Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Targeting triple-negative breast cancer cells using Dengue virus-mimicking pH-responsive framboidal triblock copolymer vesicles†
Charlotte J.
Mable‡
*a,
Irene
Canton‡
*b,
Oleksandr O.
Mykhaylyk
a,
Burcin
Ustbas Gul§
b,
Pierre
Chambon¶
a,
Efrosyni
Themistou||
a and
Steven P.
Armes‡
 *a
*a
aDepartment of Chemistry, University of Sheffield, Dainton Building, Brook Hill, Sheffield, South Yorkshire S3 7HF, UK. E-mail: C.Morse@sheffield.ac.uk; S.P.Armes@sheffield.ac.uk; Tel: +44 (0)114 222 9340 Tel: +44 (0)114 222 9342
bDepartment of Biomedical Sciences, University of Sheffield, Firth Court, Sheffield, South Yorkshire S10 2TN, UK. E-mail: I.Canton@sheffield.ac.uk; Tel: +44 (0)114 222 1093
First published on 9th April 2019
Abstract
It is well-known that the Dengue fever virus undergoes a distinct morphological transition from topologically smooth particles to ‘bumpy’ particle on increasing the temperature from that of the mosquito carrier (28 °C) to that of the human host (37 °C). This virus also possesses pH-sensitive surface domains that undergo conformational changes during infection which facilitates exit from the endosomes. Herein we take a bio-inspired approach to design synthetic Dengue virus-mimicking nanoparticles to target triple-negative (TN) breast cancer cells that overexpress SR-B1 scavenger receptors. Thus, sterile pH-responsive methacrylic ABC triblock copolymer vesicles were prepared in aqueous solution via polymerization-induced self-assembly. Microphase separation between two enthalpically-incompatible hydrophobic membrane-forming blocks produced a well-defined framboidal morphology, with surface globules of ∼28 nm diameter protruding from the membrane. The hydrophilic stabilizer block comprises 97% hydroxyl-functionalized chains and 3% phosphorylcholine-functionalized chains, with the latter being critical for selective intracellular uptake. These framboidal vesicles remain intact at neutral pH but become swollen and cationic at pH 5–6 because the tertiary amine residues in the hydrophobic C block become protonated. We demonstrate that such nanoparticles enable selective targeting of TN breast cancer cells. This is because such malignant cells overexpress SR-B1 receptors for naturally-occurring phospholipids and hence take up the phosphorylcholine-decorated framboidal vesicles preferentially. In contrast, negligible cell uptake is observed over the same time period for both human dermal fibroblasts and normal breast cancer cells that minimally express the SR-B1 receptor. Moreover, we show that genetic material within such pH-responsive framboidal vesicles can be efficiently delivered to the cell nuclei while maintaining high cell viability.
Introduction
Viruses have evolved over millions of years to become extremely effective vectors for efficient intracellular delivery.1 They are able to target specific mammalian cells, avoid subcellular barriers and deliver highly sensitive biological molecules such as proteins and large nucleic acids very efficiently.2,3 However, attempts to use viruses (or viral coats) to treat disease has led to undesirable side-effects, including patient mortality in some cases.4 Thus there has been considerable recent interest in the design of much safer synthetic analogues for viruses.5–8 Two critical properties that enable viruses to deliver their cargo are (i) their size and (ii) their ability to change their surface morphology. The smallest mammalian viruses, such as Parvoviridae and Picornaviridae, have particle diameters of approximately 20–30 nm,9 whereas the largest range from 200 to 400 nm diameter (e.g. the ovoid Poxviridae virus9) and even up to 540 nm diameter (e.g. Paramixoviruses9,10). Interestingly, Langer and co-workers recommend that a broad range of length scales should be considered when designing appropriate drug delivery carriers.11 Similarly, Zhao and Stenzel12 recently highlighted the importance of nanoparticle size, shape, modulus and surface charge on the efficacy of intracellular delivery.In some cases, viral infection is believed to involve stimulus-responsive behavior. For example, the Dengue fever virus undergoes a distinct thermally-induced morphological transition from topologically smooth, pseudo-spherical nanoparticles of around 50 nm diameter in the mosquito vector at 28 °C to slightly larger, distinctly framboidal nanoparticles in the human victim at 37 °C.13,14 This subtle change in morphology exposes the DIII domain of the E protein, which participates in receptor binding.13,14 The complete mechanism of infection for this virus has not yet been elucidated but the scavenger receptor SR-B1 appears to play an important role.15 Moreover, protruding nanodomains can induce lipid bilayer deformation, which should in turn enhance the kinetics of cell uptake.16–18 Indeed, nanoscale surface roughness is well-known to promote both protein receptor binding and the intracellular uptake of nanoparticles.19,20 Furthermore, the Dengue fever virus also possesses pH-sensitive surface domains that undergo conformational changes during infection which facilitate its timely exit from the endosomes.21 Such viruses transit through the early endosomes and fuse with late endocytic organelles at pH 5.5.22 At this lower local pH, the virus undergoes a further subtle change in its surface morphology to adopt a more ‘open’ conformation before its fusion with the endosomal membrane.21–23
Triple-negative (TN) breast cancer has a very poor prognosis because it is extremely aggressive and lacks targeted therapies.24 TN breast cancer tumors express unusually high levels of SR-B1 and reduced levels of CD-36 receptors.25 Moreover, the literature suggests that the optimal nanoparticle diameter for cell uptake mediated by scavenger receptors ranges from 20 to 30 nm.26–28 In principle, wholly synthetic Dengue virus-mimicking nanoparticles that exhibit appropriate surface roughness and stimulus-responsive behavior should enable selective targeting of TN breast cancer cells.
It is well-known that amphiphilic diblock copolymers undergo spontaneous self-assembly in aqueous solution.29–32 Moreover, recent advances in polymerization-induced self-assembly (PISA)33,34 now enable a wide range of block copolymer nanoparticles to be prepared directly in concentrated aqueous solution via reversible addition–fragmentation chain transfer (RAFT) aqueous dispersion polymerization.35–37 Of particular relevance to the present study, a poly(glycerol monomethacrylate) macromolecular chain transfer agent (PGMA macro-CTA) can be chain-extended using 2-hydroxypropyl methacrylate (HPMA).37–39 Micellar self-assembly produces sterically-stabilized PHPMA-core nanoparticles with hydrophilic PGMA stabilizer chains. Depending on the precise PISA formulation, a range of copolymer morphologies can be achieved, including spheres, worms and vesicles.38–40 Furthermore, RAFT seeded emulsion polymerization has been utilized to chain-extend PGMA–PHPMA diblock copolymer vesicles using a water-immiscible monomer (benzyl methacrylate, BzMA).41,42 This leads to microphase separation within the vesicle membrane, resulting in the formation of PGMA–PHPMA–PBzMA triblock copolymer vesicles with a distinctive framboidal morphology. More recently, Mable et al.43 reported the closely-related synthesis of pH-responsive framboidal vesicles, simply by using 2-(diisopropylamino)ethyl methacrylate (DPA) instead of BzMA. Importantly, the mean size of the globules protruding from the vesicle membrane can be tuned by systematic variation of the degree of polymerization (DP) of the PDPA block.
