Open Access Article
Open Access ArticleTelomerase and poly(ADP-ribose) polymerase-1 activity sensing based on the high fluorescence selectivity and sensitivity of TOTO-1 towards G bases in single-stranded DNA and poly(ADP-ribose)†
Haitang
Yang
a,
Fangjia
Fu
b,
Wei
Li
 b,
Wei
Wei
b,
Wei
Wei
 *a,
Yuanjian
Zhang
*a,
Yuanjian
Zhang
 a and
Songqin
Liu
a and
Songqin
Liu
 *a
*a
aJiangsu Engineering Laboratory of Smart Carbon-Rich Materials and Device, Jiangsu Province Hi-Tech Key Laboratory for Bio-medical Research, School of Chemistry and Chemical Engineering, Southeast University, Nanjing, 211189, China. E-mail: weiw@seu.edu.cn; liusq@seu.edu.cn; Fax: +86-25-52090618; Tel: +86-25-52090613
bInstitution of Theoretical and Computational Chemistry, School of Chemistry and Chemical Engineering, Nanjing University, Nanjing 210023, People's Republic of China
First published on 19th February 2019
Abstract
Telomerase and poly(ADP-ribose) polymerase-1 (PARP-1) are two potential cancer biomarkers and are closely related to tumor initiation and malignant progression. TOTO-1 is well-known for differentiating ss-DNA from ds-DNA because it is virtually non-fluorescent without DNA and exhibits very low fluorescence with ss-DNA, while it emits strong fluorescence with ds-DNA. In this paper, for the first time, it was found that TOTO-1 has high fluorescence selectivity and sensitivity towards the G bases in single-stranded DNA and poly(ADP-ribose) (PAR). Poly(dG) was used as the model target to explore its possible mechanism. Molecular dynamics (MD) simulation proved that intramolecular π–π stacking existed in TOTO-1 (in an aqueous solution), while intermolecular π–π stacking formed between TOTO-1 and poly(dG) in a similar way as that observed for dsDNA. Interestingly, telomerase and PARP-1 catalyzed the formation of G-rich DNA and PAR in vivo, respectively. Therefore, TOTO-1 was explored in detecting both of them, obtaining satisfactory results. To the best of our knowledge, no probe has been reported to recognize PAR. It is also the first time where telomerase is detected based on the specific recognition of G bases. Importantly, integrating multiple functions into one probe that can detect not only telomerase but also PARP-1 will significantly raise the specificity of screening cancer and decrease false positive proportion, which make TOTO-1 a promising candidate probe for clinical diagnosis and pharmaceutical screening.
Introduction
Telomerase and poly(ADP-ribose) polymerase-1 (PARP-1) are two potential cancer biomarkers and are closely related to tumor initiation and malignant progression.1,2 Integrating multiple functions into one probe that can detect not only telomerase but also PARP-1 will significantly raise the specificity of screening cancer and decrease false positive proportion. To the best of our knowledge, few probes have been reported to detect not only telomerase but also PARP-1.Most telomerase detection methods depend on the unique property of its extended primer, 5′-AAT CCG TCG AGC AGA GTT (TTAGGG)n-3′. First, the extended primer is a powerful displacement strand that can be used to initiate the signal “turn-on” or “turn-off” based on the hybridization of DNA.3–5 Second, the extended G-rich primer forms a G-quadruplex, which has high peroxidase-like activity and can initiate signals closely related to the catalyzed reaction.6–9 Third, the extended DNA sequence has ample negative charges that have great influence on the signal depending on the electrostatic interactions.10–12 Only few telomerase detection methods have been reported based on the specific recognition of bases.
Most detection methods for PARP-1 are based on its catalyzed production of PAR that has a large number of negative charges, which can adsorb positively charged probes by electrostatic attraction.13–15 These methods do not have good selectivity because of the high background signals. Because PAR lacks unique properties as the extended telomerase primer, very few methods have been developed for its detection even though its activity is very important for assessing cancer development.16,17
Fluorescence analysis has been widely applied in detecting biomolecules because of its inherent advantages of being simple, convenient and sensitive. The fluorescence properties of dyes differ widely when they are in different environments, and they are strongly dependent on the local environment of the macromolecules and their circumstances.18 DNA carries the genetic information of all known living organisms. The ability to specifically discriminate DNA sequences is significant for disease screening and oncology studies.19 Some fluorescence probes that specifically recognize DNA sequences have been developed.20–22 Fluorescence nanoparticles such as DNA-silver nanoclusters23–26 and small organic dyes such as thioflavin T,27–29 ethidium bromide (EtBr),30 thiazole orange (TO),31 TOTO,32 and iridium complex33 can bind to DNA chains and show various fluorescence properties depending on their manner of binding and binding ability. Some of them can distinguish ss-DNA from ds-DNA or G-quadruplex DNA. However, only a few fluorescence probes are known to effectively distinguish deoxynucleotides34 even though they are significant for DNA sequence discrimination, bio-analysis and clinical diagnosis.
TOTO-1, an unsymmetrical cyanine dye dimer, is well-known for differentiating ss-DNA from ds-DNA (Fig. S1†).35,36 It is virtually non-fluorescent without DNA and exhibits strong fluorescence when it binds to ds-DNA (Fig. S2†).37,38 In this paper, for the first time, we found that TOTO-1 has unique fluorescence selectivity and high sensitivity towards the G bases in single-stranded DNA and PAR. Its fluorescence intensity was much stronger with poly(dG) than those with poly(dA), poly(dC) and poly(dT) (Fig. S2A†). More interestingly, the fluorescence of TOTO-1 with poly(dG) was far stronger than that with dGTP even when the concentration of dGTP was 4 orders of magnitude higher than that of the G bases in poly(dG). Furthermore, the fluorescence of TOTO-1 with PAR increased sharply compared to those with higher concentrations of ADP or poly(dA).
