Open Access Article
Open Access ArticleSelective high-resolution DNP-enhanced NMR of biomolecular binding sites†
Ildefonso
Marin-Montesinos
 a,
David
Goyard
a,
David
Goyard
 b,
Emilie
Gillon
c,
Olivier
Renaudet
b,
Emilie
Gillon
c,
Olivier
Renaudet
 b,
Anne
Imberty
b,
Anne
Imberty
 c,
Sabine
Hediger
c,
Sabine
Hediger
 *a and
Gaël
De Paëpe
*a and
Gaël
De Paëpe
 *a
*a
aUniv. Grenoble Alpes, CEA, CNRS, INAC-MEM, Grenoble, France. E-mail: sabine.hediger@cea.fr; gael.depaepe@cea.fr
bUniv. Grenoble Alpes, CNRS, DCM, Grenoble, France
cUniv. Grenoble Alpes, CNRS, CERMAV, Grenoble, France
First published on 1st February 2019
Abstract
Locating binding sites in biomolecular assemblies and solving their structures are of the utmost importance to unravel functional aspects of the system and provide experimental data that can be used for structure-based drug design. This often still remains a challenge, both in terms of selectivity and sensitivity for X-ray crystallography, cryo-electron microscopy and NMR. In this work, we introduce a novel method called Selective Dynamic Nuclear Polarization (Sel-DNP) that allows selective highlighting and identification of residues present in the binding site. This powerful site-directed approach relies on the use of localized paramagnetic relaxation enhancement induced by a ligand-functionalized paramagnetic construct combined with difference spectroscopy to recover high-resolution and high-sensitivity information from binding sites. The identification of residues involved in the binding is performed using spectral fingerprints obtained from a set of high-resolution multidimensional spectra with varying selectivities. The methodology is demonstrated on the galactophilic lectin LecA, for which we report well-resolved DNP-enhanced spectra with linewidths between 0.5 and 1 ppm, which enable the de novo assignment of the binding interface residues, without using previous knowledge of the binding site location. Since this approach produces clean and resolved difference spectra containing a limited number of residues, resonance assignment can be performed without any limitation with respect to the size of the biomolecular system and only requires the production of one protein sample (e.g.13C,15N-labeled protein).
Introduction
Addressing site-specific atomic scale information of biomolecular binding sites is of primary importance.1 The main experimental methods have relied so far on the use of X-ray crystallography, cryo-electron microscopy (cryo-EM) and nuclear magnetic resonance (NMR). Beyond solving structures, NMR is also uniquely suited to perform dynamics studies at the atomic scale, does not require crystals of the bound-ligand complex and is compatible with measurements performed under physiological conditions, including membrane or cellular milieu. In addition, solid-state NMR, possibly combined with a hyperpolarization technique called Dynamic Nuclear Polarization (DNP),2 has recently emerged as a key technique to extract precise structural details of the ligand (or drug) in functional biomolecular complexes (including membrane and fibrillar proteins).3 This approach relies on the use of isotopic labeling and allows the ligand structure to be solved without requiring prior knowledge of the host biomolecular architecture.4–9 Even if such information does not give a direct insight into binding sites, it can be efficiently combined with computational docking studies.10Nevertheless there is still a lack of NMR methodologies capable of locating binding sites in biomolecular systems and solving their structures at the atomic scale. Indeed, access to this type of information requires the use of advanced non-uniform isotopic spin labeling strategies in order to label residues involved in the binding site while reducing spectral crowding.11,12 Although possible, this approach suffers from strong limitations, since it is restricted to systems that can be over-expressed accordingly, often relies on previous knowledge of the binding site location and is limited to biomolecular assemblies composed of monomeric units of 50 kDa or less.13
The present contribution provides a solution to this problem. We report a new approach that allows identification of hyperpolarized binding sites in biomolecular complexes using DNP-enhanced NMR. This method, abbreviated as Sel-DNP (Selective Dynamic Nuclear Polarization), relies on the use of a ligand functionalized polarizing agent (PA), and allows extraction of site-specific information from the interaction site without prior knowledge of its location. The application of Sel-DNP is demonstrated on the galactose-specific lectin LecA14 for which highly resolved multi-dimensional experiments are obtained that allow the determination of residues involved in the binding site. The approach does not have any limitations with respect to the size of the biomolecular system, and only requires the preparation of one protein sample.
