Open Access Article
Open Access ArticleCharacterizing gibberellin flow in planta using photocaged gibberellins†
Shira
Wexler
a,
Hilla
Schayek
a,
Kandhikonda
Rajendar
a,
Iris
Tal
a,
Eilon
Shani
 a,
Yasmine
Meroz
a,
Yasmine
Meroz
 a,
Roman
Dobrovetsky
a,
Roman
Dobrovetsky
 b and
Roy
Weinstain
b and
Roy
Weinstain
 *a
*a
aSchool of Plant Sciences and Food Security, Faculty of Life Sciences, Tel Aviv University, Tel Aviv 69978, Israel. E-mail: royweinstain@post.tau.ac.il
bSchool of Chemistry, Raymond and Beverly Sackler Faculty of Exact Sciences, Tel Aviv University, Tel Aviv 69978, Israel
First published on 20th November 2018
Abstract
Gibberellins (GAs) are ubiquitous plant hormones that coordinate central developmental and adaptive growth processes in plants. Accurate movement of GAs throughout the plant from their sources to their destination sites is emerging to be a highly regulated and directed process. We report on the development of novel photocaged gibberellins that, in combination with a genetically encoded GA-response marker, provide a unique platform to study GA movement at high-resolution, in real time and in living, intact plants. By applying this platform to the Arabidopsis thaliana endogenous bioactive gibberellin GA4, we measure kinetic parameters of its flow, such as decay length and velocity, in vivo.
Introduction
Gibberellins (GAs) are a class of tetra-cyclic di-terpenoid plant hormones that play a major role in regulation of key developmental and adaptive growth processes, including seed germination, organ elongation, transition from vegetative to reproductive growth, and flower, seed, and fruit development.1–3 Over the years, more than 130 GAs have been identified, of which less than a handful (namely GA1,3,4,7) are bioactive.4 Plants exert multiple layers of regulation (i.e. biosynthesis metabolism and perception4–7) to coordinate their GA responses in order to achieve appropriate development and growth. Recent evidences, including the identification of specific GA distribution patterns8,9 and characterization of putative GA transporters,10–12 suggest that plants also actively and directly regulate the movement of GAs via a set of protein transporters to ensure their localized activity. Consequently, understanding the dynamics of GA flow in plants is imperative for an understanding of their regulation and function.While the long-distance flow of GAs through the phloem sap is extensively being studied,13–15 the nature of their short-distance movement remains poorly characterized. This is mainly due to a lack of methods with sufficient resolution to introduce and then monitor the movement of GAs, especially in vivo. To date, the main approach to quantitatively characterize GA (as well as other plant hormones) short-distance movement has been the application of a radiolabeled GA to a whole plant, or its cut segments, and subsequently quantifying radioactivity at certain locations and times.16–20 Such experiments have enabled measuring, for example, the GA1 flux in oat coleoptiles.16 While highly sensitive and quantitative, the approach is limited by working with cut plant segments, low resolution and the need to apply the labeled hormone un-naturally from outside the plant, leading to issues of penetration efficiency and time, which complicate the analysis.
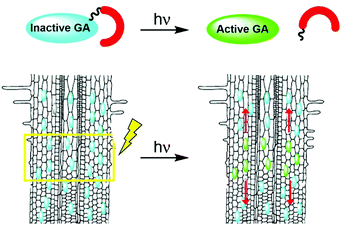
Photoactivation (photocaging21) has the potential to overcome the limitations associated with traditional applications of bioactive molecules: although caged molecules are introduced from the outside, bioactive molecules are practically released inside the organism, better mimicking endogenous conditions and overcoming penetration issues. In addition, light facilitates unprecedented spatio-temporal control over activation of a caged molecule, enabling initiation of a biological process in a highly defined area and monitoring of it from the get go. Importantly, the technique can be applied to whole, living organisms. We hypothesized that development of photoactivatable GAs will provide an effective way to introduce bioactive GAs directly in planta with high spatio-temporal control and, coupled with a fluorescent GA response marker, will facilitate visualization and quantitation of their movement in vivo (Fig. 1).
The synthesis of caged gibberellins has been reported twice in the literature.22,23 The first report describes the synthesis of several GA3 conjugates, among them to a caging group, yet it focuses on the synthetic feasibility of such molecules' preparation and does not include any evaluation of their functionalities.22 In the second report, GA3 was caged with two-photon excitable caging groups,23 to complement a GA-based chemically induced dimerization (CID) system that was “hijacked” from plants to induce protein degradation in mammalian cells.24 Understandably, its suitability to the very different physiological conditions in plants has not been evaluated, let alone, its applicability or utility in planta.