These findings urged us to develop a simple and sensitive method to assay the activities of both telomerase and PARP-1; they catalyzed the formation of a G-rich elongated TS primer and PAR in vivo, respectively, which were highly sensitive to TOTO-1. The high background signals produced by the telomerase primer and the activated dsDNA were reduced by 9 times and 38 times, respectively, under the influence of digestion by the Exo III enzyme. As a result, the detection sensitivity improved greatly. A linear relationship between the fluorescence intensities and logarithmic concentrations of telomerase was observed between 13 and 4000 cells per mL, and the limit of detection (LOD) was determined to be 13 cells per mL. Also, the fluorescence intensities showed a linear response between 0.02 U and 1.5 U for PARP-1 with LOD of 0.02 U. This simple and sensitive approach is promising for clinical diagnosis and is a powerful tool for pharmaceutical screening.
Experimental section
Molecular dynamics (MD) simulation
The initial single-stranded poly(dG) structure was constructed using the NAB module in the AmberTools16 package.39 Then, the configuration of TOTO-1 binding to poly(dG) was constructed using AutoDock 4.0 with the Lamarckian genetic algorithm.40 The ground-state geometry of free TOTO-1 molecule was first optimized at the M06-2X/6-311++G(d,p) level with the polarizable continuum model (PCM), and the RESP charge of the TOTO-1 molecule was calculated using the GAUSSIAN09 program.41 The configurations of TOTO-1 without and with poly(dG) at the center with 8000 water molecules were simulated at 298 K in cubic boxes of 56.61 × 56.61 × 56.61 Å3 and 62.46 × 62.46 × 62.46 Å3, respectively. The generalized amber force field (GAFF) and FF99BSC0 force field were used for the TOTO-1 molecule and poly(dG), respectively. All the water molecules were described by the TIP4PFB force field, which was found to be able to reproduce the experimental properties of liquid water.42 Cl− and Na+ counterions were added to neutralize the system of TOTO-1 without and with poly(dG) in aqueous solutions, respectively. The simulations of the two systems were run on the NPT ensemble at 1.0 bar with a Berendsen barostat. Periodic boundary conditions (PBC) were carried out and the long-range electrostatic interactions were taken into account using the particle mesh Ewald (PME) method. The velocity Verlet algorithm was employed to solve Newton's motion equations with the bonds of hydrogen atoms constrained. The van der Waals cutoff was set as 10 Å for the two systems. For TOTO-1 without and with poly(dG) in aqueous solutions, two simulation trajectories of 20 ns after 2 ns of equilibrium were carried out and snapshots were collected at the interval of 1 ps with every time step of 1 fs.Hybrid quantum mechanical and molecular mechanical (QM/MM) calculation
The electronic absorption spectra of TOTO-1 without and with poly(dG) in aqueous solutions were calculated using the QM/MM method at the level of time-dependent density functional theory (TDDFT) with the M06-2X functional and the 6-31G(d) basis set. TOTO-1 molecule and TOTO-1 with poly(dG) were treated in the QM part for the two systems, and the remaining molecules including the counterions and water molecules were treated as background point charges in the MM region.Procedures of telomerase activity and inhibition efficiency evaluation
Cell cultivation, telomerase extraction and primer extension reaction were performed according to previously described methods (ESI†). The extended G-rich sequence was hybridized with cDNA (complementary to the TS primer) at 95 °C for 5 min, followed by cooling down slowly to room temperature. Then, Exo III was used to digest the duplex part formed by cDNA and TS primer at 37 °C. The resulting solution (30 µL) was diluted to 200 µL with PBS buffer containing 250 nM TOTO-1. Then, the fluorescence spectra were recorded. Various concentrations of BIBR 1532 or curcumin were incubated with 750 cells per mL A549 cells to evaluate their telomerase inhibition efficiency. Heated A549 cells (95 °C, 5 min) were used for control experiments.Procedures of PARP-1 activity and inhibition efficiency evaluation
First, active dsDNA was prepared by hybridizing two complementary sequences ssDNA1 and ssDNA2 in a hybridization buffer at 95 °C for 5 min, followed by cooling to room temperature slowly. Then, dsDNA (150 nM) was incubated with PARP-1, NAD+ (250 µM) and reaction buffer (R-buffer) for 1 h at room temperature. After PARP-1 catalyzed the polymerization of PAR, dsDNA was digested by Exo III at 37 °C to reduce the background signal. The resulting solution was mixed with PBS buffer containing 250 nM TOTO-1. Fluorescence spectra were recorded. For PARP-1 inhibitor assay, the same procedure was carried out except for the treatment of PARP-1 with different concentrations of AG014699.Result and discussion
High fluorescence selectivity and sensitivity of TOTO-1 towards the G bases in single-stranded DNA and PAR
