Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
α-Conotoxin GI triazole-peptidomimetics: potent and stable blockers of a human acetylcholine receptor†‡
Astrid
Knuhtsen
 a,
Charlotte
Whitmore
b,
Fergus S.
McWhinnie
a,
Charlotte
Whitmore
b,
Fergus S.
McWhinnie
 a,
Laura
McDougall
a,
Rachel
Whiting
b,
Brian O.
Smith
a,
Laura
McDougall
a,
Rachel
Whiting
b,
Brian O.
Smith
 c,
Christopher M.
Timperley
c,
Christopher M.
Timperley
 b,
A. Christopher
Green
b,
Kenneth I.
Kinnear
b and
Andrew G.
Jamieson
b,
A. Christopher
Green
b,
Kenneth I.
Kinnear
b and
Andrew G.
Jamieson
 *a
*a
aSchool of Chemistry, University of Glasgow, Joseph Black Building, Glasgow, G12 8QQ UK. E-mail: Andrew.Jamieson.2@glasgow.ac.uk
bChemical, Biological and Radiological Division, Defence Science and Technology Laboratory, Porton Down, Salisbury, Wiltshire, SP4 0JQ UK
cInstitute of Molecular, Cell & Systems Biology, College of Medical, Veterinary & Life Sciences, University of Glasgow, Joseph Black Building, Glasgow, G12 8QQ UK
First published on 26th November 2018
Abstract
The potency and selectivity of conotoxin peptides for neuropathic receptors has made them attractive lead compounds in the development of new therapeutics. Specifically, α-conotoxin GI has been shown to be an unparalleled antagonist of the nicotinic acetylcholine receptor (nAChR). However, as with other peptidic leads, poor protease resistance and the redox instability of the conotoxin scaffold limit bioactivity. To counter this, we have employed the underutilised 1,5-disubstituted 1,2,3-triazole to act as a structural surrogate of the native disulfide bonds. Using an efficient, on-resin ruthenium azide-alkyne cycloaddition (RuAAC), each disulfide bond was replaced in turn and the biological activities quantified. One of the mimetic isomers exhibited a comparable activity to the native toxin, while the other showed no biological effect. The active mimetic isomer 11 was an order of magnitude more stable in plasma than the native GI. The NMR solution structure of the mimetic overlays extremely well with the structure for the native GI demonstrating that the triazole bridge is an exceptional surrogate for the disulfide bridge. Development of this potent and stable mimetic of GI leads us to believe that this strategy will yield many other new conotoxin-inspired probes and therapeutics.
Introduction
The conotoxin peptides have attracted a substantial amount of interest over recent years due to their properties as selective chemical probes that target ion channels and receptors.1 Conotoxin peptides are also potential therapeutic leads,2 and several have found clinical applications in the treatment of neuropathic pain, hypertension and type 2 diabetes.3 α-Conotoxin GI (GI) was originally isolated from the venom of the fish-hunting cone snail Conus geographus.4,5 This venom has been reported to have caused several human fatalities,6–8 and can pose a risk to human health.9 α-GI is a 13-residue peptide that acts as a competitive antagonist for the muscle-type nicotinic acetylcholine receptor (nAChR) with excellent selectivity for α/δ receptor subunit binding over α/γ. GI is therefore used extensively as a pharmacological tool for receptor characterisation.10Neuromuscular blocking agents mainly antagonise nAChRs at the skeletal neuromuscular junction.11 nAChRs, however, are also found in the autonomic nervous system and the action of muscle relaxants on these synapses are responsible for their major negative side-effects. Administration of GI, however, has no observed effect on blood pressure, heart rate, vagal stimulation or ganglionic transmission.12 Furthermore, GI's neuromuscular blocking effect is both fast and reversible through the administration of acetylcholinesterase inhibitors (AChEI).12 These data illustrate that GI's activity is very specific for receptors within the neuromuscular junction and is competitively reversible. Therefore, GI has great potential as a lead compound for the development of a new class of muscle relaxant. However, as with other peptides, conotoxins suffer from a number of unfavourable physicochemical properties that have thus far limited their potential as therapeutics. These include poor protease stability in some instances (resulting in short half-lives in plasma), immunogenicity and redox reactivity leading to disulfide scrambling and subsequent loss of activity.13 The general field of peptidomimetics has arisen to address such issues with peptides and provide bioavailable compounds for drug discovery. Such mimetics generally incorporate structural surrogates of problematic chemical functionalities that overcome unfavourable physicochemical properties, yet maintain binding affinity for the target receptor.14
Most conotoxins, including all α-conotoxins, incorporate a disulfide bond network that constrains the polypeptide into a bioactive conformation. This functionality presents a major synthetic challenge to produce the specific, bioactive disulfide isomer. Disulfide bond reduction by glutathione and then protease-mediated degradation generally leads to poor blood plasma stability.15 Analogues of MrIA conotoxin have been prepared incorporating a 1,4-disubstituted 1,2,3-triazole that retained biological activity.16 However, we have shown previously that a 1,5-disubstituted 1,2,3-triazole bridge more effectively mimics the orientation and geometry of a disulfide bond.17 The incorporation of the 1,5-disubstituted functionality can be easily achieved on solid support using commercially available amino acid building blocks.