Herein we report the design, synthesis and biological evaluation of pH-responsive fluorescently-labeled framboidal vesicles as a new, wholly synthetic mimic for the Dengue fever virus. Our approach utilizes the pH-responsive PGMA–PHPMA–PDPA framboidal vesicles (hereafter denoted as G–H–D for brevity) previously reported by Mable et al.43 Such nanoparticles can be prepared via PISA in aqueous solution while replacing a small fraction (3 mol%) of the PGMA stabilizer chains with a well-known biocompatible polymer, poly(2-(methacryloyloxy)ethyl phosphorylcholine) (PMPC).44–49 The pendent phosphorylcholine groups on this latter polymer act as a ligand to enabletargeting of SR-B1 scavenger receptors,50,51 which are overexpressed by TN breast cancer cells.25,52
Results and discussion
Synthesis and characterization
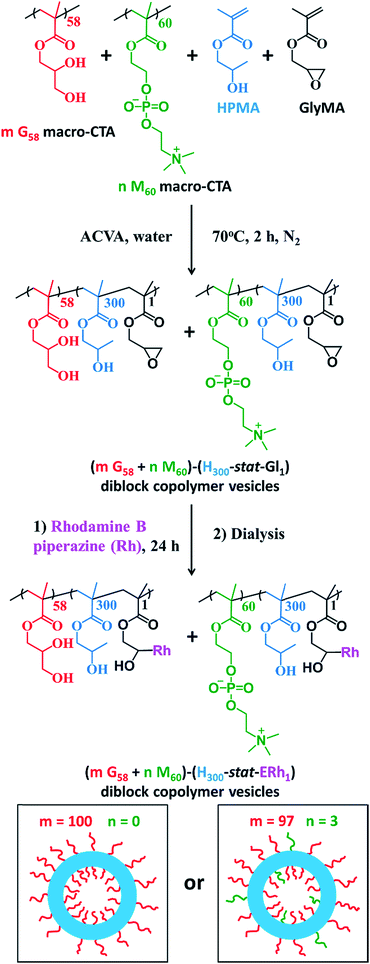
The framboidal nanoparticles prepared in this work were synthesized under rigorously sterile conditions. Firstly, PGMA58 and PMPC60 macromolecular chain transfer agents (macro-CTAs) were each synthesized on a multigram scale. A 97![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 binary mixture of these two macro-CTAs (and also a control comprising 100% PGMA58 macro-CTA) was chain-extended via RAFT aqueous dispersion copolymerization of a mixture of HPMA and GlyMA (Fig. 1) under sterile conditions, as described fully in the ESI.† Briefly, 200 nm sterile filters were used to sterilize all liquid reagents via ultrafiltration, all glassware was autoclaved and work was carried out exclusively in a fully-sterile Class II laminar flow cabinet. The epoxy groups on the GlyMA units were reacted with rhodamine B piperazine53 using an epoxy/amine molar ratio of unity to produce fluorescently-labeled diblock copolymer vesicles. Visible absorption spectroscopy studies of a dilute methanolic copolymer solution enabled quantification of the dye conjugation reaction and indicated that approximately 80% of the epoxy groups had reacted with the rhodamine B piperazine (see ESI† for further details).
3 binary mixture of these two macro-CTAs (and also a control comprising 100% PGMA58 macro-CTA) was chain-extended via RAFT aqueous dispersion copolymerization of a mixture of HPMA and GlyMA (Fig. 1) under sterile conditions, as described fully in the ESI.† Briefly, 200 nm sterile filters were used to sterilize all liquid reagents via ultrafiltration, all glassware was autoclaved and work was carried out exclusively in a fully-sterile Class II laminar flow cabinet. The epoxy groups on the GlyMA units were reacted with rhodamine B piperazine53 using an epoxy/amine molar ratio of unity to produce fluorescently-labeled diblock copolymer vesicles. Visible absorption spectroscopy studies of a dilute methanolic copolymer solution enabled quantification of the dye conjugation reaction and indicated that approximately 80% of the epoxy groups had reacted with the rhodamine B piperazine (see ESI† for further details).
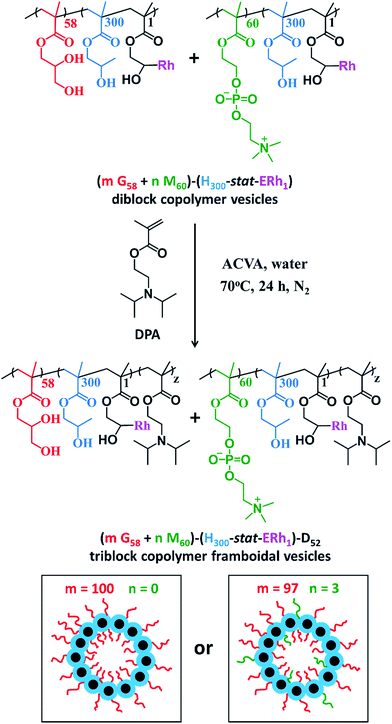
After dialysis to remove excess rhodamine B piperazine, these precursor nanoparticles were then chain-extended via RAFT aqueous seeded emulsion polymerization of DPA to obtain the desired pH-responsive framboidal triblock copolymer vesicles (Fig. 2). Triblock copolymer compositions of PGMA58–P(HPMA300-stat-GlyMARh1)–PDPA100 and (97 PGMA58 + 3 PMPC60)–P(HPMA300-stat-GlyMARh1)–PDPA100 were targeted (see Fig. 1 and 2). For the sake of brevity, these are hereafter denoted as G58–(H300-stat-ERh1)–D100 and (97 G58 + 3 M60)–(H300-stat-ERh1)–D100, respectively (where ‘E’ stands for the reactive epoxy groups within the GlyMA residues).
1H NMR studies indicated that more than 99% conversion of the HPMA and GlyMA comonomers was achieved within 2 h at 70 °C but the final DPA conversion was only 48% (for vesicles without PMPC macro-CTA) or 52% (for vesicles with PMPC macro-CTA) after 24 h at 70 °C (see Fig. S1†).
The latter conversions are relatively low compared to those reported by Mable et al.43 The only difference is that the DPA monomer was filter-sterilized prior to its addition to the reaction flask in the present study. Thus a control experiment was conducted whereby the mass of DPA monomer was determined before and after filter sterilization. This indicated that around 40% of the DPA monomer remained on the filter and therefore was not in fact added to the reaction mixture. Correcting for this mass loss of reagent, the DPA conversions are estimated to be 80 and 87% respectively, which is more comparable to those reported by Mable et al.43 In both cases unreacted DPA monomer was removed from the reaction solution by dialysis, see ESI.† In summary, the final diblock copolymer compositions obtained were G58–(H300-stat-ERh1)–D48 and (97 G58 + 3 M60)–(H300-stat-ERh1)–D52.
DMF gel permeation chromatography (GPC) studies indicated a low polydispersity (Mw/Mn = 1.13) for the PGMA58 macro-CTA, see Fig. S2a.† Similarly, aqueous GPC data obtained for the PMPC60 macro-CTA also indicated a low polydispersity (Mw/Mn = 1.09, see Fig. S2b†). DMF GPC traces indicated that both diblock copolymer precursors were obtained with low polydispersities (Mw/Mn < 1.20, see Fig. S2c†). However, a weak high molecular weight shoulder was observed, which was attributed to light branching caused by small amounts of dimethacrylate impurity within the HPMA monomer (0.07 mol% as judged by HPLC analyses).32 In the case of the triblock copolymers, the PDPA block is DMF-insoluble, while the PGMA block is THF-insoluble, rendering GPC characterization of the triblock copolymers rather problematic. To render these triblock copolymers THF-soluble for GPC analysis, the PGMA block was esterified according to a previously reported protocol (see ESI† for further details).43,54,55 THF GPC traces obtained for the modified triblock copolymers were unimodal and indicated high blocking efficiencies but somewhat broader molecular weight distributions (see Fig. S2d†).