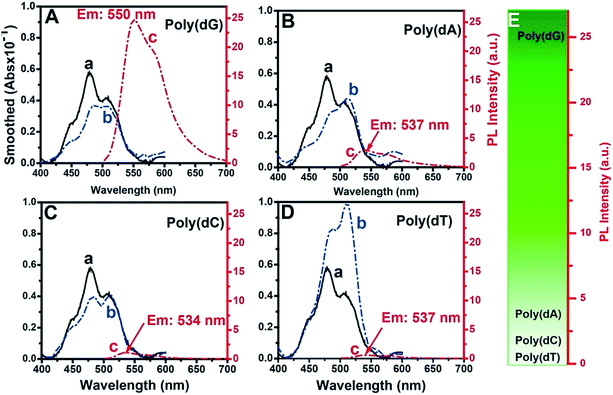
The fluorescence selectivities of TOTO-1 towards 15 mM dGTP, dATP, dCTP, dTTP and ADP were detected (Fig. S3†). The results showed that TOTO-1 has much stronger fluorescence with dGTP or ADP than that with dATP, dCTP or dTTP. Considering that TOTO-1 was used to respond to G-rich DNA sequences catalyzed by telomerase, poly(dG) was used as the model target in this study. Compared with the result for 15 mM of dGTP, the fluorescence intensity of TOTO-1 increased from 1.5 to 25 when only 86 nM poly(dG) (690 nM of G bases) was present. These data indicated that TOTO-1 is more sensitive to the G bases existing in single-stranded DNA than single dGTP, which is significant for the detection of extended DNA sequences catalyzed by telomerase. The absorbance and fluorescence spectra of TOTO-1 are shown in Fig. 1. From curves (a and b) in Fig. 1A–D, it can be seen that their absorption spectra vary significantly in the presence of different oligodeoxynucleotides, indicating that the molecular orbitals of TOTO-1 were changed by these oligodeoxynucleotides. The orbital distribution of the excited state transitions for TOTO-1/poly(dG) corresponding to the absorbance at 454 nm and 542 nm were simulated (Fig. S4†). The fluorescence intensity of TOTO-1 in the presence of different types of oligonucleotides varied greatly (curve c, Fig. 1A–D). Specifically, TOTO-1 in the presence of poly(dG) yielded much higher signals than those with the three other types of oligonucleotides. The quantum yields in the presence of poly(dG), poly(dA), poly(dC), and poly(dT) were determined to be 0.25, 0.11, 0.05 and 0.02, respectively. These data indicated that TOTO-1 showed strong fluorescence selectivity towards four types of bases: poly(dG) ≫ poly(dA) > poly(dC) > poly(dT). Fig. 1E shows the comparison of fluorescence intensities of TOTO-1 in the presence of different types of oligonucleotides.MD simulation of the interaction between TOTO-1 and poly(dG)
MD simulation indicated that intramolecular π–π stacking existed in free TOTO-1, while intermolecular π–π stacking formed between TOTO-1 and poly(dG) (Fig. 2A). As a result, a folding structure similar to that of duplex DNA was formed. The absorption spectrum calculated by M06-2X/6-31G(D) was highly consistent with the experimental spectrum (Fig. 2B), which demonstrated the intermolecular π–π stacking in poly(dG)/TOTO-1. Other fluorescent dyes such as fluorescein sodium, thioflavin T and acriflavine did not have fluorescence selectivity towards the bases (Fig. S5†). Although TO showed selectivity similar to that of TOTO-1, its PL intensity was much lower (30%) than that of TOTO-1 in the presence of poly(dG) and poly(dA) (Fig. S5C†).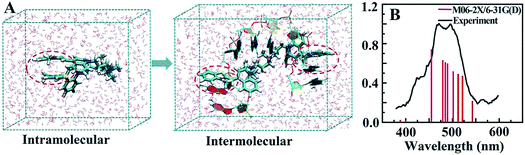 | ||
| Fig. 2 (A) Intramolecular π–π stacking of free TOTO-1 and intermolecular π–π stacking of TOTO-1/poly(dG). (B) Simulated and experimental absorption spectra of TOTO-1 in the presence of poly(dG). | ||
Fluorescence of TOTO-1 in the presence of G-rich elongated TS primer and PAR
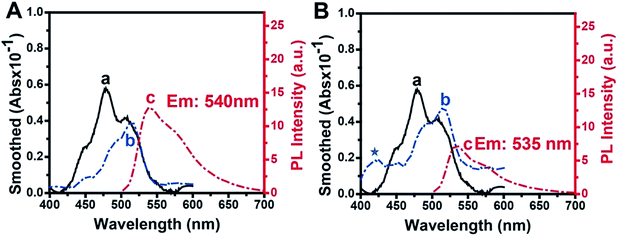
The absorbance and fluorescence spectra of TOTO-1 in the presence of G-rich elongated TS primer (Fig. 3A) and PAR (Fig. 3B) were also studied. In the presence of G-rich elongated TS primer, the absorbance and fluorescence spectra of TOTO-1 changed obviously compared with those in the presence of poly(dG) and poly(dA). Compared with the observation for poly(dG), the fluorescence intensity for the elongated TS primer decreased from 25 to 12.6, indicating that thymine and adenine certainly decreased the fluorescence emission of TOTO-1. The corresponding quantum yield decreased to 0.16. Under this circumstance, its fluorescence was still sensitive enough to distinguish the G-rich elongated TS primer, making it possible to detect telomerase activity.PAR, synthesized by PARP-1, is composed of adenosine diphosphate ribose (ADP-ribose) monomers (Fig. S6†). In the presence of PAR, the absorbance spectrum of TOTO-1 showed significant changes with a new absorbance peak at 425 nm and increased absorbance values at 480 nm and 508 nm, indicating that the molecular orbitals of TOTO-1 were also different from those in the presence of poly(dA). Although its maximum fluorescence emission wavelength changed slightly, its fluorescence intensity increased by 2.5 times. The corresponding quantum yield also increased from 0.11 to 0.32, which definitely contributed to improving PARP-1 activity detection. Furthermore, compared with the result for 15 mM of ADP, the fluorescence intensity of TOTO-1 increased from 0.7 to 7 in the presence of PAR produced by 0.8 U PARP-1. This also indicates that TOTO-1 is more sensitive to polymerized ADP than single ADP. Thus, it is possible to construct a sensitive and simple method to detect telomerase and PARP-1 activities based on the unique fluorescence selectivity and sensitivity of TOTO-1 towards the G-rich elongated primer and PAR.