Here, we optimize the chemical structure required to generate caged gibberellins that are stable in vivo in the absence of light, using GA3 as a model bioactive gibberellin. Based on the resultant insights, we synthesized a caged version of the Arabidopsis thaliana endogenous bioactive gibberellin GA4, and, in conjunction with a genetically encoded fluorescent GA-response marker, demonstrated real-time monitoring of GA4 movement in roots of intact, live plants. Furthermore, the technique enabled extraction of quantitative parameters of GA4 movement over short distances in planta.
Results and discussion
Design considerations and synthesis of photocaged GA3
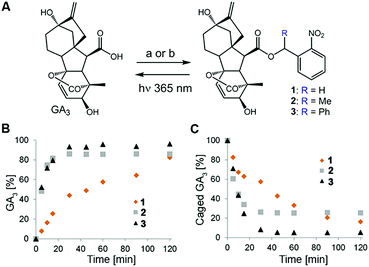
Design of the caged GA was based on our previous observation that conjugation of a bulky molecule in close proximity to bioactive gibberellins results in the latter's complete loss of function in vitro and in vivo.9 We opted for the readily available GA3 for use as a model bioactive gibberellin and nitrobenzyls as photoprotecting groups25 because their structural simplicity provides access to diverse derivatives and their small structure adds minimal hydrophobicity to the already hydrophobic GA. We hypothesized that hydrolytic enzymes (e.g. esterases) might exist in planta that will cleave the ester bond connecting GA3 to the photo-protecting group, resulting in light-independent activation of GA3. We therefore synthesized GA3 caged with three derivatives of 2-nitrobenzyl, characterized by increasing steric hindrance around the conjugating ester bond (1–3, Fig. 2A), anticipating that more steric hindrance will lead to increased stability in planta.The caged GA3 derivatives were synthesized by reacting GA3 with either the appropriate 2-nitrobenzyl bromide (1–2) or 2-nitrobenzyl alcohol (3). Upon 365 nm irradiation, all three compounds effectively released GA3 with relatively similar overall chemical yields (82%, 85% and 96% for 1–3, respectively, Fig. 2B and C) albeit at different rates (t1/2 ≈ 28, 5.5 and 5 minutes, respectively). We verified that GA3 itself does not decompose under such prolonged irradiation conditions (Fig. S1†).
In vitro and in vivo evaluation of caged GA3 stability
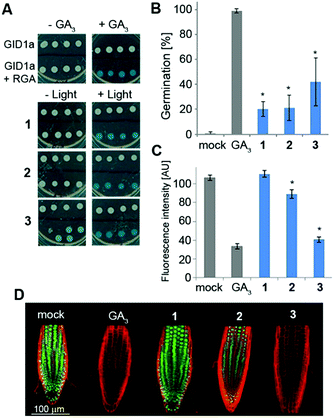
We next studied the derivatives' stability in the absence of light. This is a crucial property as inadvertent release of bioactive molecules completely undermines the utility of the caging strategy. Mechanistically, GA responses in plants are initiated by the binding of a bioactive GA to the receptor GID1 (gibberellin insensitive dwarf 1), promoting its association with repressors of GA responses called DELLA proteins. This association triggers degradation of the DELLAs, resulting in activation of the GA responses.6 In a yeast-two-hybrid (Y2H) system that expresses both GID1a and the DELLA protein RGA (repressor of GAs), we evaluated whether irradiated and non-irradiated caged GA 1–3 would promote the interaction between these two proteins as bioactive GAs do (Fig. 3A). Pre-irradiated 1–3 (25 μM) demonstrated robust promotion of the interaction between GID1a and RGA (as evidenced by blue color formation) to the same extent as native GA3, confirming that photo-activation leads to release of a functional GA. Importantly, under dark conditions, 1 and 2 did not promote such an interaction, verifying that these caged derivatives are not recognized by the GA's perception mechanism and demonstrating dark stability in this experimental system. Conversely, 3 did promote the GID1a–RGA interaction in the dark, suggesting at least partial hydrolysis of the ester bond and release of free GA3. We next evaluated dark stability in planta. The GA biosynthesis mutant ga1–3, a knockout of the first enzyme in gibberellins' biosynthetic pathway26 (CPP