In this work we have addressed this synthetic challenge by applying a 1,5-triazole bridge as a disulfide bond surrogate to GI to produce the stable and highly potent analogue 11. We report the bioactivity of this and related molecules against a human nAChR, expressed in HEK cells, for the first time. Structural determination of GI peptidomimetic 11 using NMR spectroscopy was used to determine the accuracy of the triazole bridge as a surrogate of the disulfide bridge and whether the structure of the pharmacophore is retained. Plasma stability was determined for the active GI mimetic 11 which, together with the structural requirements for bioactivity of the α-conotoxin GI will underpin future drug development.
Results and discussion
Synthesis of 1,5-disubstituted triazole mimetic α-GI conotoxins
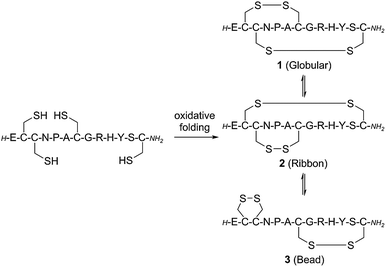
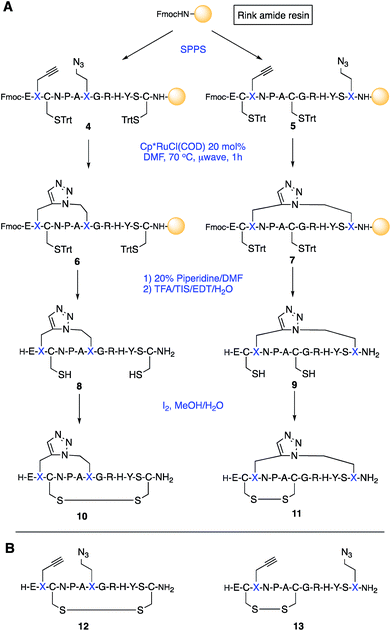
The native linear GI peptide was initially synthesised and oxidised under thermodynamic conditions providing three different disulfide isomers as a crude mixture (see ESI‡). The linear peptide is expected to fold into the thermodynamically most stable ‘globular’ isomer 1 [Cys2–Cys7, Cys3–Cys13] (Scheme 1). However, there are two other possible folding arrangements – the ‘ribbon’ fold 2 [Cys2–Cys13, Cys3–Cy7] and the ‘bead’ fold 3 [Cys2–Cys3, Cys7–Cy13] (1–3, Scheme 1). Previous literature has proposed that the major product from the oxidation of linear α-conotoxins is not necessarily the most active, thus all three isomers were isolated for biological comparison.18 To confirm which of these three peptides had the ‘globular’ fold we synthesised an authentic standard according to an adapted orthogonal protection strategy (Section S2 in the ESI‡).19Peptidomimetics incorporating a 1,5-disubstituted 1,2,3-triazole bridge in place of either the Cys2–Cys7 or the Cys3–Cys13 disulfide bonds were synthesized by microwave-assisted Fmoc solid phase peptide synthesis (SPPS) on Rink Amide resin using commercially available building blocks Fmoc-L-propargylglycine (Fmoc-Pra-OH) and Fmoc-L-azido-homoalanine (Fmoc-Aha-OH) (Scheme 2).