Results
DNP-enhanced NMR under Magic-Angle-Spinning (MAS-DNP) is a recently developed hyperpolarization technique which has been demonstrated on a large range of systems with new applications in surface chemistry, materials science and structural biology.15–17 The increase in NMR sensitivity is achieved by transferring the polarization from unpaired electrons, contained in dopant molecules called Polarizing Agents (PAs), to the surrounding nuclear spins of the system of interest. PAs are typically composed of stable organic mono- or bi-radicals. The transfer of polarization is driven by high-power high-frequency microwave irradiation of the sample, typically generated by a gyrotron.18PAs are usually dissolved in a glass-forming matrix that is used to suspend or impregnate the sample of interest.15,16,19 Where appropriate, further removal of the solvent can also be performed such as in the Matrix Free or Film Casting sample preparation approaches, as long as radical aggregation is avoided.20–22 Recently several groups have discussed the use of a localized PA, either covalently linked to the system of interest or to a bound-ligand, or just using a PA with high-affinity for the system of interest.21,23–29 The motivations behind these contributions were different from those for the work presented here, ranging from optimizing sample sensitivity through PA co-localization,21,23–26 along with a reduction of the cryo-protectant content,23,24 to the development of targeted DNP experiments in complex environments27 or the experimental investigation of the bleaching effect using specific isotopic labelled samples.28,29
The Sel-DNP approach: introducing selectivity in DNP NMR
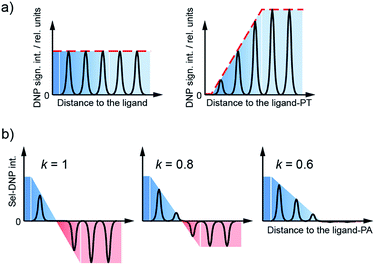
The approach reported here and dubbed Selective DNP (Sel-DNP) combines for the first time the use of localized Paramagnetic Relaxation Enhancement (PRE) and difference spectroscopy to recover high-resolution and high-sensitivity information from hyperpolarized binding sites. It is well known that the presence of paramagnetic centers in MAS-DNP samples induces signal broadening for nearby nuclear spins (so-called bleaching/quenching effect). Consequently, the introduction of a functionalized ligand, composed of the ligand moiety, a linker, and a paramagnetic species (see Fig. 1a), is expected to induce a localized PRE effect. This is not the case when paramagnets (used either to increase relaxation or as a PA) are uniformly distributed in the sample of interest.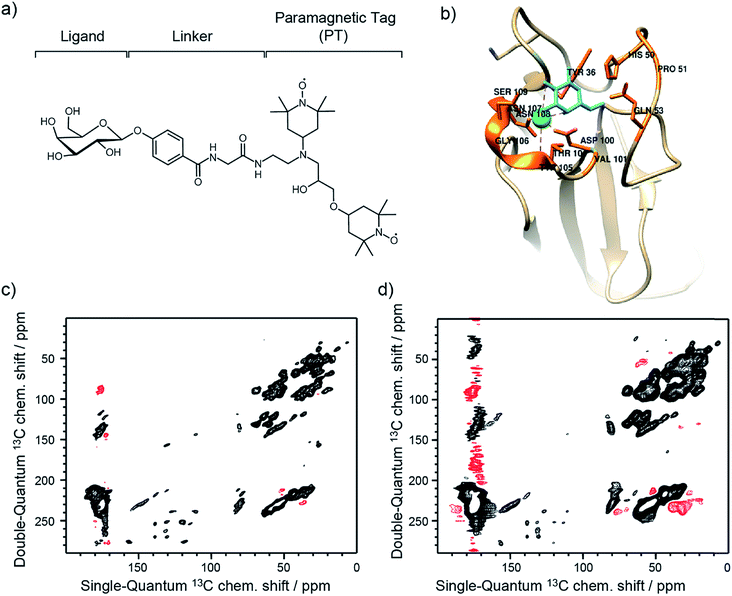 | ||
| Fig. 1 (a) Schematic of the functionalized ligand used in Sel-DNP and composed of a ligand tethered to a paramagnetic tag (PT) via a short linker. In this work, the ligand corresponds to D-galactose, the linker is made of a phenylglycine unit, and the paramagnetic tag chosen is the bis-nitroxide TOTAPOL.30 (b) Structure of the binding site of LecA highlighting the residues known (from the crystal structure) to interact with the galactose ligand (in cyan). (c) and (d) DNP-enhanced 13C–13C DQ/SQ one-bond correlation spectra of LecA, using (c) AMUPol (reference spectrum S0) and (d) the functionalized ligand of (a) (spectrum S). Positive contours are in black and negative ones are in red. Contours are set to the same level throughout the entire manuscript. | ||
Sel-DNP requires the acquisition of two datasets of a given ssNMR experiment recorded for two different sample preparations of the protein. The first spectrum, called reference (S0), is obtained for the biomolecular bound-ligand complex, for which the PA is homogeneously distributed in the sample. The corresponding NMR signals are uniformly enhanced by DNP, including the binding region. The second spectrum (S) is obtained with the same pulse sequence for the same protein, but using a specific ligand tethered to a paramagnetic tag (PT).
The proximity of the paramagnetic moiety to the binding region in the receptor–ligand–PT complex induces a broadening and attenuation of the closest resonances. Upon microwave irradiation, 1H spins from the protein are polarized by DNP, thanks to spin-diffusion of the hyperpolarized magnetization from the bleached zone (resonances broadened and attenuated beyond detection) to the distant protons.31,32 As demonstrated in this work, residues involved in the binding region can be revealed by the subtraction of the two spectra.
Application to carbohydrate-binding proteins
The approach was tested on lectins, which are carbohydrate-binding proteins that act as molecular readers to decipher information coded in glycoconjugates on the cell surface. They therefore play important biological roles in recognition processes relevant for human health. Lectins from pathogens are involved in host recognition of human tissues33 and are targets for the development of anti-infectious compounds that could provide alternatives to antibiotics. The lectin LecA from the opportunistic pathogen Pseudomonas aeruginosa consists of 121 amino acids (MW 12.75 kDa), is specific for D-galactose-containing molecules, and forms a tetramer in solution.14 The X-ray structure of the complex between LecA and galactose is presented in Fig. 1b, and unveils details of hydrogen bonds and Ca2+ bridge interactions.14 Note that no NMR study has been reported so far, and thus chemical shift assignment is unknown for this protein.Two DNP samples of LecA were prepared as described in detail in the Experimental section. The first sample, used for the reference spectrum S0, was prepared with one equivalent of D-galactose and impregnated with an AMUPol-biradical34 containing solution. For the second sample (used for spectrum S), a specific ligand–PT was synthesized (see Experimental section), composed of D-galactose tethered to the well-known polarizing agent TOTAPOL30via a short phenylglycine linker (Fig. 1a). One equivalent of this ligand–PT was added as the PA to the protein solution prior to evaporation of the excess solvent.
For each sample, a 13C–13C Double-Quantum (DQ) dipolar correlation spectrum was acquired with the mixing time of the recoupling sequence set to produce only one-bond correlations (see Experimental section for more details).35 Both spectra, recorded at 9.4 T and 100 K sample temperature, look very similar and illustrate the typical limited resolution obtained with MAS-DNP NMR experiments performed on biomolecular systems (Fig. 1c and d). Overall, linewidths in our experiment S appear broader compared to the ones in S0. This results in part from a higher overall concentration in ligand–PT (compared to the AMUPol concentration used in the reference sample), and from the fact that the paramagnetic species are not distributed the same way in both samples.