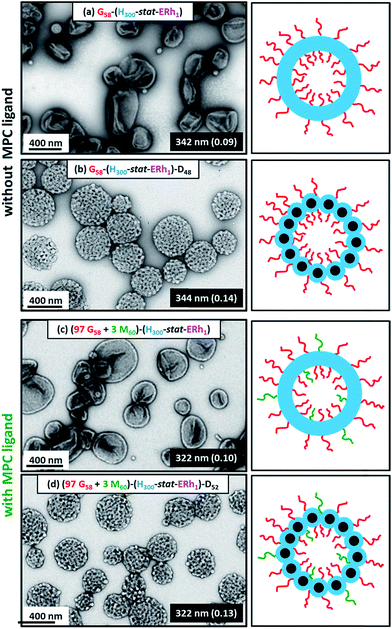
Dynamic light scattering (DLS) studies indicated comparable hydrodynamic diameters of 344 ± 128 nm and 322 ± 116 nm for the G58–(H300-stat-ERh1)–D48 and (97 G58 + 3 M60)–(H300-stat-ERh1)–D52 framboidal vesicles, respectively. Allowing for the effect of polydispersity, this is consistent with transmission electron microscopy (TEM) studies, which suggest number-average vesicle diameters of approximately 300 nm in each case (Fig. 3). TEM images also confirmed the distinctive framboidal morphology expected for the G58–(H300-stat-ERh1)–D48 and (97 G58 + 3 M60)–(H300-stat-ERh1)–D52 vesicles, with mean pseudospherical globule diameters (based on analyzing 50 globules averaged over several vesicles) of around 25 ± 6 nm and 28 ± 8 nm, respectively. Such values should be regarded as estimates because these surface globules are not as well-defined as those previously reported by Mable et al.42 This is because the DP of the globule-forming PDPA block is only 48 or 52 in the present study, so these surface features are both smaller and less prominent.
Fortunately, small-angle X-ray scattering (SAXS) provides a much more statistically meaningful estimate of the mean globule diameter.42 SAXS analysis of 1.0% w/w aqueous dispersions of (97 G58 + 3 M60)–(H300-stat-ERh1)–D52 enables rigorous assessment of the copolymer morphologies obtained at various solution pH. Synchrotron SAXS patterns (Fig. 4a) were collected at pH 7.4 (red), pH 5.5 (blue) and pH 3.0 (green). A two-population vesicle plus sphere model is required to fit such SAXS patterns, as reported previously for similar framboidal vesicles.43 Here, population 1 represents the overall vesicle morphology while population 2 accounts for the pseudo-spherical globules protruding from the vesicle membrane, see the schematic cartoons corresponding to pH 7.4 and pH 5.5 shown in Fig. 4a. A radius of gyration, Rg, of 2.4 nm and a membrane solvent fraction, xsol, of 0.40 were used as fixed parameters to fit these two SAXS patterns.
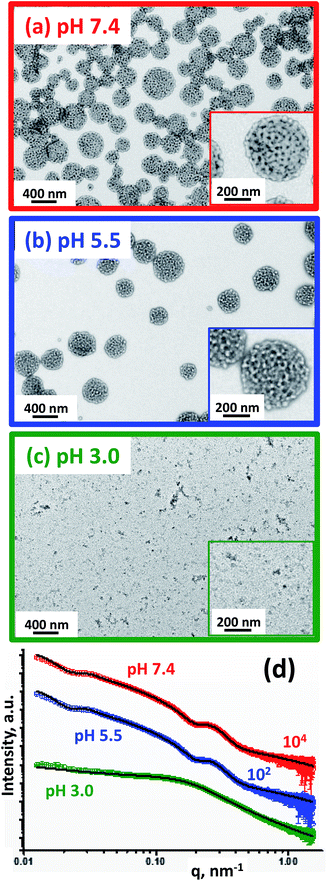 | ||
| Fig. 4 TEM images obtained for the rhodamine-labeled (97 G58 + 3 M60)–(H300-stat-ERh1)–D52 triblock copolymer nano-objects formed at (a) pH 7.4, (b) pH 5.5 and (c) pH 3.0. Inset: higher magnification TEM images and schematic cartoons representing the copolymer morphology (PGMA = red, PMPC = green, P(HPMA-stat-ERh1) = blue and PDPA = black). (d) SAXS patterns obtained for 1.0% w/w aqueous dispersions of rhodamine-labeled framboidal (97 G58 + 3 M60)–(H300-stat-ERh1)–D52 triblock copolymer vesicles prepared under sterile conditions via aqueous PISA at pH 7.4 (red), pH 5.5 (blue) and pH 3.0 (green). Open circles represent experimental data and solid black lines denote the fits to these X-ray patterns: a two-population ‘vesicle plus sphere’ model was required to fit the data recorded at pH 7.4 and pH 5.5, whereas a spherical micelle model incorporating mass fractals was required to fit the SAXS data obtained at pH 3.0 (Table S1†). | ||
At pH 7.4, SAXS analysis (Table S1†) indicates that the overall volume-average vesicle diameter, Dv, is 294 nm, which is reasonably consistent with the TEM and DLS data (Fig. 3). The mean core radius of the globules protruding from the framboidal vesicle membrane, Rs, is 9.01 ± 1.7 nm, thus the overall globule diameter, Ds, was calculated to be 27.6 nm (where Ds = 2Rs + 4Rg). This value is consistent with that estimated directly from the SAXS pattern, whereby the local minimum at q ∼ 0.226 nm leads to a mean globule diameter, Ds, of 27.7 nm (calculated using Ds = 2π/q). This is also consistent with the estimated globule diameter of 28 ± 8 nm from TEM analysis, as discussed above.
The TEM image obtained for the (97 G58 + 3 M60)–(H300-stat-ERh1)–D52 triblock copolymer vesicles dried at pH 5.5 (Fig. 4b, blue-framed image) suggests a similar framboidal morphology to that observed at pH 7.4. Indeed, DLS studies indicate a Dh of 329 ± 123 nm, which is comparable to that at pH 7.4 (Table 1). Dv increases by 4% to 305 nm at pH 5.5, which suggests slightly swollen vesicles. The mean vesicle membrane thickness, Tm, is reduced by 3% from 22.1 ± 5.0 nm at pH 7.4 to 21.5 ± 5.0 nm at pH 5.5 (see Table S1†). In addition, the relative proportion of population 2 (c2/c1) required to achieve a satisfactory fit to these SAXS patterns is reduced from 0.149 at pH 7.4 to 0.137 at pH 5.5, suggesting less framboidal character at the lower pH. These observations are consistent with protonation of some of the tertiary amine groups on the PDPA block, which consequently becomes less hydrophobic. This reduces the enthalpic incompatibility (and hence degree of microphase separation) between the two hydrophobic blocks, which leads to a discernible reduction in Rs from 9.01 ± 1.7 nm at pH 7.4 to 8.12 ± 1.6 nm at pH 5.5 (Table S1,† population 2). Aqueous electrophoresis data are also consistent with this interpretation: the zeta potential of the vesicles increases from −2.3 mV at pH 7.4 to +12 mV at pH 5.5 (Table 1). In summary, the TEM images, SAXS analysis and aqueous electrophoresis data suggest that cationic framboidal vesicles are formed at pH 5.5 (see inset in Fig. 4b). It is worth emphasizing that these framboidal vesicles remain intact at pH 5.5. Given that the pKa of the PDPA block is 6.2, this means that most of the tertiary amine groups are protonated at this pH which accounts for the appreciable cationic character for these slightly swollen vesicles. This is comparable to the exit mechanism for the Dengue virus from the endosomes at pH 5.5, whereby its overall diameter increases by approximately 10% and a more ‘open’ conformation is adopted.22,23
At pH 3.0, the PDPA blocks should be fully protonated. TEM analysis indicates that relatively small nano-objects are formed under these conditions, although it is difficult to assign the predominant copolymer morphology (Fig. 4c, green-framed image). DLS studies indicate the presence of nanoparticles with a sphere-equivalent intensity-average diameter of 39 ± 16 nm and a count rate of 4400 kcps. The latter is a measure of the light scattering intensity and should be compared to that of 106![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 kcps observed at pH 7.4 for the original framboidal vesicles. The SAXS pattern recorded for the aqueous dispersion of (97 G58 + 3 M60)–(H300-stat-ERh1)–D52 at pH 3.0 differs markedly from those collected at pH 7.4 and pH 5.5 (Fig. 4d). The former is relatively featureless, which is consistent with the anticipated pH-induced disintegration of the original framboidal vesicles to form pseudo-spherical micelles. The slight upturn in I(q) observed at low q indicates weakly-interacting particles, which is interpreted in terms of a mass fractal component.56 Indeed, a reasonably good data fit to the SAXS pattern recorded at pH 3.0 (Fig. 4d) can be achieved by employing a spherical micelle model (population 2 in Table S1†) combined with an additional population of mass fractals (population 3 in Table S1†), as recently reported by Mable et al.43 For this model, it was assumed that the protonated PDPA chains are located within the micelle corona along with the permanently hydrophilic PGMA and PMPC blocks. Thus, only the rhodamine-labeled PHPMA-rich block forms the nanoparticle cores.