The strategy for telomerase activity evaluation
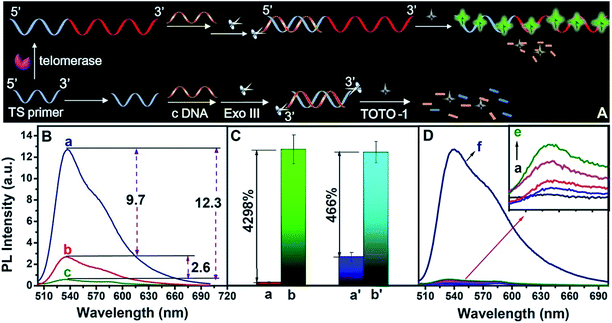
The scheme for telomerase activity detection is illustrated in Fig. 4A. The telomerase primer was incubated with dNTPs and telomerase was extracted from tumor A549 cells. The TTAGGG repeat units were continuously synthesized on the 3′ end of the TS primer to form a G-rich extended primer. In the presence of telomerase, the G-rich elongated primer led to a sharp increase in the fluorescence intensity of TOTO-1 (a, Fig. 4B). In the presence of TS primer without telomerase, the background signal was 2.9 (b, Fig. 4B). A primer was designed to form ds-DNA by incubation with complementary DNA (c-DNA), followed by digesting with Exo III; thus, the background signal decreased to 0.30 (c, Fig. 4B). The signal-to-noise ratio (S/N) increased by nearly 9 times (Fig. 4C). The control experiment results are shown in Fig. 4D. The fluorescence intensities of TOTO-1 in the presence of dNTPs, heated A549 cells and Exo III were negligible. Its fluorescence increased sharply when TOTO-1 was incubated with the primer, dNTPs and A549 cells, indicating that telomerase triggered the fluorescence of the system (f, Fig. 4D).The strategy for PARP-1 activity evaluation
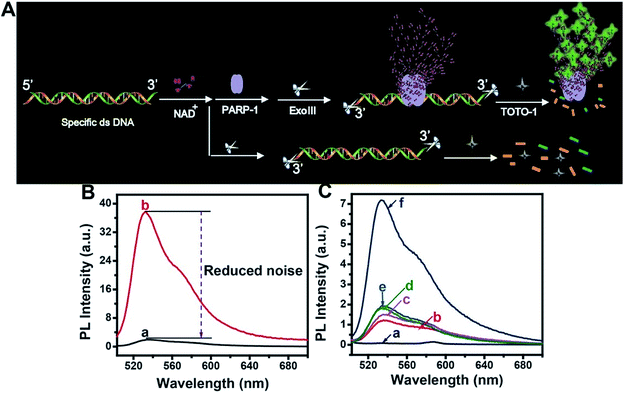
The scheme for PARP-1 activity detection is illustrated in Fig. 5A. PARP-1 was activated by specific dsDNA. Then, PARP-1 transferred the first ADP-ribose unit from nicotinamide adenine dinucleotide (NAD+) to an acceptor protein and sequentially added multiple ADP-ribose units to the preceding ones to form hyper-branched PAR. However, the background fluorescence produced by the activated dsDNA was very high because TOTO-1 showed strong fluorescence in the presence of dsDNA (curve b, Fig. 5B). It was also necessary to use Exo III to reduce the background signal. From curve a in Fig. 5B, we can see that the background signal is reduced by 38 times, which makes it possible to detect the PARP-1 activity sensitively. The control experiments for PARP-1 detection are shown in Fig. 5C. Only activated PARP-1 could trigger the fluorescence of the system, while other substances such as NAD+, heated PARP-1, and Exo III exhibited negligible effects on the fluorescence intensity.Performances for telomerase and PARP-1 analysis
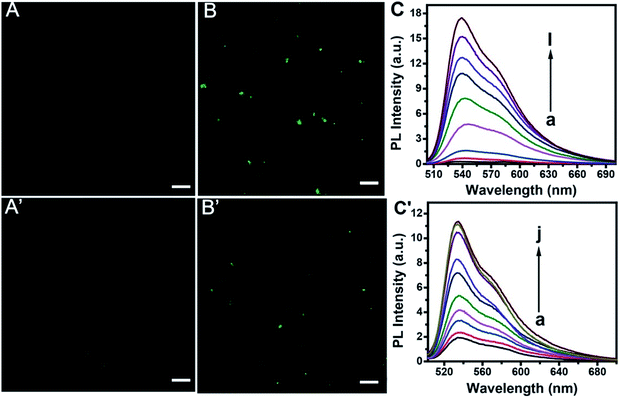
Confocal imaging gave further evidence for the feasibility of the proposed method. As is shown in Fig. 6, TOTO-1 exhibits very weak fluorescence in the presence of the telomerase and PARP-1 detection system without telomerase (A) or PARP-1 (A′). The presence of telomerase (B) or PARP-1 (B′) triggered the fluorescence of TOTO-1 sharply due to the formation of G-rich elongated primer and PAR, respectively.The fluorescence spectra for telomerase and PARP-1 detections are shown in Fig. 6C and C′, respectively. The optimization of the detection conditions is shown in Fig. S7 and S8.† PL intensity showed a linear correlation with increasing concentration of telomerase from 13 to 4000 A549 cells per mL. The regression equation is Y = −0.75 + 4.79![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lg
lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C, (R2 = 0.997). A linear relationship between the PL intensity and PARP-1 ranging from 0.02 U to 1.5 U (0.04 nM to 3 nM) was obtained Y = 2.66 + 5.23C (R2 = 0.992), where Y is the PL intensity and C is the content of A549 cells or PARP-1. The detection limit for telomerase and PARP-1 was 13 cells per mL and 0.02 U, respectively. Compared with the previously reported methods for telomerase detection listed in Table 1, it can be concluded that this method is more sensitive than the UV methods.6,43 It is also comparable to other sensitive methods such as ECL,40–47 CL,9 SERS,48 and fluorescence methods that use other probes.49–53 It can also be seen that the detection sensitivity for PARP-1 is comparable to those of other reported methods (Table S2†).