Tuning the selectivity focus using Sel-DNP
The Sel-DNP spectra can be obtained by combining the two datasets S0 and S, using the following expression:| SkSel-DNP = S0 − kS |
By tuning the relative weight of both spectra through the selectivity factor k, we can progressively adjust the focus on the binding region, selecting resonances as a function of the distance between the corresponding site and the ligand–PT. As S and S0 are obtained from different DNP samples, their respective spectral intensity is arbitrary. Prior to Sel-DNP subtraction, the spectra are relatively adjusted such that the difference using k = 1 gives rise to the least number of positive Sel-DNP peaks (highest focus). This arbitrary pre-scaling is then kept constant for all other Sel-DNP spectra produced with lower k. After subtraction, only positive peaks are kept in the Sel-DNP spectra. This Sel-DNP procedure is illustrated in Fig. 2. It is important to note that the residual positive NMR signal itself originates from the protein with its native ligand (S0).
Locating the binding site using de novo resonance assignment
Following the subtraction procedure described in the previous section, several binding-site selective spectra are produced with decreasing selectivity factor, k = 1.0, 0.8, 0.7, 0.6 and 0.4 (Fig. 3a–c, e–g, S1a–c, and S2†). They all show excellent resolution arising from the limited number of residues selected in each Sel-DNP spectrum, with linewidths at half height of 0.5 to 1 ppm. This contrasts severely with the very limited resolution obtained in the S0 spectrum (using uniformly distributed AMUPol), which contains all the 121 residues of the protein but cannot be used to perform resonance assignment. In contrast, each Sel-DNP spectrum can be used to identify and connect carbon resonances (from carbonyl to sidechain carbons) within a given residue. These 13C NMR fingerprints can be compared to tabulated values to identify and assign the residues involved in the binding site.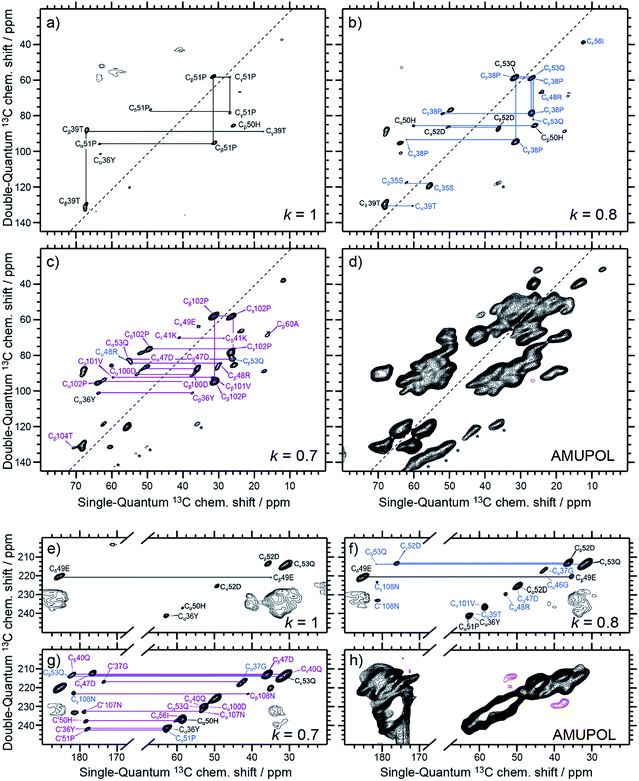 | ||
| Fig. 3 Aliphatic–aliphatic (a–d) and CO–aliphatic (e–h) regions of Sel-DNP 13C–13C DQ/SQ one-bond correlation spectra obtained with k = 1 (a and e), k = 0.8 (b and f), and k = 0.7 (c and g), compared in (d and h) to the reference spectrum S0 obtained with AMUPol as the PA. The aromatic region of the spectra is displayed in ESI Fig. S1.† Contour levels are uniform across all the spectra. Spinning sidebands are indicated by stars. Further Sel-DNP spectra for k = 0.6 and 0.4 are given in Fig. S2.† 1D traces through the different spectra can be found in Fig. 5. | ||
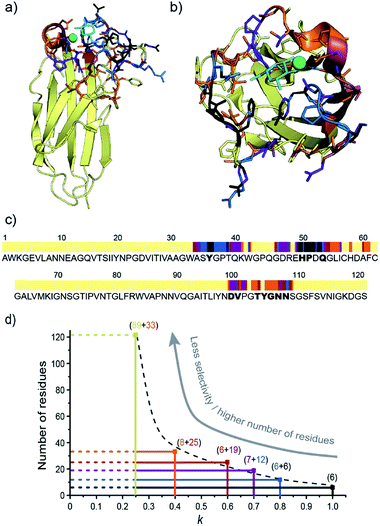
The Sel-DNP spectrum obtained with k = 1 contains cross-peaks corresponding to ∼6 residues labeled in black in Fig. 3a and e, and S1a.† For two of them, an almost complete carbon fingerprint is observed, which easily allows these residues to be assigned to a proline and a histidine. This is a very positive result since it is known from the crystal structure that 51Pro and 50His are involved in the binding of galactose in LecA (Fig. 1b).14 A second set of residues with only a partial carbon fingerprint in the k = 1 Sel-DNP spectrum is observed. With the help of Sel-DNP spectra of lower k, in which the missing resonances appear, they could be assigned to a tyrosine, a threonine, a glutamic acid, and an aspartic acid, respectively. Based on the amino acid sequence of the protein, and assuming that these residues belong to 2 or 3 groups of spatially close residues involved in the binding site, it is straightforward to identify 36Tyr (involved as well in the binding of galactose according to the crystal structure), 39Thr, 49Glu, and 52Asp, in addition to 51Pro and 50His. With k = 0.8, further cross-peaks of the 6 residues appear clearly, as well as new resonances corresponding to residues color-coded in blue in Fig. 3b and f, and S1b.†
Following the protocol described above, these carbon fingerprints can be identified and assigned to residues that are either located at neighboring positions to the black residues, or belong to a third binding region, as highlighted in Fig. 4c. The same analysis can be conducted for k = 0.7, revealing new residues that are color-coded in purple in Fig. 3c and g, and S1c,† and for k = 0.6 and 0.4, whose new residues are color-coded in burgundy and orange in Fig. S2a–c and S2d–f,† respectively. Gradually, by decreasing the selectivity factor k, the full binding site (see Fig. 1b and Table S1†) is highlighted with the appearance of 53Gln, 100Asp, 101Val, 104Thr, 105Tyr, 106Gly, 107Asn, and 108Asn, as well as other residues located outside the binding pocket (e.g.34Ala, 60Ala, and 55Leu). The list of assigned resonances is given in ESI Table S2.† Note that we were not able to use Sel-DNP spectra with k < 0.4 because of spectral crowding which prevents us from readily extracting site-specific information.
