Open Access Article
Open Access ArticleA novel strategy for facile serum exosome isolation based on specific interactions between phospholipid bilayers and TiO2†
Fangyuan
Gao‡
a,
Fenglong
Jiao‡
 ab,
Chaoshuang
Xia
a,
Yang
Zhao
a,
Wantao
Ying
a,
Yuping
Xie
a,
Xiaoya
Guan
c,
Ming
Tao
d,
Yangjun
Zhang
*a,
Weijie
Qin
*a and
Xiaohong
Qian
*a
ab,
Chaoshuang
Xia
a,
Yang
Zhao
a,
Wantao
Ying
a,
Yuping
Xie
a,
Xiaoya
Guan
c,
Ming
Tao
d,
Yangjun
Zhang
*a,
Weijie
Qin
*a and
Xiaohong
Qian
*a
aState Key Laboratory of Proteomics, National Center for Protein Sciences (Beijing), Beijing Institute of Lifeomics, Beijing Proteome Research Center, China. E-mail: qianxh1@163.com; 13683167093@163.com; aunp_dna@126.com
bSchool of Life Science and Technology, Beijing Institute of Technology, China
cPeking University Cancer Hospital, China
dPeking University Third Hospital, China
First published on 3rd December 2018
Abstract
Exosomes are cell-derived, phospholipid bilayer-enclosed vesicles that play important roles in intercellular interactions and regulate many biological processes. Accumulating evidence suggests that serum exosomes are potential biomarkers for the early diagnosis of cancer. To aid the downstream molecular analyses of tumour-secreted exosomes, purified exosomes are highly desirable. However, current techniques for exosome isolation are time-consuming and highly instrument-dependent, with limited specificity and recovery. Thus, rapid and efficient methods are strongly needed for both basic research and clinical applications. Here, we present a novel strategy for facile exosome isolation from human serum by taking advantage of the specific interaction of TiO2 with the phosphate groups on the lipid bilayer of exosomes. Due to their simplicity and highly affinitive binding, model exosomes can be reversibly isolated with a high recovery (93.4%). Downstream characterization and proteome profiling reveal that high-quality exosomes can be obtained from human serum by this TiO2-based isolation method in 5 min, which is a fraction of the time required for the commonly used ultracentrifugation method. We identified 59 significantly up-regulated proteins by comparing the serum exosomes of pancreatic cancer patients and healthy donors. In addition to the 30 proteins that were reported to be closely related to pancreatic cancer, we found an additional 29 proteins that had not previously been shown to be related to pancreatic cancer, indicating the potential of this novel method as a powerful tool for exosome isolation for health monitoring and disease diagnosis.
Introduction
Exosomes are lipid bilayer-enclosed biological nanoparticles with sizes ranging from 30 to 200 nm.1,2 They are cell-derived and present in a variety of body fluids, including serum, urine, saliva, milk, bile and other fluids.3–6 In recent years, exosomes have gained much attention, since they have emerged as intercellular communication carriers for nucleic acids, proteins and other biomolecules. Furthermore, they play important roles in many biological processes such as cell-to-cell communication, tumourigenesis, signal transduction and immune response.7–11 It is clear that exosomes can be transported and exchanged between cells and body fluids, while the intact vesicle structure of exosomes protects the contained biomarkers from degradation and enzymatic hydrolysis.12,13 On the other hand, liquid biopsy targeting tumour-associated exosomes in body fluids provides a minimally invasive approach for determining the progression and metastasis of tumours. Thus, they are ideal drug delivery carriers and potential targets for cancer diagnosis.4,13–17The isolation of exosomes from body fluids is an essential step before subsequent analysis. Currently, the most widely used method for exosome isolation is ultracentrifugation.2 Cells, cellular debris and exosomes in the fluids are separated sequentially based on their different densities and sizes. However, this method requires multiple steps and a long separation time (up to 8–10 hours), which limits its throughput and recovery. Recent studies using immunoaffinity methods have enabled the isolation of exosomes based on their specific markers, such as CD63, CD81, and CD9.4,18,19 However, these methods frequently suffer from drawbacks such as long processing time (4–20 h), low reproducibility, and low exosome yield. Microfluidic chip-based methods have also been developed to isolate exosomes from blood and cell culture medium.20–24 The isolation time is shortened to tens of minutes and the purity of obtained exosomes reaches 98%.21,25 However, the complicated manufacturing procedure of microfluidic chips and the low throughput of these methods limit their large-scale clinical applications.26 Other techniques including filtration, size-exclusion chromatography and coprecipitation suffer from problems such as complicated procedures, long processing times, requirement of sophisticated instruments and limited specificity.2 Thus, a rapid, efficient and convenient isolation technique is in high demand to facilitate the routine analysis of exosomes.