C, (R2 = 0.997). A linear relationship between the PL intensity and PARP-1 ranging from 0.02 U to 1.5 U (0.04 nM to 3 nM) was obtained Y = 2.66 + 5.23C (R2 = 0.992), where Y is the PL intensity and C is the content of A549 cells or PARP-1. The detection limit for telomerase and PARP-1 was 13 cells per mL and 0.02 U, respectively. Compared with the previously reported methods for telomerase detection listed in Table 1, it can be concluded that this method is more sensitive than the UV methods.6,43 It is also comparable to other sensitive methods such as ECL,40–47 CL,9 SERS,48 and fluorescence methods that use other probes.49–53 It can also be seen that the detection sensitivity for PARP-1 is comparable to those of other reported methods (Table S2†).
| Method | System | Detection range | Detection limit | Reference |
|---|---|---|---|---|
| UV-vis | Hemin-graphene conjugate-based biosensor | 100–2300 cells per mL | 60 cells per mL | 10 |
| UV-vis | Primer-modified GNPs | 0–200 cells per mL | 29 cells per mL | 43 |
| ECL | DNA tetrahedral scaffold-based platform | 0–32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL 000 cells per mL |
70 cells per mL | 44 |
| ECL | Porphyrin-functionalized graphene | 10–750 cells per mL | 10 cells per mL | 45 |
| ECL | Ruthenium polyethylenimine complex-doped ZIF-8 | 50–106 cells | 11 cells | 46 |
| ECL | Polyluminol-PtNPs composite | 15–9000 cells | 15 cells | 47 |
| CL | Hemin-G-quadruplex | 1–1000 cells | 1 cell | 9 |
| SERS | ABT modified AuNPs | 5–100 cells | 1 cell | 48 |
| Fluorescence | TS primer-modified AuNPs | 10–1000 cells | 1 cell | 49 |
| Fluorescence | Cy3-labeled DNA | 10–2000 cells | 10 cells | 50 |
| Fluorescence | AIEgens | 5–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells 000 cells |
5 cells | 51 |
| Fluorescence | GNRs-based FRET | 1–500 cells | 1 cell | 52 |
| Fluorescence | FRET-based TS-FAM/CCP platform | 5–2000 cells | 5 cells | 53 |
| Fluorescence | TOTO-1 | 2–4000 cells per mL | 13 cells per mL | This work |
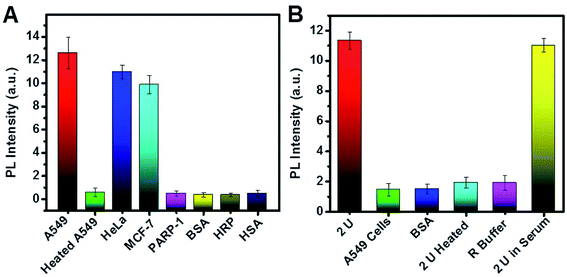
The practicality and selectivity for telomerase activity detection were verified by using other kinds of cell lines, proteins and enzymes (Fig. 7A). The PL intensity increased significantly for A549, HeLa, and MCF-7 cells and was negligible for heated A549 cells, PARP-1, HRP, BSA, and HSA, indicating that high PL resulted from telomerase in these cancer cell lines. As shown in Fig. 7B, we also studied the selectivity of the biosensor for PARP-1. The experimental results indicate no obvious change in PL when PARP-1 was replaced by BSA, heated PARP-1, and telomerase, suggesting satisfactory selectivity for PARP-1 detection.