Discussion
Relating selectivity factor and distance to the paramagnetic tag
The X-ray structure of the LecA/Gal complex (PDB ID 1OKO)14 was then used to check whether residues assigned using the Sel-DNP methodology are indeed located in or close to the binding region. Fig. 4a and b show the crystal structure of the protein–ligand complex with the assigned residues color-coded in black, blue, purple, burgundy and orange, depending on in which Sel-DNP spectrum (with k = 1, 0.8, 0.7, 0.6 or 0.4, respectively) their NMR resonances appear. This confirms that all residues assigned with Sel-DNP are in the proximity of the ligand-binding site. This is also clearly seen in Fig. 4c which uses the same color code in combination with the sequence of the protein, in which the residues identified in the crystal structure to be in direct interaction with the ligand or the calcium ion are highlighted in bold (see Table S1†). Note that the residues from the binding site that are located behind the calcium atom (residue numbers 100 to 108) appear in Sel-DNP spectra with k = 0.7–0.4, and not with k = 1–0.8, since they are further away from the ligand (and thus from the PT) compared to residues (and their direct neighbors) that have either a direct interaction with the ligand, or interactions with both the calcium and the ligand.We then tried to relate the selectivity factor k to a distance cutoff from the center of the paramagnetic tag. The position of the latter was estimated by averaging three possible conformations of the functionalized ligand inside the binding pocket (as represented in Fig. S3†). Using the X-ray structure of the binding site, it is possible to then relate the changes in the selectivity factor k to an approximate distance cut-off from the PT. For instance, k = 1, 0.8 and 0.6 correspond to distance cut-offs of ∼10, ∼17 and ∼25 Å from the PT, respectively. This translates into a gradual increase in the number of residues appearing in the Sel-DNP spectra, from 6 residues for k = 1 to about 33 residues for k = 0.4 (Fig. 4d). The Sel-DNP spectra obtained with k < 0.4 and corresponding to a distance cut-off larger than 30 Å do contain a number of residues, which precludes further assignment under our experimental conditions due to spectral crowding.
Paramagnetic relaxation enhancement of NMR resonances at low temperature
Although PRE has been well described for solution and solid-state NMR experiments at ambient temperature,36,37 it has not yet been thoroughly investigated under the specific experimental conditions of DNP (e.g. 100 K, MAS, nitroxide-based PA). As demonstrated here, the presence of paramagnetic centers affects the signal of nuclear spins in two ways: first a broadening of the nearby resonances (i.e. increased signal dephasing during detection,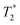 ), and second an attenuation of the detected peak intensities, resulting from the paramagnetic relaxation occurring during each block of pulses and delays present before the detection period (i.e. CP step, dipolar recoupling period, etc.). This can be seen from the 1D traces through the double-quantum frequency of spectra S and S0 shown in Fig. 5.
), and second an attenuation of the detected peak intensities, resulting from the paramagnetic relaxation occurring during each block of pulses and delays present before the detection period (i.e. CP step, dipolar recoupling period, etc.). This can be seen from the 1D traces through the double-quantum frequency of spectra S and S0 shown in Fig. 5.
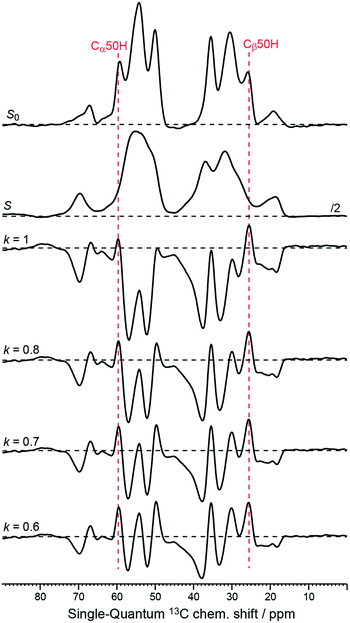 | ||
| Fig. 5 1D-traces through the double-quantum frequency at 85 ppm corresponding to the 50His Cα–Cβ cross peaks from the original data S0 and S (see Fig. 1c and d), as well as from the Sel-DNP spectra with k = 1, 0.8, 0.7, and 0.6 (see Fig. 3a–c and S2a†). Black horizontal dotted lines are plotted to help the reader distinguish between positive and negative intensities. Positions of the 50His Cα and Cβ in the single-quantum dimension are given by the red dotted lines. | ||
In depth quantitative study of the PRE effect at low temperature is beyond the scope of this article but will be investigated in the near future. Nevertheless, we have demonstrated with this work that the presence of localized paramagnetic centers as found for ligand–PT induces an intensity gradient of the detected signal around the position of the electron spin, as shown in Fig. 2. According to our DQ–SQ 2D data, this effect is noticeable up to at least 30 Å from the paramagnetic center, in agreement with DNP bleaching radii reported by other groups.28,29 When the PA is uniformly distributed in the sample, partial broadening and signal attenuation statistically affect all resonances.