After incorporation of the non-natural amino acids, the linear N-terminally Fmoc protected peptides (4 and 5) were cyclised on-resin using a Ruthenium-catalysed Azide-Alkyne Cycloaddition (RuAAC). This strategy has been shown to produce specifically 1,5-disubstituted 1,2,3-triazole functionalities.20 As the azide functionality exhibits a strong IR absorption at 2100 cm−1, while the triazole does not, the cyclisation could be followed on-resin without a cleavage step.21 Subsequent HPLC analysis confirmed complete conversion of the linear species to the cyclised products 6 and 7. The removal of the terminal Fmoc, cleavage from the solid support and global side-chain deprotection afforded triazole-containing peptides 8 and 9. Final oxidation using catalytic iodine produced the cyclised, disulfide containing α-GI mimetics 10 and 11. Two linear, non-cyclised peptides, 12 and 13, were also synthesised to act as control substrates.
In order to confirm the 1,5-disubstitution pattern of the 1,2,3-triazoles, we acquired non-proton decoupled 1H–13C HSQC spectra of 10 and 11. Creary et al. previously demonstrated a significant difference in non-substituted carbon shifts between 1,4- vs. 1,5-disubstituted 1,2,3-triazoles.22 The C5 signal was shown to occur at a chemical shift of δ ∼ 120 ppm in a 1,4-triazole, while a 1,5-triazole exhibits a C4 shift of δ ∼ 133 ppm. As expected the non-substituted carbon shifts for both isomers (10 and 11) were above 133 ppm, indicative of both being the desired 1,5-triazole product (Fig. S4‡).
Biological evaluation of native α-GI and triazole mimetics
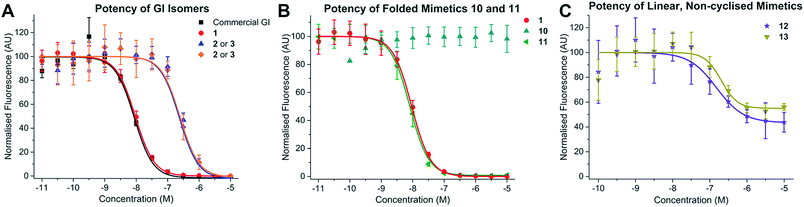
Previous conotoxin-derived drug leads have failed in clinical trials due to a lack of efficacy.23,24 This was due in part to the biological assessment of the compounds using rat rather than human receptors. We chose to assess the biological activity of these peptides and peptidomimetics using human cells expressing human nAChR receptors. Peptides 1–3 and peptidomimetics 10–13 were incubated with human muscle-type nAChRs and their activity determined in vitro by assessing antagonism of the nAChR mediated increase of [Ca2+] in CN21 cells (Fig. 1).The CN21 nAChR line was derived from the TE671 human rhabdomyosarcoma cell line by stable transfection with the adult ε subunit of nAChR. This cell line expresses both the human foetal and adult muscle nAChR25 and is a well validated platform in which to assess the activity of nAChR antagonists.26
Addition of the native ‘globular’ α-conotoxin compound 1 to the CN21 cells produced a concentration-dependent decrease in the ACh-induced response, indicative of inhibition at the nAChR (Fig. 1A). This effect was duplicated using commercially sourced GI peptide. The ‘ribbon’ GI 2 and ‘bead’ GI 3 were also biologically active, although approximately 100-fold less potent than the ‘globular’ isomer, with IC50 values in the high nM range (Table 1). In an in vivo mouse toxicity assay the bead and ribbon GI isomers have also been reported to be an order of magnitude less potent than the globular GI isomer.27 As the synthesised compounds corresponding to ribbon and bead (2 and 3, respectively) were equipotent, the definitive structures of the two isolated peptides were not determined.