synthase), germinates poorly due to the absence of an endogenous GA whereas application of an exogenous bioactive GA fully restores its germination competency (Fig. 3B). When ga1–3 seeds were treated with 1–3 (25 μM) for 4 days under light devoid of UV, significantly lower germination levels were observed for 1 and 2 (20% and 21%, respectively) in comparison to 3 (42%), albeit higher than that of the control (0%). Very similar results were observed in a root elongation assay (Fig. S2†), implying that 1 and 2 are more resistant to hydrolysis in planta than 3. In an effort to tease out a stability difference between 1 and 2, we utilized the genetically encoded GA marker GFP-RGA. The fused green fluorescent protein (GFP) and RGA serves as a “turn-off” marker for GAs; in the absence of GAs, GFP-RGA is stable and can be readily detected while in the presence of GAs, the fusion protein is degraded and the fluorescence signal diminishes.27 Treatment of transgenic Arabidopsis seedlings expressing GFP-RGA under RGA's native promoter (pRGA:GFP-RGA) with 1–3 (25 μM) revealed a significant difference; while 2 led to an ∼20% reduction in the fluorescence signal, 1 was completely stable at this time point and 3 showed the lowest fluorescence signal (Fig. 3C and D).Susceptibility to acidic hydrolysis correlates with caged GA stability in planta
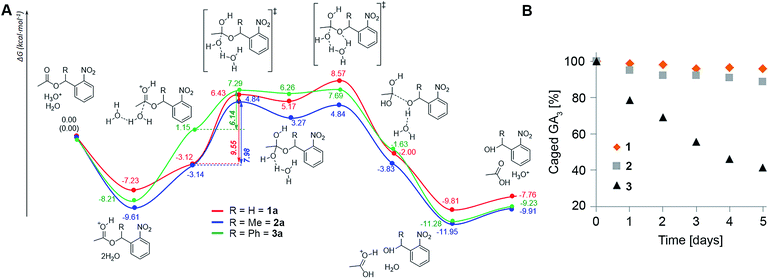
The assays described above reveal a clear trend of in planta stability with an inverse correlation to steric hindrance around the ester bond (1 > 2 > 3), suggesting that enzymatic cleavage is not the cause for degradation of the conjugates as was initially hypothesized. Since the apoplast (extracellular space) in plants is a low pH environment (ca. 5.5 (ref. 28)), we speculated that ester hydrolysis through an acidic mechanism could be responsible for degradation of the conjugates. To explore this assumption, we performed density functional theory (DFT) calculations of the potential energy surface (PES) of the hydrolysis of the model systems 1a–3a at the B3LYP/6-31(d) level of theory in water using a conductor-like polarizable continuum model (CPCM) (Fig. 4A). The results show that formation of the orthoesters is the rate determining step (RDS) of the hydrolysis reaction and is exergonic for all three model compounds (ΔG = 8.29, 7.98 and 5.11 kcal mol−1, respectively). The calculated barriers of the RDS for the hydrolysis of 1a is 1.57 kcal mol−1 higher than that for 2a and 3.41 kcal mol−1 higher than that for 3a. Thus, the barriers of the RDSs for the hydrolysis of esters of the order 1a > 2a > 3a clearly reflect the difference in hydrolysis rates (1 > 2 > 3) observed in planta. Moreover, incubation of 1–3 in liquid MS medium (pH 5.5) further corroborated the differences in the hydrolysis rate: 1 being slightly more resistant than 2 while 3 hydrolyses comparatively quickly (Fig. 4B). Collectively, both DFT calculations and experimental results point towards susceptibility to acidic hydrolysis as the determinant of GA conjugate stability in planta. This insight should be highly valuable in designing controlled-release conjugates of GAs.Caged GA4 can be effectively photolyzed in planta
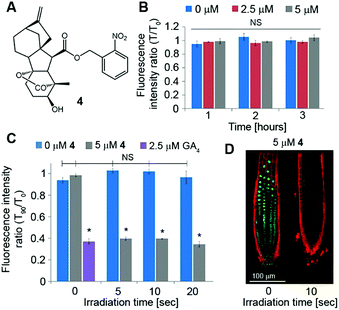
Although GA3 is a bioactive gibberellin, it is not endogenous to Arabidopsis thaliana. Previous studies have shown that putative GA transporters have differential specificity and activity towards distinct GAs, and therefore, GA variants are expected to be transported differentially in planta.29,30 Thus, in order to study the short-distance movement of a bioactive GA in its proper context, we synthesized a caged version of GA4, the endogenous and most abundant bioactive GA in Arabidopsis,31 in a similar manner to 1 (4, Fig. 5A). We first evaluated the stability of 4 in