Experimental section
Expression and purification of the LecA protein
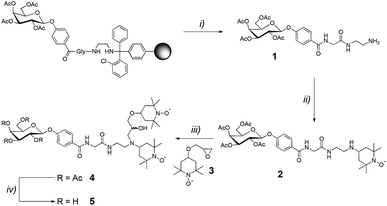
E. coli BL21 STAR (DE3) carrying the LecA plasmid was progressively adapted in four stages over 48 h to a minimal M9/D2O medium (5.3 g L−1 Na2HPO4, 3 g L−1 KH2PO4, 0.5 g L−1 NaCl, 1 g L−1 15NH4Cl, 5 mg thiamine, 1 mM MgSO4, 0.1 mM CaCl2, 50 μM ZnSO4, 100 μM FeCl3 and a vitamin cocktail) with 100 mg L−1 ampicillin and 2 g L−1 U-[13C, 2H]-D-glucose. Eight 500 mL cultures in minimal M9/D2O medium were inoculated from the previously described preculture and grown at 37 °C. When the OD600 reached 0.75, protein expression was induced by the addition of 0.25 mM IPTG and the culture was incubated at 20 °C overnight. Cells were harvested by centrifugation (6000g, 15 min) and the pellet was re-suspended in 80 mL of buffer A (20 mM Tris/HCl, 100 mM NaCl, pH 7.5, 100 μM CaCl2). Purification proceeded through affinity to D-galactose resin (Sigma) with elution with buffer B (20 mM Tris/HCl, 100 mM NaCl, pH 7.5, 100 mM EDTA). After 5 days' dialysis in MilliQ H2O supplemented with 100 μM CaCl2, and 3 days' dialysis in H2O alone, the protein was lyophilized. A final yield of 238 mg for 4 L of culture was obtained. The triple-labelled protein samples were characterized by 15% SDS gel electrophoresis. Activity was checked by titration microcalorimetry and the triple-labelled protein displayed the same affinity for galactose as the non-labelled one.Chemical synthesis of the ligand–PT 5 (Scheme 1)
The glyco-amino acid 1 was obtained through solid-phase peptide synthesis, following the Fmoc/tBu strategy and using a 2-chlorotrityl resin (m = 1.000 g, loading = 1.04 mmol g−1). Coupling reactions were performed using, relative to the resin loading, 2 eq. of ethylenediamine, N-Fmoc-protected L-glycine and 1-(4-carboxyphenyl)-2,3,4,6-tetra-O-acetyl-β-D-galactopyranoside activated in situ with PyBOP (2 eq.) and DIPEA (4 eq.) in DMF (10 mL) for 30 min. N-Fmoc protecting groups were removed by treatment with piperidine/DMF (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, 10 mL) for 10 min. This process was repeated three times. The resin was then washed five times with DMF (10 mL) for 1 min. The glyco-amino acid was cleaved from the resin by treatment with DCM/TFA (99
4, 10 mL) for 10 min. This process was repeated three times. The resin was then washed five times with DMF (10 mL) for 1 min. The glyco-amino acid was cleaved from the resin by treatment with DCM/TFA (99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (5 × 10 mL × 10 min). The combined cleavage fractions were concentrated under reduced pressure and the crude mixture was purified by semi-preparative RP-HPLC (5–40% B in 15 min) to obtain the glyco-amino acid 1.
1) (5 × 10 mL × 10 min). The combined cleavage fractions were concentrated under reduced pressure and the crude mixture was purified by semi-preparative RP-HPLC (5–40% B in 15 min) to obtain the glyco-amino acid 1.
Acetic acid was added to a solution of 4-oxo-TEMPO (50 mg, 0.29 mmol, 1 eq.) and glyco-amino acid 1 (1.1 eq.) in dry THF (10 mL) in order to reach pH = 4. The mixture was stirred at room temperature for 4 h. Then sodium cyanoborohydride (93 mg, 0.44 mmol, 1.5 eq.) was added and the mixture was stirred overnight. After concentration under reduced pressure, the crude mixture was purified by semi-preparative RP-HPLC (C18, λ = 214 nm, 5–40% B in 15 min) yielding the glyco-amino acid 2.
A solution of 2 (0.1 mmol, 1 eq.), epoxide 3 (25 mg, 0.11 mmol, 1.1 eq.), and lithium perchlorate (106 mg, 1 mmol, 10 eq.) in dry acetonitrile (10 mL) was stirred at reflux for 72 h. Additional lithium perchlorate (10 eq.) was added after 24 h and 48 h. After concentration under reduced pressure, the crude mixture was purified by semi-preparative RP-HPLC (C18, λ = 214 nm, 5–40% B in 15 min) yielding compound 4 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 diastereomeric mixture).
1 diastereomeric mixture).
A solution of tetra-O-acetylated compound 4 (0.020 mmol, 1 eq.) in a mixture of methanol, triethylamine and water (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) (8 mL) was stirred at room temperature overnight. After concentration under reduced pressure, the crude mixture was purified by semi-preparative RP-HPLC (C18, λ = 214 nm, 5–40% B in 15 min), obtaining the ligand–PT 5.
2) (8 mL) was stirred at room temperature overnight. After concentration under reduced pressure, the crude mixture was purified by semi-preparative RP-HPLC (C18, λ = 214 nm, 5–40% B in 15 min), obtaining the ligand–PT 5.
The chemical analysis of 1, 2, 4 and 5 is given in the ESI (Fig. S5 to S18†).