| Peptide/mimetic | IC50 (nM) (95% CI) | Max inhibition at 10 μM (% ±SEM) |
|---|---|---|
| a Calculated from partial IC50 curve. IC50 = half maximal inhibitory concentration. N.D. = not determined. | ||
| 1 (Globular GI) | 9.8 (7.4 to 12.8) | 100.0 |
| Commercial GI | 8.8 (6.6 to 11.6) | 100.0 |
| 2 or 3 (Ribbon/Bead GI) | 857 (548 to 1344) | 100.0 |
| 2 or 3 (Ribbon/Bead GI) | 969 (713 to 1317) | 100.0 |
| 10 | N.D. | 1.5 ± 9.7 |
| 11 | 8.2 (6.4 to 10.5) | 100.0 |
| 12 | 140 (33.4 to 587) | 56.2 ± 7.9 |
| 13 | 203 (84.4 to 487) | 43.9 ± 5.6 |
Mimetic 11, which had the [Cys3–Cys13] disulfide exchanged for a triazole bridge, had comparable biological activity to the native ‘globular’ GI toxin 1. Both 1 and 11 exhibited a concentration-dependent inhibition of the ACh-mediated response that reached complete inhibition at higher concentrations, with IC50 values in the low nM range (9.8 nM and 8.2 nM, respectively) (Fig. 1B and Table 1). Both linear uncyclised control peptides 12 and 13 exhibited activity at muscle-type nAChRs, but complete inhibition was not achieved in the concentration range tested (Fig. 1C). However in the mimetic 10, with the triazole functionality moved to the [Cys2–Cys7] position, no inhibition was observed (Fig. 1B).
Analysing the peptides/peptidomimetics at a fixed concentration (10 μM) allows their normalised percentage inhibition values to be compared. Native globular GI 1 and peptidomimetic 11 exhibited 100% inhibition, while linear controls 12 and 13 displayed 56 and 44% inhibition, respectively, suggesting they are only partial antagonists of the nAChR. Increasing the concentration to either 30 μM (n = 2) or 100 μM (n = 1) of peptide did not further decrease the fluorescence response (data not shown).
These data show that the disulfide [Cys3–Cys13] in GI is amenable to modification to a 1,5-triazole mimetic while retaining comparable activity to the native toxin 1. However, substitution of the other disulfide position [Cys2–Cys7] abolishes any activity towards nAChRs. Furthermore, the need for the scaffold to contain two restraints (either two disulfides, 1–3, or a mixed system, 11) is evident by the poor activity of the linear control peptides. Since 11 is both a peptidomimetic and retains analogous activity to the native toxin it has the potential to be developed as a bioavailable drug lead.
Serum plasma stability of native α-GI and triazole mimetics
Blood plasma stability of conotoxin-derived peptidomimetics is an important consideration in drug discovery efforts. Folded conotoxins possess a relatively rigid peptide backbone that limits their ability to adopt the extended conformations required for protease degradation. However, the disulfide bond network is susceptible to reduction and/or scrambling leading to inactivity.16The blood plasma stability of native ‘globular’ GI toxin 1 was thus compared to the most active 1,5-triazole bridged peptidomimetic 11 at pH 7.4 (Fig. 2). These data demonstrate that the incorporation of the triazole bond in place of the native disulfide significantly improves its plasma half-life by ∼10 fold c.f. the native 1. The inhibition of both proteolytic degradation and disulfide scrambling greatly increases the applicability of mimetic 11 as a stable, bioavailable drug lead. However, the stability assay used rat plasma, not human plasma, and some minor differences in the stability of the conotoxins in the two types of plasma is plausible.
 | ||
Fig. 2 Stability comparison of native ‘globular’ GI 1 compared to the 1,5-triazole mimetic 11 in rat plasma. Points are the mean of three independent experiments ±SEM. Calculated half-lives are 1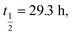 11 11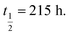 | ||
Conformational analysis by NMR spectroscopy
Currently there are >12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 structures in the Protein Data Bank derived from NMR restraints, constituting ∼8% of the database. However, these structures are almost wholly comprised of natural amino acid moieties, with only a select few containing non-canonical sidechains.28 The need to computationally describe a non-natural amino acid is a significant roadblock in the development of accurate peptidomimetic structures. Therefore, to better understand the structural effect of using a 1,2,3-triazole as a disulfide surrogate, we produced modified CNS topology/parameter files to allow the description of a 1,5-disubstituted triazole. Using these descriptors, we then determined the NMR solution structure of the most potent GI mimetic 11 so as to provide a physical basis to rationalise the disulfide discrimination identified previously.