Arabidopsis seedlings. Transgenic pRGA:GFP-RGA seedlings were treated for 15 minutes with an increasing concentration of 4 (5–50 μM) and imaged 90 minutes later. At concentrations above 10 μM, a significant decrease in the GFP signal was observed, suggesting partial hydrolysis of 4 and release of GA4 in sufficient amounts to be detected (Fig. S3†). At lower concentrations however (2.5 and 5 μM), no change in the GFP fluorescence signal was observed for at least 3 hours (Fig. 5B and S4†), indicating the hydrolysis rate below the detection limit of the marker. These results provide a time-window of at least 3 hours for in vivo experiments using the optimized concentration of 4. We next tested photoactivation of 4in planta. Transgenic pRGA:GFP-RGA seedlings were treated for 15 minutes with 4 (2.5 and 5 μM); their root tips were irradiated using a confocal lamp (365/10 nm, 2 mW cm−2) for increasing periods of time and imaged 90 minutes later. Light exposure for >5 seconds resulted in a decrease in the GFP fluorescence signal to the same level observed in seedlings treated directly with GA4 (2.5 μM) (Fig. 5C and D and S5†). Importantly, seedlings that were exposed to similar irradiation conditions but in the absence of 4 showed no decrease in their GFP-RGA signal and revealed no signs of phototoxicity even at the highest light dosage, as determined by a follow-up root elongation assay (Fig. S6†). The response of the GA marker to the photoreleased GA4 could be monitored in real time (Movies S1, S2†), providing a platform to study its movement at high-resolution. Collectively, these results establish that a tunable pulse of bioactive GA4 can be successfully generated in planta by light control and that its movement from the activation point can be monitored in real time. This is the first demonstration of in vivo light-mediated control over this central plant hormone.Quantitation of GA4 movement in Arabidopsis roots
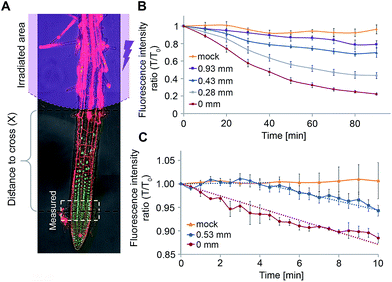
We next sought to utilize our technique's optimized parameters to extract kinetic properties of GA4 movement in roots of intact Arabidopsis plants. We first turned to evaluate the GA decay length in the root, a parameter that provides information about the remoteness at which these hormones can act. As weak acids, GAs tend to get trapped inside cells, where the relatively high pH (∼7.0) limits them from crossing into the apoplast. This “ion-trap” mechanism29 restricts the distance to which GAs can diffuse. The fraction of the GA that can cross a distance x through the apoplast f(x) was previously defined32 as| f(x) = 10−x/Lapo | (1) |
We postulated that our experimental system will allow us to directly measure f(x) for different distances x and thus, extract Lapo based on experimental observations. For this, transgenic pRGA:GFP-RGA seedlings were treated with 4 (5 μM) and irradiated (365/10 nm, 2 mW cm−2, 10 s) at increasing distances from the root tip (x = 0.28–0.93 ± 0.05 mm, Fig. 6A). The GFP-RGA signal was monitored at the root tip and quantified. The results showed that both the extent and rate of the marker response at the root tip decreases as the distance of the photoactivation location from the root tip increases (Fig. 6B). Since the intensity of the marker response is correlated with the amount of GA present, we substituted f(x) with the marker response for each distance x in eqn (1) thus extracting Lapo = 1.6 ± 0.4 mm (for a detailed calculation, see the ESI†). The finding that the observed Lapo is 1.5–2 orders of magnitude higher than the theoretically calculated one (which assumes no active transport) provides additional, experimental support to the assumption that active transport of GAs takes place, enabling GAs to overcome the “ion-trap” mechanism and travel relatively long distances in planta.