Sample preparation for Sel-DNP experiments
Two samples (of the same protein) were prepared in order to record the Sel-DNP spectra. For the first sample, a solution of 1.5 mM 2H,13C,15N-uniformly labeled LecA was prepared in a protonated buffer containing 50 mM ammonium acetate (pH = 4.7), 1 mM calcium chloride, 1 mM magnesium chloride and 1.6 mM D-galactose. This solution was incubated for 24 h at 5 °C, followed by evaporation of the excess solvent using a speed-vac until a slightly wet solid was obtained. The resulting sample was impregnated with a solution of 60% 2H-glycerol, 30% D2O and 10% H2O (v/v) containing 10 mM AMUPol.34 Approximately 5 mg was packed into a 3.2 mm sapphire NMR rotor using a silicone plug and it was closed with a zirconia cap. The final concentration of AMUPol in the sample is estimated to be about one molecule for five LecA units. This sample was used to record the reference spectrum S0 (Fig. 1c).For the second sample, a solution of 1.5 mM 2H,13C,15N-uniformly labeled LecA was prepared in a protonated buffer containing 50 mM ammonium acetate (pH = 4.7), 1 mM calcium chloride, 1 mM magnesium chloride and 1.6 mM of the ligand–PT compound 5 (Fig. 1a). This solution was incubated for 24 h at 5 °C, followed by evaporation of the excess solvent with a speed-vac. Approximately 14 mg of the resulting sample was packed into a 3.2 mm sapphire NMR rotor using a silicone plug and it was closed with a zirconia cap. This sample was used to record the spectrum S (Fig. 1d).
DNP experiments
All the experiments reported here were carried out on a Bruker 263 GHz DNP-NMR Avance III spectrometer (400 MHz 1H frequency) equipped with a low temperature 3.2 mm wide-bore MAS probe. Experiments were conducted at ∼100 K sample temperature and 9.5 kHz MAS frequency, to prevent both rotational resonance effects38 and spinning sidebands from overlapping with the NMR peaks. The ratio of the 1H–13C CP MAS signals with and without microwave irradiation, εon/off, was evaluated for the two DNP samples, returning 70 and 43 for the AMUPol and the ligand–PT samples, respectively. The DNP buildup times (TB), measured using a saturation-recovery experiment, were 5.3 s and 0.7 s for the AMUPol and ligand–PT samples, respectively. 13C–13C DQ–SQ dipolar correlation experiments were performed for each sample using the pulse sequence given in ESI Fig. S4.† The hyperpolarized 1H magnetization is first transferred to carbons using a 1H–13C cross polarization (CP) step. The dipolar recoupling symmetry-based sequence R1672 is then applied to generate double-quantum (DQ) coherences.35 The excitation period was set to τDQ = 0.4 ms to maximize one-bond polarization transfer. After free evolution during t1 (indirect dimension) of the generated 13C–13C DQ coherences (at frequencies corresponding to the sum of the chemical shifts of the coupled 13C spins), a second block of R1672 recoupling of the same duration τDQ is applied to reconvert DQ coherences into detectable single-quantum (SQ) coherences. No 1H decoupling is required during the R1672 recoupling periods.39 SPINAL64 1H decoupling at a field strength of 100 kHz was applied during both indirect (t1) and direct (t2) detection times. Each two-dimensional spectrum was acquired with a maximum t1 evolution time of 5.2 ms using states-TPPI for quadrature detection (256 complex points). For each increment, 16 transients were added with interscan delays of 4 s and 0.9 s for the AMUPol and the ligand–PT sample, respectively. Acquisition time in the direct t2 dimension was 10 ms. Spectral widths were 492 ppm in the DQ dimension and 405 ppm in the SQ dimension. Cosine apodization was applied in both dimensions prior to Fourier transformation, followed by baseline correction with a polynomial function of grade 5.Sel-DNP spectra
Sel-DNP spectra with k = 1, 0.8, 0.7, and 0.6 have been used to address the NMR fingerprint of the LecA binding site. To avoid baseline artefacts, both experiments S and S0 need to be acquired with similarly good signal-to-noise ratios. The assignment of the Sel-DNP spectra for k = 1, 0.8, and 0.7 was possible without pre-knowledge of the crystal structure: the type of amino acid was identified using chemical shift statistics for amino acid residues published by the Biological Magnetic Resonance Data Bank (BMRB)40 and their position in the sequence was obtained considering that all residues have to be located close to each other in only a few regions of the sequence (see Fig. 4c). For k = 0.6 and 0.4, calculations based on the crystal structure using the software SHIFTX2 (ref. 41) were used as well, in particular for ambiguous residues.Molecular modeling
A three-dimensional model of glycoconjugate 5 was built with Sybyl (Certara, Princeton, NJ) graphical software using the atomic coordinates of TOTAPOL proposed from the PubMed site (https://pubchem.ncbi.nlm.nih.gov). The molecule was graphically extended by adding a glycine, a benzene and a galactose residue from the Sybyl molecular libraries. The galactose was docked on the calcium ion in two adjacent binding sites of the LecA structure by superimposition on an equivalent sugar in the crystal structure with PDB ID 1OKO.14 In order to illustrate the extension that the TOTAPOL group can attain on the surface of LecA, three conformations were generated by varying the torsion angles in the phenylglycine linker (Fig. S3†).Conclusions
The Sel-DNP approach enables the extraction of site-specific information from native biomolecular binding sites. Since only a limited number of resonances are sequentially highlighted, this approach yields highly resolved multi-dimensional spectra that allow resonance assignment and identification of residues in close spatial proximity to the paramagnetic tag, and therefore location of the binding site.Sel-DNP does not have any intrinsic limitation in the size of the biomolecular system that can be studied, since only residues close to the ligand–PT are detected. The approach only requires one protein sample, either uniformly isotopically 13C, 15N labeled or not, but can of course be combined with advanced site-specific labeling strategies. In addition, the protein–ligand complex does not need to be crystallized. It is important to stress that this approach can be implemented without any previous knowledge of the binding site location. Nevertheless, resonance assignment is greatly simplified if a global fold or a structure of the protein alone is available. Extension to 15N–13C correlation experiments using Sel-DNP is currently being implemented in our laboratory, as well as correlation experiments that can provide long-range distance contacts. This should help in performing sequential resonance assignment and extracting specific structural constraints from the binding site.