Application of the strategy to detect telomerase in urine and PARP-1 in cells
Telomerase was detected by the proposed method in 11 early morning urine samples provided by the Nanjing General Hospital of Chinese People's Liberation Army. A threshold value of 0.59 was calculated from the mean fluorescence intensity of 100 blank samples plus 3 times the relative standard deviation. The detection results are shown in Table 2. Sample 1 is the blank solution and samples 2–8 were obtained from normal, vesical calculus, kidney stone and inflammation patients. All PL intensities were lower than the threshold value. Samples 9–12 were obtained from bladder cancer patients. All their results were far higher than the threshold value. Therefore, this painless method has great potential to be used for bladder cancer diagnosis.| Telomerase | Patient ID | Clinical outcome | TRAP | PL intensity |
|---|---|---|---|---|
| a “−” or “+” means negative or positive for telomerase, respectively. | ||||
| 1 | Water | − | 0.25 | |
| 2 | Normal | Normal | − | 0.3 |
| 3 | 1003295831 | Vesical calculus | − | 0.3 |
| 4 | 1008455348 | Bladder cancer | − | 10.5 |
| 5 | 1002693440 | Kidney stone | − | 0.35 |
| 6 | 1008455719 | Kidney stone | − | 0.31 |
| 7 | 1008461668 | Kidney stone | − | 0.29 |
| 8 | 1007144483 | Inflammation | − | 0.25 |
| 9 | 1007677148 | Bladder cancer | + | 5 |
| 10 | 1007922178 | Bladder cancer | + | 4 |
| 11 | 1007045669 | Bladder cancer | + | 12 |
| 12 | 1007924814 | Bladder cancer | + | 7 |
| PARP-1 | Cells | Nucleus (U) | Plasma (U) | |
| 13 | IOSE80 | 0.05 | 0.02 | |
| 14 | MCF-7 | 0.67 | 0.53 | |
| 15 | SK-BR-3 | 0.74 | 0.5 | |
The PARP-1 levels in normal cells of IOSE80 and breast cancer cells of MCF-7 and SK-BR-3 were evaluated. The detection results for samples 13–15 showed that PARP-1 was present at a low level in normal human cells, while it was significantly up-regulated in the malignant SK-BR-3 cells and MCF-7 cells. PARP-1 was higher in the nucleus than that in the cytoplasm of both SK-BR-3 cells and MCF-7 cells, which was consistent with previous results.54,55 Recovery experiments were conducted in human serum, normal cells IOSE80 and human ovarian cancer cells A2780 to prove the accuracy of the sensor in complex biological matrices. Recoveries were from 94% to 109% and the relative standard derivations were in the range of 2.48–8.21%, indicating that the method has good accuracy and high precision (Table S4†).
Detection of inhibition efficiency on telomerase and PARP-1
To test the inhibitor screening function of the proposed sensor, BIBR1532 and curcumin were chosen as the model inhibitors for telomerase and AG014699 was selected as the model inhibitor for PARP-1. The PL intensities decreased continually to a stable value when the amounts of BIBR 1532 and curcumin increased (Fig. S9A and C†). Decreased percentages of the PL intensity in the presence of various concentrations of BIBR 1532 and curcumin are shown in Fig. S9B and D.† A549 without an inhibitor was used as the control (750 cells per mL). The IC50 (concentration of inhibitor producing 50% inhibition) values for BIBR1532 and curcumin against telomerase were 251 nM and 8.64 µM, respectively. These results were in accordance with previously reported values.56,57 The fluorescence spectra in the presence of PARP-1 inhibited by different concentrations of AG014699 are shown in Fig. S9E.† The decreased percentages of the PL intensity are shown in Fig. S9F.† The IC50 value of AG014699 was determined to be 8.2 nM; it was also in good agreement with the reported IC50 value in literature.58Conclusions
In summary, the unique fluorescence selectivity and sensitivity of TOTO-1 towards the G bases in single-stranded DNA and PAR were found for the first time and studied elaborately. MD simulation proved that intermolecular π–π stacking formed between TOTO-1 and poly(dG) and TOTO-1/poly(dG) folded in a way analogous to that of duplex DNA, which explained why TOTO-1 has high selectivity towards poly(dG). TOTO-1 is also the first probe that has been reported to have high selectivity and sensitivity towards PAR. A label-free, simple and sensitive fluorescence biosensor was constructed for telomerase and PARP-1 sensing based on TOTO-1 and Exo III-assisted background noise reduction. This strategy shows high sensitivity and selectivity towards telomerase and PARP-1. Integrating multiple functions into one simple probe, i.e., detecting not only telomerase but also PARP-1 can significantly raise the specificity of screening cancer and decrease false positive proportion. This makes TOTO-1 a promising candidate probe for clinical diagnosis. However, the probe cannot be used to image telomerase or PARP-1 in vivo because Exo III is necessary to decrease the background signals and it is difficult to be transferred into cells.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We genuinely express our gratitude to Prof. Jing Ma, Institution of Theoretical and Computational Chemistry, Nanjing University, for her MD simulation of the interaction between TOTO-1 and poly (dG). This work was supported by the National Natural Science Foundation of China (No. 21775019, 21475020, 21635004, 81730087). Fundamental Research Funds for the Central Universities and A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (No. 2242018K3DN04), The Open Project of The Key Laboratory of Modern Toxicology of Ministry of Education, Nanjing Medical University (NMUMT201804).References
- L. J. Wang, F. Ma, B. Tang and C. Y. Zhang, Chem. Sci., 2017, 8, 2495–2502 RSC.
- M. F. Langelier, J. L. Planck, S. Roy and J. M. Pascal, Science, 2012, 336, 728 CrossRef CAS PubMed.
- R. C. Qian, L. Ding, L. Yan, M. Lin and H. X. Ju, J. Am. Chem. Soc., 2014, 136, 8205–8208 CrossRef CAS PubMed.
- M. Hong, L. D. Xu, Q. W. Xue, L. Li and B. Tang, Anal. Chem., 2016, 88, 12177–12182 CrossRef CAS PubMed.
- X. W. Xu, L. Wang, K. Li, Q. H. Huang and W. Jiang, Anal. Chem., 2018, 90, 3521–3530 CrossRef CAS PubMed.
- H. T. Yang, A. R. Liu, M. Wei, Y. J. Liu, B. J. Lv, W. Wei, Y. J. Zhang and S. Q. Liu, Anal. Chem., 2017, 89, 12094–12100 CrossRef CAS PubMed.