000 structures in the Protein Data Bank derived from NMR restraints, constituting ∼8% of the database. However, these structures are almost wholly comprised of natural amino acid moieties, with only a select few containing non-canonical sidechains.28 The need to computationally describe a non-natural amino acid is a significant roadblock in the development of accurate peptidomimetic structures. Therefore, to better understand the structural effect of using a 1,2,3-triazole as a disulfide surrogate, we produced modified CNS topology/parameter files to allow the description of a 1,5-disubstituted triazole. Using these descriptors, we then determined the NMR solution structure of the most potent GI mimetic 11 so as to provide a physical basis to rationalise the disulfide discrimination identified previously.
1H,1H NOESY data were collected at four mixing times (80, 120, 200, 300 ms) together with 1H,1H TOCSY, DQF-COSY and 1H,13C HSQC spectra.29 The peaks assigned using CCPN analysis and the structure computed using ARIA2.3/CNS1.2 (output statistics can be found in Table S2‡).30,31 From 100 calculated structures, the 20 lowest energy structures were refined in explicit solvent and the structure closest to the mean selected as the representative structure (Fig. 3A and S6‡). To assess the structural similarity between mimetic 11 and the native globular form 1, the two were superimposed using their backbone heavy atoms for alignment and the RMSD between them quantified (Fig. 3B).
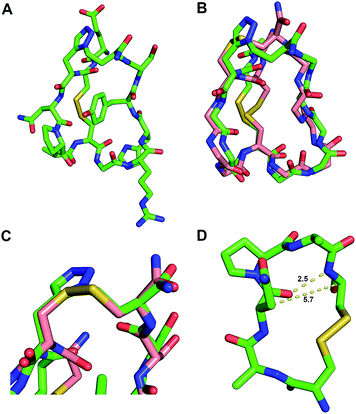 | ||
| Fig. 3 Solution structure of peptide mimetic 11 determined by NMR spectroscopy. (A) Representative structure of the calculated NMR ensemble. (B) Superposition of the native ‘globular’ GI 1 (PDB: 1NOT) (pink) with the mimetic structure 11 (green) exhibits an excellent backbone RMSD of 0.442 Å. (C) Disulfide and triazole bridge overlay with Cα–Cβ overlay RMSD of 0.256 Å. (D) Residues 2–7 of mimetic 11 form a β-turn with an intramolecular hydrogen-bond (i, i + 3) and Cα–Cα distance < 7 Å. | ||
Comparing the solution structure of mimetic 11 with the NMR and crystal structures of native GI 1 reveals that the structures are very similar.32,33 The triazole bridge appears to act as a very good structural surrogate for the Cys3–Cys13 disulfide bridge when comparing the orientation and spacing of the two Cα–Cβ bonds. The α-subunit binding face of GI 1 incorporates residues Cys2, Asn4, Pro5, Ala6 and Cys7 in a previously proposed model for binding of GI to the mammalian nAChR.32 Mimetic 11 has a highly constrained β-turn (i, i + 3 CO to HN H-bond (2.5 Å) and Cα–Cα distance of 5.7 Å) in this region and adopts a very similar structure (0.442 Å backbone RMSD) to the native GI conformation. This indicates that the conformation of the β-turn pharmacophore is retained in mimetic 11 resulting in similar bioactivity. It is apparent from the NMR structure that the second loop (residues Gly8-Ser12) of mimetic 11 adopts a slightly different conformation to the native globular GI 1. This may enable the mimic 11 to bind to other nAChR subtypes, as has previously been observed with dicarba α-conotoxin Vc1.1 analogues.34
An explanation for the lack of efficacy of disulfide isomer 10 is not immediately obvious if the triazole linkage is a faithful surrogate of a disulfide bridge. However, as noted above, while the 1,5-disubstituted triazole linkage mimics the disulfide bridge well, it also introduces additional steric bulk. Replacement of the [Cys2–Cys7] disulfide with the triazole linkage is likely to mimic the disulfide bridge, but would also introduce severe steric clash within this highly constrained turn and appears to abolish its activity. In addition, the NMR spectra of compound 10 are characterised by the presence of multiple resonances for many of its chemical groups and spectra recorded over a wide range of temperatures suggest that 10 exchanges between multiple conformations at a rate that is intermediate to slow on the NMR timescale.