We expect Sel-DNP to become an important method for the study of protein binding sites, e.g. membrane proteins (possibly in their cellular environment), fibrillar assemblies, etc. Multi-dimensional difference spectroscopy and DNP, which was recently used to improve the spectral resolution of the various cell-wall components of bacteria,42 are likely to become important tools for the study of complex systems.
Finally, it is worth noting that on-going instrument development has recently demonstrated the feasibility of improving the sensitivity by several orders of magnitude compared to the current state of the art. This approach which relies on the use of sustainable cryogenic helium spinning at much lower temperatures,43 should allow Sel-DNP to be used for the investigation of interaction interfaces in very large biologically relevant complexes.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Isabel Ayala for her help in producing LecA, Diego Gauto, Daniel Lee and Serge Pérez for fruitful discussions and manuscript revision, and Fabien Aussenac (Bruker BioSpin) for providing spectrometer time at the initial stage of the work. This work was supported by the French National Research Agency (ARCANE ANR-11-LABX-0003-01, Glyco@Alps ANR-15-IDEX-02, and ANR-16-CE11-0030-03) and the European Research Council (ERC-CoG-2015, No. 682895; ERC-CoG-2014, No. 647938). We used the platforms of the Grenoble Instruct Center (ISBG; UMS 3518 CNRS-CEA-UJF-EMBL) with support from FRISBI (ANR-10-INSB-05-02) and GRAL (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology (PSB).References
- J. Drews, Science (80-), 2000, 287, 1960–1964 CrossRef CAS.
- D. A. Hall, D. C. Maus, G. J. Gerfen, S. J. Inati, L. R. Becerra, F. W. Dahlquist and R. G. Griffin, Science (80-), 1997, 276, 930–932 CrossRef CAS.
- A. Watts, Nat. Rev. Drug Discovery, 2005, 4, 555–568 CrossRef CAS PubMed.
- F. Creuzet, A. McDermott, R. Gebhard, K. van der Hoef, M. Spijker-Assink, J. Herzfeld, J. Lugtenburg, M. Levitt and R. Griffin, Science (80-), 1991, 251, 783–786 CrossRef CAS.
- S. Luca, J. F. White, A. K. Sohal, D. V. Filippov, J. H. van Boom, R. Grisshammer and M. Baldus, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 10706–10711 CrossRef CAS PubMed.
- V. S. Bajaj, M. L. Mak-Jurkauskas, M. Belenky, J. Herzfeld and R. G. Griffin, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 9244–9249 CrossRef CAS PubMed.
- A. H. Linden, S. Lange, W. T. Franks, Ü. Akbey, E. Specker, B.-J. van Rossum and H. Oschkinat, J. Am. Chem. Soc., 2011, 133, 19266–19269 CrossRef CAS PubMed.
- R. Tycko and R. B. Wickner, Acc. Chem. Res., 2013, 46, 1487–1496 CrossRef CAS PubMed.
- H. Yang, D. Staveness, S. M. Ryckbosch, A. D. Axtman, B. A. Loy, A. B. Barnes, V. S. Pande, J. Schaefer, P. A. Wender and L. Cegelski, ACS Cent. Sci., 2018, 4, 89–96 CrossRef CAS PubMed.
- R. Marchetti, S. Perez, A. Arda, A. Imberty, J. Jimenez-Barbero, A. Silipo and A. Molinaro, ChemistryOpen, 2016, 5, 274–296 CrossRef CAS PubMed.
- T. Sugiki, K. Furuita, T. Fujiwara and C. Kojima, Molecules, 2018, 23, 148 CrossRef PubMed.
- R. Kerfah, M. J. Plevin, R. Sounier, P. Gans and J. Boisbouvier, Curr. Opin. Struct. Biol., 2015, 32, 113–122 CrossRef CAS PubMed.
- J.-P. Demers, P. Fricke, C. Shi, V. Chevelkov and A. Lange, Prog. Nucl. Magn. Reson. Spectrosc., 2018, 109, 51–78 CrossRef CAS PubMed.
- G. Cioci, E. P. Mitchell, C. Gautier, M. Wimmerová, D. Sudakevitz, S. Pérez, N. Gilboa-Garber and A. Imberty, FEBS Lett., 2003, 555, 297–301 CrossRef CAS PubMed.
- A. B. Barnes, G. De Paëpe, P. C. A. van der Wel, K.-N. Hu, C.-G. Joo, V. S. Bajaj, M. L. Mak-Jurkauskas, J. R. Sirigiri, J. Herzfeld, R. J. Temkin and R. G. Griffin, Appl. Magn. Reson., 2008, 34, 237–263 CrossRef CAS PubMed.
- A. J. Rossini, A. Zagdoun, M. Lelli, A. Lesage, C. Copéret and L. Emsley, Acc. Chem. Res., 2013, 46, 1942–1951 CrossRef CAS PubMed.
- D. Lee, S. Hediger and G. De Paëpe, in Modern Magnetic Resonance, ed. G. A.Webb, Springer International Publishing, Cham, 2017, pp. 1–17 Search PubMed.
- M. Rosay, L. Tometich, S. Pawsey, R. Bader, R. Schauwecker, M. Blank, P. M. Borchard, S. R. Cauffman, K. L. Felch, R. T. Weber, R. J. Temkin, R. G. Griffin and W. E. Maas, Phys. Chem. Chem. Phys., 2010, 12, 5850–5860 RSC.
- D. Lee, S. Hediger and G. De Paëpe, Solid State Nucl. Magn. Reson., 2015, 66–67, 6–20 CrossRef CAS PubMed.
- H. Takahashi, D. Lee, L. Dubois, M. Bardet, S. Hediger and G. De Paëpe, Angew. Chem., Int. Ed., 2012, 51, 11766–11769 CrossRef CAS PubMed.