We utilized a similar experimental setup to measure the GA4 velocity in the root. This is an important parameter in defining which GA biosynthesis sites contribute to a GA response in a specific location. To be relevant, a GA must be able to cross the distance from its source to destination within the time-frame of a response initiation. Photoactivation of 4 at a known distance from the root tip (0.53 ± 0.05 mm) and monitoring of the marker at higher frequency (every 30 seconds) revealed a delay of 4 ± 1 minute in the marker's response compared to its photoactivation at the site of monitoring (0 mm, Fig. 6C). The observed lag time mostly represents the time required for GA4 to cross this distance, thus enabling determining a minimal velocity of 8.0 ± 2.0 mm h−1 for GA4 in the root tip. To the best of our knowledge, this is the first measurement of GA4 velocity in whole, live plants. The observed velocity is comparable to that measured for auxin in Arabidopsis roots (9 ± 1 mm h−1 (ref. 20 and 34)) and higher than that previously reported for GA3 in cut Coleus petioles (1.4 mm h−1 (ref. 35)).
Conclusions
In summary, we have established a conceptually novel platform to study the movement of bioactive GAs in whole, living plants. Development of photocaged gibberellins and their combination with a genetically encoded GA-response marker, enabled monitoring of the hormone's movement at high-resolution and in real time in living, intact plants. This technique was applied to quantitate kinetic properties of an endogenous gibberellin flow in roots, such as its decay length and velocity. These data will facilitate a better understanding of GA regulation and function in plants as well as provide concrete, experimental information to support development of computational models for GA flow. We expect that improving the spatial-resolution of caged GA photoactivation by switching to visible light-excitable photocages, along with replacement of the GA marker with a faster and more sensitive one,33 would significantly enhance the technique's accuracy and its capacity to unveil higher-resolution details of GA flow. We further anticipate that a similar approach could be taken to study other hormones and bioactive small-molecules in plants.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to thank the German-Israel Foundation (I-2387-302.5/2015), the European Research Council (679189) and the Israeli Science Foundation (1832/14) for funding this research.Notes and references
- P. Achard and P. Genschik, J. Exp. Bot., 2009, 60, 1085–1092 CrossRef CAS PubMed
.
- J. M. Daviere and P. Achard, Development, 2013, 140, 1147–1151 CrossRef CAS PubMed
.
- P. Hedden and V. Sponsel, J. Plant Growth Regul., 2015, 34, 740–760 CrossRef CAS PubMed
.
- S. Yamaguchi, Annu. Rev. Plant Biol., 2008, 59, 225–251 CrossRef CAS PubMed
.
- K. Murase, Y. Hirano, T. P. Sun and T. Hakoshima, Nature, 2008, 456, 459–463 CrossRef CAS PubMed
.
- T. P. Sun, Plant Physiol., 2010, 154, 567–570 CrossRef CAS PubMed
.
- M. Ueguchi-Tanaka, M. Ashikari, M. Nakajima, H. Itoh, E. Katoh, M. Kobayashi, T. Y. Chow, Y. I. Hsing, H. Kitano, I. Yamaguchi and M. Matsuoka, Nature, 2005, 437, 693–698 CrossRef CAS PubMed
.
- L. Matias-Hernandez, A. E. Aguilar-Jaramillo, M. Osnato, R. Weinstain, E. Shani, P. Suarez-Lopez and S. Pelaz, Plant Physiol., 2016, 170, 1624–1639 CAS
.
- E. Shani, R. Weinstain, Y. Zhang, C. Castillejo, E. Kaiserli, J. Chory, R. Y. Tsien and M. Estelle, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 4834–4839 CrossRef CAS PubMed
.
- Y. Chiba, T. Shimizu, S. Miyakawa, Y. Kanno, T. Koshiba, Y. Kamiya and M. Seo, J. Plant Res., 2015, 128, 679–686 CrossRef CAS PubMed
.
- Y. Kanno, T. Oikawa, Y. Chiba, Y. Ishimaru, T. Shimizu, N. Sano, T. Koshiba, Y. Kamiya, M. Ueda and M. Seo, Nat. Commun., 2016, 7, 13245 CrossRef CAS PubMed
.
- I. Tal, Y. Zhang, M. E. Jorgensen, O. Pisanty, I. C. Barbosa, M. Zourelidou, T. Regnault, C. Crocoll, C. E. Olsen, R. Weinstain, C. Schwechheimer, B. A. Halkier, H. H. Nour-Eldin, M. Estelle and E. Shani, Nat. Commun., 2016, 7, 11486 CrossRef CAS PubMed
.
