, M. Pennell, M. Troiani and J. B. Haun, Lab Chip, 2015, 15, 339–350 RSC.
- A. I. Martínez-Banderas, A. Aires, F. J. Teran, J. E. Perez, J. F. Cadenas, N. Alsharif, T. Ravasi, A. L. Cortajarena and J. Kosel, Sci. Rep., 2016, 6, 35786 CrossRef PubMed.
- R. Mansell, T. Vemulkar, D. C. M. C. Petit, Y. Cheng, J. Murphy, M. S. Lesniak and R. P. Cowburn, Sci. Rep., 2017, 7, 4257 CrossRef PubMed.
- Y. Shen, C. Wu, T. Q. P. Uyeda, G. R. Plaza, B. Liu, Y. Han, M. S. Lesniak and Y. Cheng, Theranostics, 2017, 7, 1735–1748 CrossRef CAS PubMed.
- A. N. Gasparski, S. Ozarkar and K. A. Beningo, J. Cell Sci., 2017, 130, 1965 CrossRef CAS PubMed.
- H. Wang, X. Yang, W. Shao, S. Chen, J. Xie, X. Zhang, J. Wang and Y. Xie, J. Am. Chem. Soc., 2015, 137, 11376–11382 CrossRef CAS PubMed.
- L. Cheng, C. Yuan, S. Shen, X. Yi, H. Gong, K. Yang and Z. Liu, ACS Nano, 2015, 9, 11090–11101 CrossRef CAS PubMed.
- G. Song, J. Hao, C. Liang, T. Liu, M. Gao, L. Cheng, J. Hu and Z. Liu, Angew. Chem., Int. Ed., 2016, 55, 2122–2126 CrossRef CAS PubMed.
- D. Chimene, D. L. Alge and A. K. Gaharwar, Adv. Mater., 2015, 27, 7261–7284 CrossRef CAS PubMed.
- S. Kapri, R. Majee and S. Bhattacharyya, ACS Sustainable Chem. Eng., 2018, 6, 8503–8514 CrossRef CAS.
- S. Kapri and S. Bhattacharyya, ACS Appl. Nano Mater., 2018, 1, 5237–5245 CrossRef CAS.
- S. Kapri and S. Bhattacharyya, Chem. Sci., 2018, 9, 8982–8989 RSC.
- N. Parker, M. J. Turk, E. Westrick, J. D. Lewis, P. S. Low and C. P. Leamon, Anal. Biochem., 2005, 338, 284–293 CrossRef CAS PubMed.
- R. A. Dixon and I. Chopra, J. Antimicrob. Chemother., 1986, 18, 557–563 CrossRef CAS PubMed.
- M. Mueller, U. Grauschopf, T. Maier, R. Glockshuber and N. Ban, Nature, 2009, 459, 726–730 CrossRef CAS PubMed.
- R. Joshi, S. Adhikari, B. S. Patro, S. Chattopadhyay and T. Mukherjee, Free Radical Biol. Med., 2001, 30, 1390–1399 CrossRef CAS PubMed.
- J. C. Phillips, R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, E. Villa, C. Chipot, R. D. Skeel, L. Kalé and K. Schulten, J. Comput. Chem., 2005, 26, 1781–1802 CrossRef CAS PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed.
- M. Kajstura, H. D. Halicka, J. Pryjma and Z. Darzynkiewicz, Cytometry, Part A, 2007, 71, 125–131 CrossRef PubMed.
- L. Henderson, D. S. Bortone, C. Lim and A. C. Zambon, Am. J. Physiol., 2013, 304, C927 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental section; zeta potential of Si-NSs; loading concentrations of ligands; AFM height profile of modified NSs; TEM images of U3 and antibody conjugated U1 NSs; FTIR spectra; HAADF-STEM image and EDS mapping of U3 NS; ICP-MS analysis; temporal internalization of Si-NSs by FESEM and EDS; FESEM images; epifluorescence microscopy; MD simulation data of U3–U7 NSs; still images from a movie file of the MD simulation revealing phospholipid extraction; apoptosis assay; TUNEL assay; DNA ladder test; ROS generation; cytotoxicity through MTT assay; annexin-PI staining; and cell cycle analysis by flow cytometry. See DOI: 10.1039/c9sc00076c |
| This journal is © The Royal Society of Chemistry 2019 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 440, and the dimension of the membrane along the
440, and the dimension of the membrane along the