Conclusions
The α-conotoxins, particularly the skeletal nAChR specific GI, present an opportunity to create a safe, selective and reversible muscle relaxant for use in anaesthesia. However, in their native form, peptides are unlikely to become lead drug compounds due to their unfavourable physicochemical properties.We therefore used a peptidomimetic triazole disulfide bridge surrogate to replace each disulfide bridge in turn within GI. This produced a mimetic with an order of magnitude increase in blood plasma stability, whilst retaining full biological activity. Crucially, [Cys3–Cys13] is amenable to this type of modification, whereas replacement of [Cys2–Cys7] eliminates antagonist activity towards the nAChR. Unlike previous studies performed in rodent assays, our studies were performed in human CN21 cells expressing human AChRs, making them far more applicable towards drug development in humans.
After developing bespoke force field descriptions of the triazole mimetic that allowed us to determine the solution structure by NMR spectroscopy, we discovered significant similarities in conformation between the mimetic and the peptidic bioactive toxin. It is evident that in peptidomimetic 11 the conformation of the β-turn pharmacophore of the native peptide 1 is preserved. This supports the hypothesis that the pharmacophore of the GI toxin is located within the first half of the peptide as a β-turn. This possibly explains the contrasting activities of mimetics 10 and 11, as a triazole linkage [aa2–aa7] may well interfere with binding to the nAChR.
Given the effectiveness of the strategy described in this article, ease of synthesis and that the required reagents are commercially available, we believe this approach is broadly applicable to other peptides containing disulfide bonds that have the potential for development as novel probes and therapeutics.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the University of Glasgow and the Defence Science and Technology Laboratory (Research Project Grant DSTL/AGR/R/CBRN/01) for financial support of this research. L. M. thanks EPSRC for a studentship (EP/N509668/1). The authors also thank Andrew Monaghan (high-resolution mass spectrometry) for technical assistance.References
- K. B. Akondi, M. Muttenthaler, S. Dutertre, Q. Kaas, D. J. Craik, R. J. Lewis and P. F. Alewood, Chem. Rev., 2014, 114, 5815–5847 CrossRef CAS PubMed.
- R. S. Norton, Expert Opin. Drug Discovery, 2017, 12, 611–623 CrossRef CAS PubMed.
- C. Netirojjanakul and L. P. Miranda, Curr. Opin. Chem. Biol., 2017, 38, 70–79 CrossRef CAS PubMed.
- W. R. Gray, A. Luque, B. M. Olivera, J. Barrett and L. J. Cruz, J. Biol. Chem., 1981, 256, 4734–4740 CAS.
- O. B. McManus, J. R. Musick and C. Gonzalez, Neurosci. Lett., 1981, 25, 57–62 CrossRef CAS PubMed.
- W. J. Clench and Y. Kondo, Am. J. Trop. Med. Hyg., 1943, 1, 105–121 CrossRef.
- D. Fegan and D. Andresen, Lancet, 1997, 349, 1672 CrossRef CAS.
- R. D. Rice and B. W. Halstead, Toxicon, 1968, 5, 223–224 CrossRef CAS PubMed.
- P. D. Anderson and G. Bokor, J. Bioterror. Biodef., 2012, 3, 1–4 Search PubMed.
- E. K. M. Lebbe, S. Peigneur, I. Wijesekara and J. Tytgat, Mar. Drugs, 2014, 12, 2970–3004 CrossRef CAS PubMed.
- A. Taly, P. J. Corringer, D. Guedin, P. Lestage and J. P. Changeux, Nat. Rev. Drug Discovery, 2009, 8, 733–750 CrossRef CAS PubMed.
- I. G. Marshall and A. L. Harvey, Toxicon, 1990, 28, 231–234 CrossRef CAS PubMed.
- R. T. Layer and J. M. McIntosh, Mar. Drugs, 2006, 4, 119–142 CrossRef CAS.
- M. J. Adler, A. G. Jamieson and A. D. Hamilton, Curr. Top. Microbiol. Immunol., 2011, 348, 1 CAS.
- A. Meister and M. E. Anderson, Annu. Rev. Biochem., 1983, 52, 711–716 CrossRef CAS PubMed.
- A. Gori, C. I. A. Wang, P. J. Harvey, K. J. Rosengren, R. F. Bhola, M. L. Gelmi, R. Longhi, M. J. Christie, R. J. Lewis, P. F. Alewood and A. Brust, Angew. Chem., Int. Ed., 2015, 54, 1361–1364 CrossRef CAS PubMed.