300 kcps observed at pH 7.4 for the original framboidal vesicles. The SAXS pattern recorded for the aqueous dispersion of (97 G58 + 3 M60)–(H300-stat-ERh1)–D52 at pH 3.0 differs markedly from those collected at pH 7.4 and pH 5.5 (Fig. 4d). The former is relatively featureless, which is consistent with the anticipated pH-induced disintegration of the original framboidal vesicles to form pseudo-spherical micelles. The slight upturn in I(q) observed at low q indicates weakly-interacting particles, which is interpreted in terms of a mass fractal component.56 Indeed, a reasonably good data fit to the SAXS pattern recorded at pH 3.0 (Fig. 4d) can be achieved by employing a spherical micelle model (population 2 in Table S1†) combined with an additional population of mass fractals (population 3 in Table S1†), as recently reported by Mable et al.43 For this model, it was assumed that the protonated PDPA chains are located within the micelle corona along with the permanently hydrophilic PGMA and PMPC blocks. Thus, only the rhodamine-labeled PHPMA-rich block forms the nanoparticle cores.
The Rg and xsol were fixed parameters for fitting the SAXS pattern recorded at pH 3.0. It is physically reasonable to use the same Rg value as that required to fit the SAXS patterns observed at pH 7.4 and 5.5 because the PDPA block is actually shorter (DP = 52) than the PGMA and PMPC blocks (DP = 58 and 60, respectively). Consequently, when the PDPA block becomes hydrophilic, the coronal chain density increases, but the corona thickness remains essentially constant. Mable et al.42 reported that, for vesicle membranes comprising PHPMA chains with a fixed DP plus PBzMA chains of variable DP, the corresponding SAXS patterns are best fitted using a constant xsol value, rather than allowing xsol to vary. Thus, it is reasonable to use the same xsol value as that used for fitting the SAXS patterns recorded at pH 7.4 and 5.5.
Similarly, the cores of the pseudo-spherical nanoparticles formed at pH 3.0 comprise mainly HPMA units, with just one unit of GlyMA and rhodamine B piperazine per copolymer chain. Warren et al. used a similar solvent volume fraction parameter for the SAXS analysis of PHPMA-based vesicles.57 To summarize, an Rg of 2.4 nm and an xsol of 0.40 were used as fixed parameters for fitting the SAXS pattern recorded at pH 3.0. The mean micelle diameter Ds obtained from SAXS analysis [Ds = 2(Rs + 2Rg) = 25 nm] was consistent with DLS and TEM results (Table 1). A reduction in the micelle core diameter at pH 3.0 (15.0 nm, Table S1†) compared to the mean diameter of the globules within the framboidal vesicle membranes at pH 5.5 (16.2 nm, Table S1†) supports the suggested location of the PDPA chains within the micelle corona at the former pH. The mass fractal dimension d obtained from SAXS analysis is relatively low (d ∼ 1.76, Table S1†), indicating that the framboidal vesicles disintegrate to form weakly-interacting spherical micelles as suggested by TEM (Fig. 4c) and DLS studies.
The aqueous vesicle dispersions produced via the ‘sterile synthesis’ protocol were assessed for the presence of bacteria. Bioburden analysis confirmed that these dispersions were indeed sterile after sample inoculation in liquid media (no turbidity) and on agar plates (no colony growth), see Fig. S3.† In contrast, aqueous vesicle dispersions prepared using a standard non-sterile protocol tested for positive for the presence of bacteria, as judged by the greater turbidity of the bacterial broth and also colony growth on agar plates (Fig. S3†). Sterile aqueous dispersions were then evaluated for their biocompatibility via MTT assays using human dermal fibroblast (HDF) cells for 24 h (Fig. S4†). No detrimental effect on the HDF cells (>95% viability) was observed for copolymer concentrations ranging from 0.25 to 5.00% w/v when compared to a control group. Such concentrations were significantly greater than the 0.1% w/v copolymer concentration used for the cell uptake experiments (Fig. S4†). These cell viability data confirmed that highly biocompatible vesicles were obtained using the sterile PISA synthesis protocol, which were suitable for subsequent cell uptake studies.
Cell uptake experiments
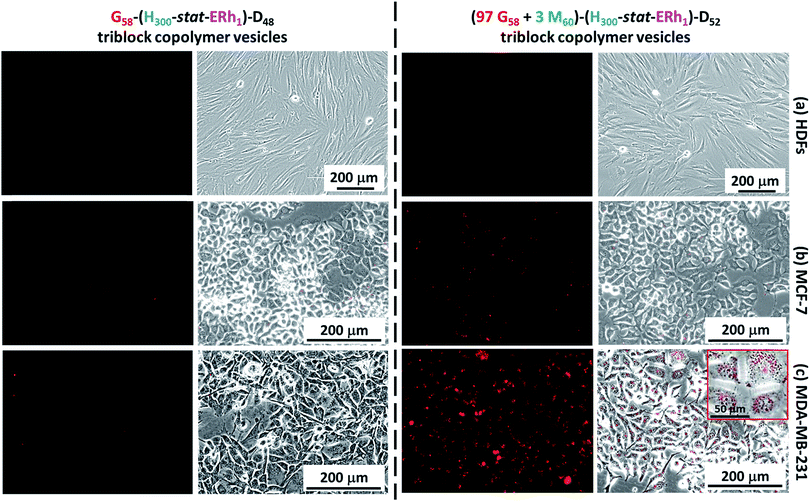
Cell uptake experiments were performed using non-cancerous HDF cells, the human breast cancer cell line MCF-7 and the TN breast cancer cell line MDA-MB-231 in conjunction with the framboidal G58–(H300-stat-ERh1)–D48 and (97 G58 + 3 M60)–(H300-stat-ERh1)–D52 triblock copolymer vesicles. Flow cytometry studies confirmed that all three cell types express SR-B1 and/or CD-36 scavenger receptors (see Fig. S5†), which are known to facilitate uptake of phosphorylcholine-decorated nanoparticles.50,51 More specifically, both HDFs and MCF-7 overexpress CD-36, whereas the MDA-MB-231 cells express minimal levels of this receptor. Conversely, MDA-MB-231 cells overexpress SR-B1 but both HDFs and MCF-7 exhibit negligible expression of SR-B1.Experiments performed over 12 h using the G58–(H300-stat-ERh1)–D48 framboidal vesicles revealed that their uptake was negligible for all three cell lines (Fig. 5). This is attributed to their all-hydroxyl surface functionality. However, fluorescence microscopy studies using the (97 G58 + 3 M60)–(H300-stat-ERh1)–D52 triblock copolymer vesicles conducted in parallel over the same time scale revealed that MDA-MB-231 cells (which overexpress SR-B1 receptors) were internalized but uptake of these nanoparticles was negligible for both the MCF-7 and HDF cells (Fig. 5).