- R. Freeman, E. Sharon, C. Teller, A. Henning, Y. Tzfati and I. Willner, ChemBioChem, 2010, 11, 2362–2367 CrossRef CAS PubMed.
- C. L. Wang, H. T. Yang, S. S. Wu, Y. J. Liu, W. Wei, Y. J. Zhang, M. Wei and S. Q. Liu, TrAC, Trends Anal. Chem., 2018, 105, 404–412 CrossRef CAS.
- L. J. Wang, Y. Zhang and C. Y. Zhang, Anal. Chem., 2013, 85, 11509–11517 CrossRef CAS PubMed.
- X. L. Xu, M. Wei, Y. J. Liu, X. Liu, W. Wei, Y. J. Zhang and S. Q. Liu, Biosens. Bioelectron., 2017, 87, 600–606 CrossRef CAS PubMed.
- X. Liu, M. Wei, E. S. Xu, H. T. Yang, W. Wei, Y. J. Zhang and S. Q. Liu, Biosens. Bioelectron., 2017, 91, 347–353 CrossRef CAS PubMed.
- X. Liu, M. Wei, Y. J. Liu, B. J. Lv, W. Wei, Y. J. Zhang and S. Q. Liu, Anal. Chem., 2016, 88, 8107–8114 CrossRef CAS PubMed.
- S. Y. Tang, Z. Nie, W. Li, D. Q. Li, Y. Huang and S. Z. Yao, Chem. Commun., 2015, 51, 14389–14392 RSC.
- Y. Y. Xu, L. Liu, Z. Y. Wang and Z. H. Dai, ACS Appl. Mater. Interfaces, 2016, 8, 18669–18674 CrossRef CAS PubMed.
- S. S. Wu, M. Wei, H. T. Yang, J. H. Fan, W. Wei, Y. J. Zhang and S. Q. Liu, Sens. Actuators, B, 2018, 259, 565–572 CrossRef CAS.
- X. J. Yang, K. Zhang, T. T. Zhang, J. J. Xu and H. Y. Chen, Anal. Chem., 2017, 89, 4216–4222 CrossRef CAS PubMed.
- R. Krishnakumar and W. L. Kraus, Mol. Cell, 2010, 39, 8–24 CrossRef CAS PubMed.
- N. Dai and E. T. Kool, Chem. Soc. Rev., 2011, 40, 5756–5770 RSC.
- J. Liang, B. Z. Tang and B. Liu, Chem. Soc. Rev., 2015, 44, 2798–2811 RSC.
- H. Zhou, J. Liu, J. J. Xu, S. S. Zhang and H. Y. Chen, Chem. Soc. Rev., 2018, 47, 1996–2019 RSC.
- H. Zhang, F. Li, B. Dever, X. F. Li and X. C. Le, Chem. Rev., 2013, 113, 2812–2841 CrossRef CAS PubMed.
- H. Peng, A. M. Newbigging, Z. Wang, J. Tao, W. Deng, X. C. Le and H. Zhang, Anal. Chem., 2018, 90, 190–207 CrossRef CAS PubMed.
- H. C. Yeh, J. Sharma, J. J. Han, J. S. Martinez and J. H. Werner, Nano Lett., 2010, 10, 3106–3110 CrossRef CAS PubMed.
- H. C. Yeh, J. Sharma, M. Shih Ie, D. M. Vu, J. S. Martinez and J. H. Werner, J. Am. Chem. Soc., 2012, 134, 11550–11558 CrossRef CAS PubMed.
- W. Zhou, J. Zhu, D. Fan, Y. Teng, X. Zhu and S. Dong, Adv. Funct. Mater., 2017, 27, 1704092 CrossRef.
- Y. Zhang, C. Zhu, L. Zhang, C. Tan, J. Yang, B. Chen, L. Wang and H. Zhang, Small, 2015, 11, 1385–1389 CrossRef CAS PubMed.
- J. Mohanty, N. Barooah, V. Dhamodharan, S. Harikrishna, P. I. Pradeepkumar and A. C. Bhasikuttan, J. Am. Chem. Soc., 2013, 135, 367–376 CrossRef CAS PubMed.
- A. Renaud de la Faverie, A. Guedin, A. Bedrat, L. A. Yatsunyk and J. L. Mergny, Nucleic Acids Res., 2014, 42, e65 CrossRef CAS PubMed.
- S. Liu, P. Peng, H. Wang, L. Shi and T. Li, Nucleic Acids Res., 2017, 45, 12080–12089 CrossRef CAS PubMed.
- S. Ding, X. Qiao, J. Suryadi, G. S. Marrs, G. L. Kucera and U. Bierbach, Angew. Chem., Int. Ed., 2013, 52, 3350–3354 CrossRef CAS PubMed.
- H. S. Rye, M. A. Quesada, K. Peck, R. A. Mathies and A. N. Giazer, Nucleic Acids Res., 1991, 19, 327–333 CrossRef CAS PubMed.
- Y. Liu, M. Wei, Y. Li, A. Liu, W. Wei, Y. Zhang and S. Liu, Anal. Chem., 2017, 89, 3430–3436 CrossRef CAS PubMed.
- D. L. Ma, S. Lin, K. H. Leung, H. J. Zhong, L. J. Liu, D. S. H. Chan, A. Bourdoncle, J. L. Mergny, H.-M. D. Wang and C.-H. Leung, Nanoscale, 2014, 6, 8489–8494 RSC.
- N. Jan, S. Nicke and K. Mikael, Biopolymers, 1998, 46, 39–51 CrossRef.