- S. Pacifico, A. Kerckhoffs, A. J. Fallow, R. E. Foreman, R. Guerrini, J. McDonald, D. G. Lambert and A. G. Jamieson, Org. Biomol. Chem., 2017, 15, 4704–4710 RSC.
- J. L. Dutton, P. S. Bansal, R. C. Hogg, D. J. Adams, P. F. Alewood and D. J. Craik, J. Biol. Chem., 2002, 277, 48849–48857 CrossRef CAS PubMed.
- E. Atherton, R. C. Sheppard and P. Ward, J. Chem. Soc., Perkin Trans. 1, 1985, 2065–2073 RSC.
- B. C. Boren, S. Narayan, L. K. Rasmussen, L. Zhang, H. Zhao, Z. Lin, G. Jia and V. Fokin, J. Am. Chem. Soc., 2008, 130, 8923–8930 CrossRef CAS PubMed.
- M. Empting, O. Avrutina, R. Meusinger, S. Fabritz, M. Reinwarth, M. Biesalski, S. Voigt, G. Buntkowsky and H. Kolmar, Angew. Chem., Int. Ed., 2011, 50, 5207–5211 CrossRef CAS PubMed.
- X. Creary, A. Anderson, C. Brophy, F. Crowell and Z. Funk, J. Org. Chem., 2012, 77, 8756–8761 CrossRef CAS PubMed.
- M. W. Pennington, A. Czerwinski and R. S. Norton, Bioorg. Med. Chem., 2018, 26, 2738–2758 CrossRef CAS PubMed.
- L. Azam and J. M. McIntosh, J. Neurochem., 2012, 122, 1137–1144 CrossRef CAS PubMed.
- D. Beeson, M. Amar, I. Bermudez, A. Vincent and J. Newsom-Davis, Neurosci. Lett., 1996, 207, 57–60 CrossRef CAS PubMed.
- A. Ring, B. O. Strom, S. R. Turner, C. M. Timperley, M. Bird, A. C. Green, J. E. Chad, F. Worek and J. E. H. Tattersall, PLoS One, 2015, 10, e0135811 CrossRef PubMed.
- Y. Nishiuchi and S. Sakakibara, FEBS Lett., 1982, 148, 260–262 CrossRef CAS PubMed.
- H. M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov and P. E. Bourne, Nucleic Acids Res., 2000, 28, 235–242 CrossRef CAS PubMed.
- M. J. Thrippleton and J. Keeler, Angew. Chem., Int. Ed. Engl., 2003, 42, 3938–3941 CrossRef CAS PubMed.
- W. F. Vranken, W. Boucher, T. J. Stevens, R. H. Fogh, A. Pajon, A. Linas, E. L. Ulrich, J. L. Markley, J. Ionides and E. D. Laue, Proteins, 2005, 59, 687–697 CrossRef CAS PubMed.
- W. Rieping, M. Habeck, B. Bardiaux, A. Bernard, T. E. Malliavin and M. Nilges, Bioinformatics, 2007, 23, 381–382 CrossRef CAS PubMed.
- J. Gehrmann, P. F. Alewood and D. J. Craik, J. Mol. Biol., 1998, 278, 401–415 CrossRef CAS PubMed.
- L. W. Guddat, J. A. Martin, L. Shan, A. B. Edmundson and W. R. Gray, Biochemistry, 1996, 35, 11329–11335 CrossRef CAS PubMed.
- B. J. van Lierop, S. D. Robinson, S. N. Kompella, A. Belgi, J. R. McArthur, A. Hung, C. A. MacRaild, D. J. Adams, R. S. Norton and A. J. Robinson, ACS Chem. Biol., 2013, 8, 1815–1821 CrossRef CAS PubMed.
Footnotes |
| † Content includes material subject to © Crown copyright (2018), Dstl. This material is licensed under the terms of the Open Government Licence except where otherwise stated. To view this licence, visit http://www.nationalarchives.gov.uk/doc/open-government-licence/version/3 or write to the Information Policy Team, the National Archives, Kew, London TW9 4DU, or email: E-mail: psi@nationalarchives.gsi.gov.uk. |
| ‡ Electronic supplementary information (ESI) available: Synthetic procedures, NMR and LC characterisation, triazole force field description and peptide solution structure (.pdb). See DOI: 10.1039/c8sc04198a |
| This journal is © The Royal Society of Chemistry 2019 |