This was confirmed by flow cytometry experiments (see Fig. S6†). This suggests that the PMPC targeting ligand is essential for the rapid intracellular uptake of framboidal vesicles by cells that overexpress SR-B1 receptors. Moreover, these studies confirmed that some internalized vesicles were retained at the periphery of the nuclear region in the endolysosomal compartments while others are located within the nuclear region (see inset shown in Fig. 5c).
To investigate this observation further, internalized rhodamine-labeled nanoparticles (red) were co-localized with the MDA-MB-231 cell nuclei, which were stained blue using Hoechst 33342. Co-staining within the nuclear region was confirmed (see pink regions in Fig. S7†), indicating that a significant proportion of the copolymer escapes from the endocytic pathway and reaches the cell nuclei, as expected for virus-mimicking nanoparticles (Fig. S7†).
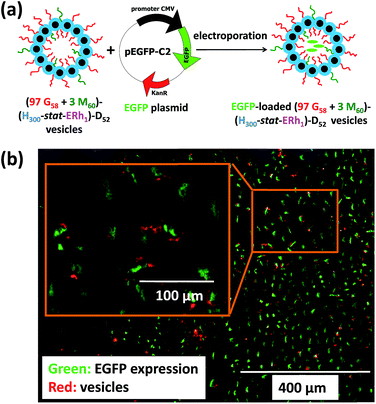
To further evaluate the ability of these new virus-mimicking framboidal vesicles to target the nuclear region, electroporation was used to load pEGFP DNA within (97 G58 + 3 M60)–(H300-stat-ERh1)–D52 framboidal vesicles (Fig. 6a). Gel shift assay analysis indicated that 87% of supercoiled pEGFP was encapsulated within the framboidal vesicles, which corresponds to approximately 70% of the total amount of plasmid DNA (see Fig. S8†).
Then cell uptake experiments were performed for 16 h using MDA-MB-231 cells. Delivery of this plasmid nucleic acid payload to the cell nuclei was confirmed via expression of fluorescent EGFP within the cells (see Fig. 6b and S9†). This confirmed the endolysosomal release of fully-functional biomolecules (in this case, DNA) with no significant toxicity (Fig. S4†). It is perhaps worth emphasizing that the framboidal vesicle morphology remained intact after electroporation, as confirmed by TEM studies (see Fig. S10†).
Herein, we utilize a schematic cartoon to discuss the in situ acidification of (97 G58 + 3 M60)–(H300-stat-ERh1)–D52 framboidal vesicles during their endocytic internalization by MDA-MB-231 breast cancer cells (Fig. 7a). At physiological pH, the framboidal vesicle morphology is stable (Fig. 4a and 7b, and Table 1). At the relevant endosomal pH (pH 5.5 to pH 6.5), the hydrophobic PDPA chains become partially protonated so the vesicles begin to swell but remain intact (Fig. 4b and 7c, and Table 1). However, further reduction in solution pH (such as that typically found within the lysosomal compartment) leads to rapid vesicle dissociation (Fig. 4c and 7d, and Table 1).
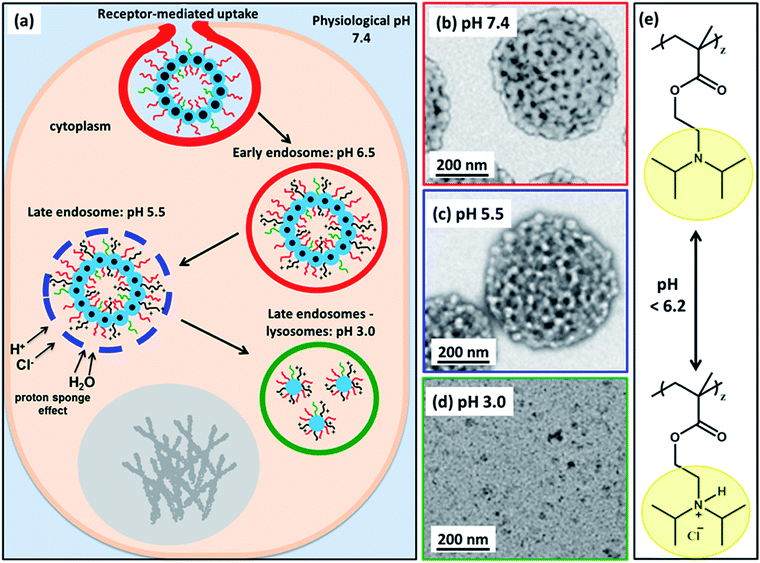 | ||
| Fig. 7 (a) Schematic cartoon depicting the likely effect of intracellular acidification of (97 G58 + 3 M60)–(H300-stat-ERh1)–D52 nanoparticles during their endocytic internalization by MDA-MB-231 triple-negative breast cancer cells. After internalization by these cells, the vesicles are expected to remain intact at physiological pH, see TEM image (b). As the pH drops below pH 6.2 in the early endosomes, the PDPA block becomes partially protonated, see chemical structure shown in (e), and thus less hydrophobic, which leads to vesicle swelling (and perhaps greater porosity). At pH 5.5, the vesicle membrane becomes increasingly porous as the PDPA block loses its hydrophobic character, see TEM image (c). The gradual increase in cationic charge density is believed to trigger a ‘proton sponge’ effect64 as water swells the late endosomes to counteract the increase in ionic strength. This should cause temporary rupture of the endosomes and release of its cargo into the nuclear region.64,65 In the lysosomes the local pH is 3.0. The PDPA block is expected to be fully protonated at this pH, leading to the formation of weakly-interacting cationic spherical micelles, see TEM image (d). These pH-induced morphological transitions can be studied by TEM for dilute aqueous vesicle dispersions dried at (b) pH 7.4, (c) pH 5.5 and (d) pH 3.0. (e) Chemical structure of the PDPA block when fully deprotonated above pH 6.2 and when full protonated below pH 6.2 (this pH approximately corresponds to the pKa of the PDPA block, although this parameter is dependent on the ionic strength). | ||
This is because the pendent tertiary amine groups on the block become fully protonated and, as a consequence, the PDPA hydrophobic chains become hydrophilic.58,59 The copolymer chains do not become molecularly dissolved but instead form weakly-interacting cationic micelles with mass fractal character. These micelles comprise a mixture of PGMA, PMPC and PDPA chains in the coronal layer and HPMA-rich cores.