- H. S. Rye and A. N. Glazer, Nucleic Acids Res., 1995, 23, 1215–1222 CrossRef CAS PubMed.
- V. L. Singer, L. J. Jones, S. T. Yue and R. P. Haugland, Anal. Biochem., 1997, 249, 228–238 CrossRef CAS PubMed.
- H. S. Rye, S. Yue, D. E. Wemmer, M. A. Quesada, R. P. Haugland, R. A. Mathies and A. N. Glazer, Nucleic Acids Res., 1992, 20, 2803–2812 CrossRef CAS PubMed.
- A. Castro, F. R. Fairfield and E. B. Shera, Anal. Chem., 1993, 65, 849–852 CrossRef CAS.
- D. A. Case, R. M. Betz, D. S. Cerutti, T. E. Cheatham III, T. A. Darden, R. E. Duke, T. J. Giese, H. Gohlke, A. W. Goetz, N. Homeyer, et al., AMBER, University of California, San Francisco, 2016 Search PubMed.
- G. M. Morris, D. S. Goodsell, R. S. Halliday, R. Huey, W. E. Hart, R. K. Belew and A. J. Olson, J. Comput. Chem., 1998, 19, 1639–1662 CrossRef CAS.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. V. Marenich, et al., Gaussian 09, revision D.01, Gaussian, Inc., Wallingford, CT, 2016 Search PubMed.
- L. P. Wang, T. J. Martinez and V. S. Pande, J. Phys. Chem. Lett., 2014, 5, 1885–1891 CrossRef CAS PubMed.
- L. Zhang, S. Zhang, W. Pan, Q. Liang and X. Song, Biosens. Bioelectron., 2016, 77, 144–148 CrossRef CAS PubMed.
- Q. M. Feng, Z. Zhou, M. X. Li, W. Zhao, J. J. Xu and H. Y. Chen, Biosens. Bioelectron., 2017, 90, 251–257 CrossRef CAS PubMed.
- L. Wu, J. S. Wang, L. Y. Feng, J. S. Ren, W. L. Wei and X. G. Qu, Adv. Mater., 2012, 24, 2447–2452 CrossRef CAS PubMed.
- C. Y. Xiong, W. B. Liang, Y. N. Zheng, Y. Zhuo, Y. Q. Chai and R. Yuan, Anal. Chem., 2017, 89, 3222–3227 CrossRef CAS PubMed.
- H. R. Zhang, B. X. Li, Z. M. Sun, H. Zhou and S. S. Zhang, Chem. Sci., 2017, 8, 8025–8029 RSC.
- M. L. Shi, J. Zheng, C. H. Liu, G. X. Tan, Z. H. Qing, S. Yang, J. F. Yang, Y. J. Tan and R. H. Yang, Biosens. Bioelectron., 2016, 77, 673–680 CrossRef CAS PubMed.
- Y. F. Gao, J. Xu, B. X. Li and Y. Jin, ACS Appl. Mater. Interfaces, 2016, 8, 13707–13713 CrossRef CAS PubMed.
- X. Su, Z. H. Li, X. Z. Yan, L. Wang, X. Zhou, L. Wei, L. H. Xiao and C. Y. Yu, Anal. Chem., 2017, 89, 3576–3582 CrossRef CAS PubMed.
- Y. Zhuang, M. Zhang, B. Chen, R. Duan, X. Min, Z. Zhang, F. Zheng, H. Liang, Z. Zhao, X. Lou and F. Xia, Anal. Chem., 2015, 87, 9487–9493 CrossRef CAS PubMed.
- Y. Wang, L. Yang, B. Li and Y. Jin, Analyst, 2016, 141, 6133–6139 RSC.
- C. Chen, M. Wei, Y. Liu, E. Xu, W. Wei, Y. Zhang and S. Liu, Microchim. Acta, 2017, 184, 3453–3460 CrossRef CAS.
- P. Domagala, T. Huzarski, J. Lubinski, K. Gugala and W. Domagala, Breast Cancer Res. Treat., 2011, 127, 861–869 CrossRef CAS PubMed.
- F. Rojo, J. García-Parra, S. Zazo, I. Tusquets, J. Ferrer-Lozano, S. Menendez, P. Eroles, C. Chamizo, S. Servitja, N. Ramírez-Merino, F. Lobo, B. Bellosillo, J. M. Corominas, J. Yelamos, S. Serrano, A. Lluch, A. Rovira and J. Albanell, Ann. Oncol., 2012, 23, 1156–1164 CrossRef CAS PubMed.
- E. Pascolo, C. Wenz, J. Lingner, N. Hauel, H. Priepke, I. Kauffmann, P. Garin-Chesa, W. J. Rettig, K. Damm and A. Schnapp, J. Biol. Chem., 2002, 277, 15566–15572 CrossRef CAS PubMed.
- S. Chakraborty, U. Ghosh, N. P. Bhattacharyya, R. K. Bhattacharya and M. Roy, Mutat. Res., Fundam. Mol. Mech. Mutagen., 2006, 596, 81–90 CrossRef CAS PubMed.
- R. Plummer, C. Jones, M. Middleton, R. Wilson, J. Evans, A. Olsen, N. Curtin, A. Boddy, P. McHugh, D. Newell, A. Harris, P. Johnson, H. Steinfeldt, R. Dewji, D. Wang, L. Robson and H. Calvert, Clin. Cancer Res., 2008, 14, 7917–7923 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8sc05770b |
| This journal is © The Royal Society of Chemistry 2019 |