- J. Dayan, N. Voronin, F. Gong, T. P. Sun, P. Hedden, H. Fromm and R. Aloni, Plant Cell, 2012, 24, 66–79 CrossRef CAS PubMed
.
- L. Ragni, K. Nieminen, D. Pacheco-Villalobos, R. Sibout, C. Schwechheimer and C. S. Hardtke, Plant Cell, 2011, 23, 1322–1336 CrossRef CAS PubMed
.
- T. Regnault, J. M. Daviere, M. Wild, L. Sakvarelidze-Achard, D. Heintz, E. Carrera Bergua, I. Lopez Diaz, F. Gong, P. Hedden and P. Achard, Nat. Plants, 2015, 1, 15073 CrossRef CAS PubMed
.
- G. A. Drake, J. Exp. Bot., 1979, 30, 429–437 CrossRef CAS
.
- G. A. Drake and D. J. Carr, J. Exp. Bot., 1979, 30, 439–447 CrossRef CAS
.
- W. P. Jacobs and P. E. Pruett, Am. J. Bot., 1973, 60, 896–900 CrossRef CAS
.
- M. Kwiatkowska, Planta, 1991, 183, 294–299 CrossRef CAS PubMed
.
- E. M. Kramer, H. L. Rutschow and S. S. Mabie, Trends Plant Sci., 2011, 16, 461–463 CrossRef CAS PubMed
.
- J. H. Kaplan, B. Forbush IIIrd and J. F. Hoffman, Biochemistry, 1978, 17, 1929–1935 CrossRef CAS PubMed
.
- J. L. Ward and M. H. Beale, Phytochemistry, 1995, 38, 811–816 CrossRef CAS
.
- K. M. Schelkle, T. Griesbaum, D. Ollech, S. Becht, T. Buckup, M. Hamburger and R. Wombacher, Angew. Chem., Int. Ed., 2015, 54, 2825–2829 CrossRef CAS PubMed
.
- T. Miyamoto, R. DeRose, A. Suarez, T. Ueno, M. Chen, T. P. Sun, M. J. Wolfgang, C. Mukherjee, D. J. Meyers and T. Inoue, Nat. Chem. Biol., 2012, 8, 465–470 CrossRef CAS PubMed
.
- P. Klan, T. Solomek, C. G. Bochet, A. Blanc, R. Givens, M. Rubina, V. Popik, A. Kostikov and J. Wirz, Chem. Rev., 2013, 113, 119–191 CrossRef CAS PubMed
.
- T. Sun, H. M. Goodman and F. M. Ausubel, Plant Cell, 1992, 4, 119–128 CrossRef CAS PubMed
.
- A. L. Silverstone, H. S. Jung, A. Dill, H. Kawaide, Y. Kamiya and T. P. Sun, Plant Cell, 2001, 13, 1555–1566 CrossRef CAS
.
- Q. Yu, C. Tang and J. Kuo, Plant Soil, 2000, 219, 29–40 CrossRef CAS
.
- J. Binenbaum, R. Weinstain and E. Shani, Trends Plant Sci., 2018, 23, 410–421 CrossRef CAS PubMed
.
- A. Rizza and A. M. Jones, Curr. Opin. Plant Biol., 2018, 47, 9–15 CrossRef PubMed
.
- M. Talon, M. Koornneef and J. A. Zeevaart, Proc. Natl. Acad. Sci. U. S. A., 1990, 87, 7983–7987 CrossRef CAS
.
- E. M. Kramer, Plant Physiol., 2006, 141, 1233–1236 CrossRef CAS PubMed
.
- A. Rizza, A. Walia, V. Lanquar, W. B. Frommer and A. M. Jones, Nat. Plants, 2017, 3, 803–813 CrossRef CAS PubMed
.
- A. M. Rashotte, J. Poupart, C. S. Waddell and G. K. Muday, Plant Physiol., 2003, 133, 761–772 CrossRef CAS PubMed
.
-
W. P. Jacobs, The Movement of Plant Hormones: Auxins, Gibberellins, and Cytokinins, Berlin, Heidelberg, 1972 Search PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures, characterization data, movies and additional figures. See DOI: 10.1039/c8sc04528c |
| This journal is © The Royal Society of Chemistry 2019 |