Discussion
Many cancer cell types use scavenger receptors in addition to de novo lipogenesis to acquire lipids, fatty acids etc. Subsequent lipolysis provides additional energy that fuels their growth.60–62 Thus cancer cells overexpress scavenger receptors at their surface to maximize their lipid uptake.61Both SR-B1 and CD-36 scavenger receptors are highly expressed by many rapidly-growing malignant tumors, including breast cancer.60,62,63 In view of the high metabolic activity that is linked to the overexpression of SR-B1/CD-36 by cancer cells, such receptors were considered to be attractive targets for developing selective cancer therapies based on the new virus-mimicking framboidal vesicles reported herein. More specifically, TN breast cancer cells (i.e. ER-negative, PR-negative and those for which HER2/Neu is not overexpressed) lack suitable targeted therapies24 and both overexpress SR-B1 and underexpress CD-36.25 Not surprisingly, such aggressive malignant tumors are usually associated with very poor prognoses. Interestingly, the same pattern of expression (i.e. relatively high levels of SR-B1 and low levels of CD-36 in TN cell line MDA-MB-231) was observed in our study. Furthermore, de Gonzalo-Calvo and collaborators25 reported that such behaviour led more effective accumulation of intratumoral lipids in such tumors, which suggests that targeting the SR-B1 receptor could provide an alternative therapy for TN breast cancers.
However, such targeting is non-trivial, as scavenger receptors are notoriously promiscuous in their affinity for many ligands.64,65 Nevertheless, it is increasingly apparent that members of the Flaviviridae family of viruses (i.e. the Hepatitis C virus66 and, more recently, the Dengue virus15) can hijack the SR-B1 cell entry pathway to facilitate efficient infection with high cell specificity. In particular, the relatively smooth surface of the Dengue virus observed in mosquitos at 28 °C acquires distinctly framboidal character in its human host at 37 °C.13 This thermally-induced morphological transition is believed to be strongly associated with the mechanism of infection, most likely because it exposes critical domains.13,14 The full entry path of this virus has not yet been elucidated, but it is known that the SR-B1 scavenger receptor plays an important role.15 The wholly synthetic Dengue virus-mimicking framboidal vesicles described herein express phosphorylcholine-functionalized PMPC chains at their surface in order to target SR-B1/CD-36 receptors. Interestingly, such framboidal vesicles proved to be very effective for targeting human breast cancer cells that overexpress SR-B1 receptors along with very low CD-36 receptor expression. In contrast, essentially no uptake was observed within 12 h for (i) non-cancerous HDF cells expressing low levels of such receptors and (ii) for breast cancer cells that do not express the SR-B1 receptor. Particle diameters of 20–30 nm have been reported to be optimal for CD36 and SRB-1 receptors when targeting ‘natural’ endogenous particles within the cell that are involved in the transport and metabolism of cholesterol, phospholipids and triglycerides.27,28 Such receptors are also involved in the uptake of many exogenous particles greater than 20–30 nm diameter,69 such as the Dengue virus.15 Moreover, small surface domains of PMPC ligand on smooth vesicles of up to 400 nm diameter are known to enhance their intracellular uptake.70 However, to the best of our knowledge the use of framboidal synthetic vesicles is novel and the unexpected selectivity observed for SRB1 is also new. Interestingly, the Dengue virus acquires ‘framboidal’ character as part of its infection strategy, which is known to involve the SRB1 receptor. Moreover, the dimensions of this virus exceeds that of the ‘natural’ targets for this receptor. Thus such behavior mimics the efficient targeting of SR-B1 displayed by the Dengue virus. This was somewhat unexpected, because it is well-known that ‘smooth’ (i.e. non-framboidal) phosphorylcholine-functionalized liposomes40 or vesicles41 are avidly taken up by a wide range of cell lines that express type B scavenger receptors. In the latter study, receptor blocking experiments confirmed that both SR-B1 and CD36 are involved in the uptake of PMPC-decorated vesicles by both normal and cancer cell lines. Thus it seems likely that the distinctive framboidal vesicle morphology may facilitate more efficient targeting of cancer cells that express SR-B1. In this context, it is perhaps noteworthy that nanodomains protruding from synthetic block copolymer vesicles have also been found to perform better than conventional ‘smooth’ vesicles. The former exhibit extended blood circulation times and faster cellular uptake than the latter, even when the former vesicles are considerably larger.7 Physical models also suggest that protruding nanoscale surface domains on nanoparticles can induce sufficient membrane deformation to promote their more efficient engulfment.16–18 Thus, framboidal surface character could be advantageous not only for targeting cancer cells that express SR-B1 but also for enhanced endocytosis. Furthermore, unlike the equivalent smooth nanoparticles, Hu and co-workers7 demonstrated that nanoparticles comprising protruding nanodomains could bypass endolysosomal compartments and efficiently deliver drugs to the nuclear region.7 However, the precise mechanism of endocytosis and release remains unclear for such systems. Nevertheless, the Dengue virus similarly utilizes nanoscale surface nanodomains along with critical pH-dependent activation to ensure efficient transport within the endocytic organelles and exit at the perinuclear area.71 The surface domains formed by the E glycoprotein become disentangled at pH 5.5, exposing an ‘open’ conformation that enables effective fusion with the endosomal membrane.22,23 Like the Dengue virus, a subtle change in morphology (and surface charge) is observed for these framboidal vesicles as the tertiary amine groups on the PDPA chains become partially protonated at around pH 5.5. It is well-known67,68 that polymeric transfection reagents such as poly(ethylene imine) have primary, secondary and tertiary amines with pKa values covering the whole physiological pH range, thus enabling buffering capacity within the endosomes and high levels of gene transfer to the cell nucleus. The high buffering capacity of such synthetic polyamines enhances proton pumping into the endosomes, which also increases the influx of chloride ions to maintain charge neutrality.67 As a result, the higher ionic strength within the endosomes causes osmotic swelling, temporary rupture of the endosomal membrane and hence release of cargo at the periphery of the nuclear region.67,68 We hypothesize that the PDPA block similarly buffers acidification of the endosomes and, although part of the cargo is retained within the endolysosomes, sufficient plasmid DNA can be delivered to the nuclear region to enable EGFP expression by the targeted cells.
Conclusions
Herein we report the efficient synthesis of rhodamine-labeled pH-responsive framboidal triblock copolymer vesicles in sterile aqueous media. TEM and SAXS studies indicate that such vesicles comprise surface globules of approximately 28 nm and their pH-responsive character enables the intracellular release of biologically-relevant cargoes (e.g. plasmid DNA) within the nuclear region, suggesting release via an endocytic pathway. In vitro studies confirm that introducing a phosphorylcholine-based targeting ligand for the SR-B1 receptor is essential for the selective intracellular uptake by MDA-MB-231 breast cancer cells compared to non-cancerous HDF cells, because the former cells overexpress such surface receptors. In summary, these new synthetic vectors suggest new therapeutic approaches for treating TN breast cancer, for which existing treatments offer only relatively poor prognoses.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Christopher Hill and Svetomir Tzokov at the University of Sheffield Biomedical Science Electron Microscopy Suite for their assistance with the TEM studies. Diamond Light Source synchrotron (Didcot, UK) is acknowledged for providing synchrotron beamtime at I22. SPA thanks the European Research Council for an ERC Advanced Investigator grant (PISA 320372) which provided a PhD studentship for CJM. SPA also thanks EPSRC for postdoctoral support of IC (EP/L024160) and for an Established Career Particle Technology Fellowship (EP/R003009).References
- N. F. Steinmetz, Nanomedicine, 2010, 6(5), 634–641 CrossRef CAS PubMed.