- H. Takahashi, S. Hediger and G. De Paëpe, Chem. Commun., 2013, 49, 9479–9481 RSC.
- D. Le, G. Casano, T. Phan, F. Ziarelli, O. Ouari, F. Aussenac, P. Thureau, G. Mollica, D. Gigmes, P. Tordo and S. Viel, Macromolecules, 2014, 47, 3909–3916 CrossRef CAS.
- C. Fernández-de-Alba, H. Takahashi, A. Richard, Y. Chenavier, L. Dubois, V. Maurel, D. Lee, S. Hediger and G. De Paëpe, Chem. - Eur. J., 2015, 21, 4512–4517 CrossRef PubMed.
- A. N. Smith, U. T. Twahir, T. Dubroca, G. E. Fanucci and J. R. Long, J. Phys. Chem. B, 2016, 120, 7880–7888 CrossRef CAS PubMed.
- M. A. Voinov, D. B. Good, M. E. Ward, S. Milikisiyants, A. Marek, M. A. Caporini, M. Rosay, R. A. Munro, M. Ljumovic, L. S. Brown, V. Ladizhansky and A. I. Smirnov, J. Phys. Chem. B, 2015, 119, 10180–10190 CrossRef CAS PubMed.
- R. Rogawski, I. V. Sergeyev, Y. Li, M. F. Ottaviani, V. Cornish and A. E. McDermott, J. Phys. Chem. B, 2017, 121, 1169–1175 CrossRef CAS PubMed.
- T. Viennet, A. Viegas, A. Kuepper, S. Arens, V. Gelev, O. Petrov, T. N. Grossmann, H. Heise and M. Etzkorn, Angew. Chem., Int. Ed., 2016, 55, 10746–10750 CrossRef CAS PubMed.
- E. A. W. van der Cruijsen, E. J. Koers, C. Sauvée, R. E. Hulse, M. Weingarth, O. Ouari, E. Perozo, P. Tordo and M. Baldus, Chem.–Eur. J., 2015, 21, 12971–12977 CrossRef CAS PubMed.
- R. Rogawski, I. V. Sergeyev, Y. Zhang, T. H. Tran, Y. Li, L. Tong and A. E. McDermott, J. Phys. Chem. B, 2017, 121, 10770–10781 CrossRef CAS PubMed.
- C. Song, K. Hu, C. Joo, T. M. Swager and R. G. Griffin, J. Am. Chem. Soc., 2006, 128, 11385–11390 CrossRef CAS PubMed.
- F. Mentink-Vigier, Ü. Akbey, H. Oschkinat, S. Vega and A. Feintuch, J. Magn. Reson., 2015, 258, 102–120 CrossRef CAS PubMed.
- F. Mentink-Vigier, S. Vega and G. De Paëpe, Phys. Chem. Chem. Phys., 2017, 19, 3506–3522 RSC.
- A. Imberty and A. Varrot, Curr. Opin. Struct. Biol., 2008, 18, 567–576 CrossRef CAS PubMed.
- C. Sauvée, M. Rosay, G. Casano, F. Aussenac, R. T. Weber, O. Ouari and P. Tordo, Angew. Chem., Int. Ed., 2013, 52, 10858–10861 CrossRef PubMed.
- M. H. Levitt, Encycl. Nucl. Magn. Reson. Vol. 9, Adv. NMR., 2002, pp. 165–196 Search PubMed.
- G. M. Clore, in Electron Paramagnetic Resonance Investigations of Biological Systems by Using Spin Labels, Spin Probes, and Intrinsic Metal Ions, Part B, ed. P. Z.Qin and K.Warncke, Academic Press, 2015, vol. 564, pp. 485–497 Search PubMed.
- C. P. Jaroniec, Solid State Nucl. Magn. Reson., 2012, 43–44, 1–13 CrossRef CAS PubMed.
- D. P. Raleigh, M. H. Levitt and R. G. Griffin, Chem. Phys. Lett., 1988, 146, 71–76 CrossRef CAS.
- I. Marin-Montesinos, D. H. Brouwer, G. Antonioli, W. C. Lai, A. Brinkmann and M. H. Levitt, J. Magn. Reson., 2005, 177, 307–317 CrossRef PubMed.
- E. L. Ulrich, H. Akutsu, J. F. Doreleijers, Y. Harano, Y. E. Ioannidis, J. Lin, M. Livny, S. Mading, D. Maziuk, Z. Miller, E. Nakatani, C. F. Schulte, D. E. Tolmie, R. Kent Wenger, H. Yao and J. L. Markley, Nucleic Acids Res., 2008, 36, D402–D408 CrossRef CAS PubMed.
- B. Han, Y. Liu, S. W. Ginzinger and D. S. Wishart, J. Biomol. NMR, 2011, 50, 43–57 CrossRef CAS PubMed.
- H. Takahashi, I. Ayala, M. Bardet, G. De Paëpe, J.-P. Simorre and S. Hediger, J. Am. Chem. Soc., 2013, 135, 5105–5110 CrossRef CAS PubMed.
- E. Bouleau, P. Saint-Bonnet, F. Mentink-Vigier, H. Takahashi, J.-F. Jacquot, M. Bardet, F. Aussenac, A. Purea, F. Engelke, S. Hediger, D. Lee and G. De Paëpe, Chem. Sci., 2015, 6, 6806–6812 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Aromatic region of Sel-DNP spectra of LecA for k = 1, 0.8, and 0.7. Sel-DNP spectra of LecA for k = 0.6 and 0.4. Conformations of the PA–ligand in the LecA binding site. DQ/SQ dipolar recoupling sequence. Table of residues involved in the binding of D-galactose according to the crystal structure. Table of assigned residues of LecA based on Sel-DNP spectra. Chemical analysis of compounds 1, 2, 4, and 5. See DOI: 10.1039/c8sc05696j |
| This journal is © The Royal Society of Chemistry 2019 |