- J. Grove and M. Marsh, J. Cell Biol., 2011, 195(7), 1071–1082 CrossRef CAS PubMed.
- H. E. van Kan-Davelaar, J. C. M. van Hest, J. J. L. M. Cornelissen and M. S. T. Koay, Br. J. Pharmacol., 2014, 171(17), 4001–4009 CrossRef CAS PubMed.
- C. E. Thomas, A. Ehrhardt and M. A. Kay, Nat. Rev. Genet., 2003, 4, 346 CrossRef CAS PubMed.
- J.-W. Yoo, D. J. Irvine, D. E. Discher and S. Mitragotri, Nat. Rev. Drug Discovery, 2011, 10(7), 521–535 CrossRef CAS PubMed.
- M. Soliman, R. Nasanit, S. R. Abulateefeh, S. Allen, M. C. Davies, S. S. Briggs, L. W. Seymour, J. A. Preece, A. M. Grabowska, S. A. Watson and C. Alexander, Mol. Pharm., 2012, 9(1), 1–13 CrossRef CAS PubMed.
- X. Hu, J. Hu, J. Tian, Z. Ge, G. Zhang, K. Luo and S. Liu, J. Am. Chem. Soc., 2013, 135(46), 17617–17629 CrossRef CAS PubMed.
- Y. Cheng, R. C. Yumul and S. H. Pun, Angew. Chem., Int. Ed., 2016, 55(39), 12013–12017 CrossRef CAS PubMed.
- D. O. White and F. F. Fenner, Medical Virology, Academic Press, United States of America San Diego, 4th edn, 1994, p. 603 Search PubMed.
- C. Loney, G. Mottet-Osman, L. Roux and D. Bhella, J. Virol., 2009, 83(16), 8191–8197 CrossRef CAS PubMed.
- A. Z. Wang, R. Langer and O. C. Farokhzad, Annu. Rev. Med., 2012, 63(1), 185–198 CrossRef CAS PubMed.
- J. Zhao and M. H. Stenzel, Polym. Chem., 2018, 9(3), 259–272 RSC.
- X. Zhang, J. Sheng, P. Plevka, R. J. Kuhn, M. S. Diamond and M. G. Rossmann, Proc. Natl. Acad. Sci. U. S. A., 2013, 110(17), 6795–6799 CrossRef CAS PubMed.
- F. A. Rey, Nature, 2013, 497(7450), 443–444 CrossRef CAS PubMed.
- Y. Li, C. Kakinami, Q. Li, B. Yang and H. Li, PLoS One, 2013, 8(7), e70390 CrossRef CAS PubMed.
- H. Gao, W. Shi and L. B. Freund, Proc. Natl. Acad. Sci. U. S. A., 2005, 102(27), 9469–9474 CrossRef CAS PubMed.
- S. Zhang, J. Li, G. Lykotrafitis, G. Bao and S. Suresh, Adv. Mater., 2009, 21(4), 419–424 CrossRef CAS PubMed.
- H. Yuan, J. Li, G. Bao and S. Zhang, Phys. Rev. Lett., 2010, 105(13), 138101 CrossRef PubMed.
- Y. Niu, M. Yu, S. B. Hartono, J. Yang, H. Xu, H. Zhang, J. Zhang, J. Zou, A. Dexter, W. Gu and C. Yu, Adv. Mater., 2013, 25(43), 6233–6237 CrossRef CAS PubMed.
- W. Wang, P. Wang, X. Tang, A. A. Elzatahry, S. Wang, D. Al-Dahyan, M. Zhao, C. Yao, C.-T. Hung, X. Zhu, T. Zhao, X. Li, F. Zhang and D. Zhao, ACS Cent. Sci., 2017, 3(8), 839–846 CrossRef CAS PubMed.
- X. Zhang, P. Ge, X. Yu, J. M. Brannan, G. Bi, Q. Zhang, S. Schein and Z. H. Zhou, Nat. Struct. Mol. Biol., 2013, 20(1), 105–110 CrossRef CAS PubMed.
- X. Zhang, J. Sheng, S. K. Austin, T. E. Hoornweg, J. M. Smit, R. J. Kuhn, M. S. Diamond and M. G. Rossmann, J. Virol., 2015, 89(1), 743–750 CrossRef PubMed.
- Y. Modis, S. Ogata, D. Clements and S. C. Harrison, Proc. Natl. Acad. Sci. U. S. A., 2003, 100(12), 6986–6991 CrossRef CAS PubMed.
- W. J. Irvin and L. A. Carey, Eur. J. Cancer, 2008, 44(18), 2799–2805 CrossRef CAS PubMed.
- D. de Gonzalo-Calvo, L. López-Vilaró, L. Nasarre, M. Perez-Olabarria, T. Vázquez, D. Escuin, L. Badimon, A. Barnadas, E. Lerma and V. Llorente-Cortés, BMC Cancer, 2015, 15(1), 460 CrossRef PubMed.
- P. C. Patel, D. A. Giljohann, W. L. Daniel, D. Zheng, A. E. Prigodich and C. A. Mirkin, Bioconjugate Chem., 2010, 21(12), 2250–2256 CrossRef CAS PubMed.
- K. K. Ng, J. F. Lovell and G. Zheng, Acc. Chem. Res., 2011, 44(10), 1105–1113 CrossRef CAS PubMed.
- R. Kuai, D. Li, Y. E. Chen, J. J. Moon and A. Schwendeman, ACS Nano, 2016, 10(3), 3015–3041 CrossRef CAS PubMed.
- A. K. Brannan and F. S. Bates, Macromolecules, 2004, 37(24), 8816–8819 CrossRef CAS.
- A. Blanazs, S. P. Armes and A. J. Ryan, Macromol. Rapid Commun., 2009, 30(4–5), 267–277 CrossRef CAS PubMed.
- J. Du and R. K. O'Reilly, Soft Matter, 2009, 5(19), 3544–3561 RSC.
- Y. Mai and A. Eisenberg, Chem. Soc. Rev., 2012, 41(18), 5969–5985 RSC.
- G. Delaittre, M. Save and B. Charleux, Macromol. Rapid Commun., 2007, 28(15), 1528–1533 CrossRef CAS.
- B. Charleux, G. Delaittre, J. Rieger and F. D'Agosto, Macromolecules, 2012, 45(17), 6753–6765 CrossRef CAS.
- Z. An, Q. Shi, W. Tang, C.-K. Tsung, C. J. Hawker and G. D. Stucky, J. Am. Chem. Soc., 2007, 129(46), 14493–14499 CrossRef CAS PubMed.
- J. Rieger, C. Grazon, B. Charleux, D. Alaimo and C. Jérôme, J. Polym. Sci., Part A: Polym. Chem., 2009, 47(9), 2373–2390 CrossRef CAS.
- Y. T. Li and S. P. Armes, Angew. Chem., Int. Ed., 2010, 49(24), 4042–4046 CrossRef CAS PubMed.
- A. Blanazs, J. Madsen, G. Battaglia, A. J. Ryan and S. P. Armes, J. Am. Chem. Soc., 2011, 133(41), 16581–16587 CrossRef CAS PubMed.
- A. Blanazs, A. J. Ryan and S. P. Armes, Macromolecules, 2012, 45(12), 5099–5107 CrossRef CAS.
- K. L. Thompson, P. Chambon, R. Verber and S. P. Armes, J. Am. Chem. Soc., 2012, 134(30), 12450–12453 CrossRef CAS PubMed.
- P. Chambon, A. Blanazs, G. Battaglia and S. P. Armes, Macromolecules, 2012, 45(12), 5081–5090 CrossRef CAS.
- C. J. Mable, N. J. Warren, K. L. Thompson, O. O. Mykhaylyk and S. P. Armes, Chem. Sci., 2015, 6(11), 6179–6188 RSC.
- C. J. Mable, L. A. Fielding, M. J. Derry, O. O. Mykhaylyk, P. Chambon and S. P. Armes, Chem. Sci., 2018, 9(6), 1454–1463 RSC.
- K. Ishihara, T. Ueda and N. Nakabayashi, Polym. J., 1990, 22, 355 CrossRef CAS.
- A. L. Lewis, Colloids Surf., B, 2000, 18(3), 261–275 CrossRef CAS PubMed.
- Y. Iwasaki and K. Ishihara, Anal. Bioanal. Chem., 2005, 381(3), 534–546 CrossRef CAS PubMed.
- J. Du, Y. Tang, A. L. Lewis and S. P. Armes, J. Am. Chem. Soc., 2005, 127(51), 17982–17983 CrossRef CAS PubMed.
- N. Bhuchar, Z. Deng, K. Ishihara and R. Narain, Polym. Chem., 2011, 2(3), 632–639 RSC.
- S. Morozova, G. Hu, T. Emrick and M. Muthukumar, ACS Macro Lett., 2016, 5(1), 118–122 CrossRef CAS.
- A. Boullier, P. Friedman, R. Harkewicz, K. Hartvigsen, S. R. Green, F. Almazan, E. A. Dennis, D. Steinberg, J. L. Witztum and O. Quehenberger, J. Lipid Res., 2005, 46(5), 969–976 CrossRef CAS PubMed.
- H. E. Colley, V. Hearnden, M. Avila-Olias, D. Cecchin, I. Canton, J. Madsen, S. MacNeil, N. Warren, K. Hu, J. A. McKeating, S. P. Armes, C. Murdoch, M. H. Thornhill and G. Battaglia, Mol. Pharm., 2014, 11(4), 1176–1188 CrossRef CAS PubMed.
- W. M. Cao, K. Murao, H. Imachi, X. Yu, H. Abe, A. Yamauchi, M. Niimi, A. Miyauchi, N. C. W. Wong and T. Ishida, Cancer Res., 2004, 64(4), 1515–1521 CrossRef CAS PubMed.
- C. G. Clarkson, J. R. Lovett, J. Madsen, S. P. Armes and M. Geoghegan, Macromol. Rapid Commun., 2015, 36(17), 1572–1577 CrossRef CAS PubMed.
- H. Zhang and E. Ruckenstein, Macromolecules, 2000, 33(13), 4738–4744 CrossRef CAS.
- M. Save, J. V. M. Weaver, S. P. Armes and P. McKenna, Macromolecules, 2002, 35(4), 1152–1159 CrossRef CAS.
- G. Beaucage, J. Appl. Crystallogr., 1995, 28, 717–728 CrossRef CAS.
- N. J. Warren, O. O. Mykhaylyk, D. Mahmood, A. J. Ryan and S. P. Armes, J. Am. Chem. Soc., 2013, 136(3), 1023–1033 CrossRef PubMed.
- A. S. Lee, A. P. Gast, V. Bütün and S. P. Armes, Macromolecules, 1999, 32(13), 4302–4310 CrossRef CAS.
- H. Lomas, I. Canton, S. MacNeil, J. Du, S. P. Armes, A. J. Ryan, A. L. Lewis and G. Battaglia, Adv. Mater., 2007, 19(23), 4238–4243 CrossRef CAS.
- S.-I. Yamashita, J.-I. Yamashita, K. Sakamoto, K. Inada, Y. Nakashima, K. Murata, T. Saishoji, K. Nomura and M. Ogawa, Cancer, 1993, 71(10), 3058–3064 CrossRef CAS PubMed.
- N. B. Kuemmerle, E. Rysman, P. S. Lombardo, A. J. Flanagan, B. C. Lipe, W. A. Wells, J. R. Pettus, H. M. Froehlich, V. A. Memoli, P. M. Morganelli, J. V. Swinnen, L. A. Timmerman, L. Chaychi, C. J. Fricano, B. L. Eisenberg, W. B. Coleman and W. B. Kinlaw, Mol. Cancer Ther., 2011, 10(3), 427 CrossRef CAS PubMed.
- N. Zaidi, L. Lupien, N. B. Kuemmerle, W. B. Kinlaw, J. V. Swinnen and K. Smans, Prog. Lipid Res., 2013, 52(4), 585–589 CrossRef CAS PubMed.
- M. M. K. Shahzad, L. S. Mangala, H. D. Han, C. Lu, J. Bottsford-Miller, M. Nishimura, E. M. Mora, J.-W. Lee, R. L. Stone, C. V. Pecot, D. Thanapprapasr, J.-W. Roh, P. Gaur, M. P. Nair, Y.-Y. Park, N. Sabnis, M. T. Deavers, J.-S. Lee, L. M. Ellis, G. Lopez-Berestein, W. J. McConathy, L. Prokai, A. G. Lacko and A. K. Sood, Neoplasia, 2011, 13(4), 309–319 CrossRef CAS PubMed.
- A. Akinc, M. Thomas, A. M. Klibanov and R. Langer, J. Gene Med., 2005, 7, 657–663 CrossRef CAS PubMed.
- J. P. Behr, Chimia, 1997, 51, 34–36 CAS.
- N. Platt and S. Gordon, Chem. Biol., 1998, 5(8), R193–R203 CrossRef CAS PubMed.
- J. Canton, D. Neculai and S. Grinstein, Nat. Rev. Immunol., 2013, 13, 621 CrossRef CAS PubMed.
- C. Voisset, N. Callens, E. Blanchard, A. Op De Beeck, J. Dubuisson and N. Vu-Dac, J. Biol. Chem., 2005, 280(9), 7793–7799 CrossRef CAS PubMed.
- I. A. Zani, S. L. Stephen, N. A. Mughal, D. Russell, S. Homer-Vanniasinkam, S. B. Wheatcroft and S. Ponnambalam, Cells, 2015, 4, 178–201 CrossRef PubMed.
- C. LoPresti, M. Massignani, C. Fernyhough, A. Blanazs, A. J. Ryan, J. Madsen, N. J. Warren, S. P. Armes, A. L. Lewis, S. Chirasatitsin, A. J. Engler and G. Battaglia, ACS Nano, 2011, 5, 1775–1784 CrossRef CAS PubMed.
- H. M. van der Schaar, M. J. Rust, C. Chen, H. van der Ende-Metselaar, J. Wilschut, X. Zhuang and J. M. Smit, PLoS Pathog., 2008, 4, e1000244 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Full experimental details for the synthesis and characterization of the copolymer vesicles, assigned 1H NMR spectra, GPC traces, bioburden and MTT test results, flow cytometry scavenger receptor expression studies, co-localization studies and a table of structural parameters obtained from SAXS analysis are provided. See DOI: 10.1039/c8sc05589k |
| ‡ The corresponding authors contributed equally. |
| § Present address: Faculty of Engineering and Natural Sciences, Sabancı University, Orta Mahalle, 34956 Tuzla, Istanbul, Turkey. |
| ¶ Present address: Department of Chemistry, University of Liverpool, Crown Street, Liverpool, L69 7ZD, UK. |
| || Present address: School of Chemistry and Chemical Engineering, Queen's University Belfast, Belfast BT9 5AG, UK. |
| This journal is © The Royal Society of Chemistry 2019 |