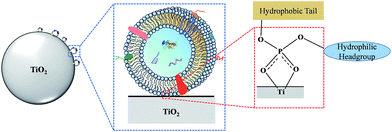
Here, we present a novel strategy for facile isolation of exosomes from human serum by taking advantage of the specific interaction between titanium oxide and the phosphate groups on the lipid bilayer of exosomes. The lipid bilayer is composed of amphiphilic phospholipids with hydrophobic tails and hydrophilic phosphate heads. In biological systems, the hydrophilic phosphate head of the phospholipids is exposed on the outer surface of the lipid bilayer. It is well known that some metal oxides, such as titanium oxide (TiO2), can reversibly bind with phosphate groups with high specificity. By exploiting this property, TiO2 has been widely used for highly selective enrichment of phosphorylated peptides,27,28 water-soluble organic phosphates29 and organophosphorus pesticides.30 In view of this, we make the first attempt to use micron-sized TiO2 particles for enrichment of exosomes via the bidentate binding between the phosphate groups on the surface of the lipid-bilayer and TiO2 (Fig. 1). Due to its simplicity and highly affinitive binding, a few advantages can be expected of this coordinate-based selective enrichment, such as improved isolation efficiency, reduced nonspecific adsorption and shorter sample processing time.
Results and discussion
Characterization of TiO2-based exosome isolation
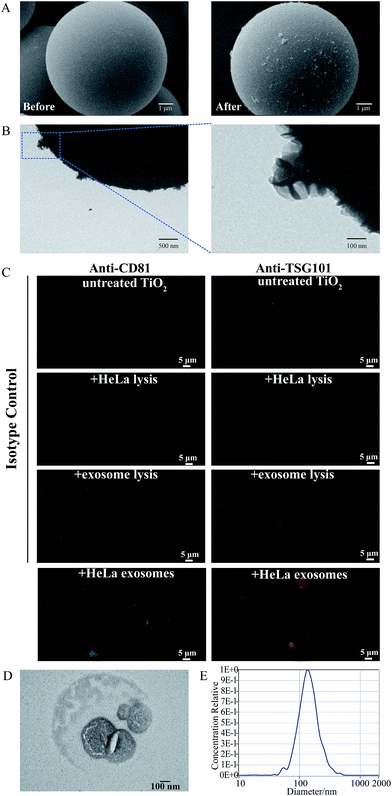
To evaluate the feasibility of our proposed strategy, the well-characterized HeLa cells' exosomes, isolated by the ultra-centrifugation strategy (Fig. S1†), were chosen as the model sample to incubate with TiO2. As shown in the scanning electron microscopy (SEM) images (Fig. 2A), before incubation with the exosome solution, the TiO2 microspheres have a clean surface and uniform spherical structure with a diameter of 5 μm. After incubation and repeated washing, numerous vesicles with a particle size of approximately 100 nm are found adsorbed on the surface of the TiO2 microspheres. Transmission electron microscopy (TEM) images show that the captured vesicles have a typical cup-shaped structure (Fig. 2B).We also performed immunofluorescence experiments using antibodies that specifically recognized the commonly used exosomal marker proteins, namely, cytosolic TSG101 and transmembrane CD81. As shown in Fig. 2C, strong fluorescence signals are observed at the surface of the TiO2 microspheres after incubation with the exosome solution using fluorescent antibodies targeting CD81 and TSG101, while only minimal background fluorescence is found in the isotype controls using untreated TiO2 microspheres or TiO2 incubated with cell lysis and exosome lysis solutions, indicating that only exosomes with intact vesicle structures can be captured by TiO2.
Next, we evaluated the elution conditions and recovery of the adsorbed exosomes from TiO2 microspheres. Alkaline solvents (pH 10–12) are usually used to disrupt the binding interaction between phosphate groups and TiO2.31 Therefore, we used 10% NH3·H2O to elute the isolated exosomes. To protect the vesicle structure and the lipid bilayer of the exosomes from the high pH environment, the eluent was quickly replaced by PBS by ultrafiltration. The eluted exosomes were characterized by TEM and NTA, and an intact vesicle structure and typical cup-shaped morphology are found, indicating that the lipid bilayer is retained during the isolation and elution process (Fig. 2D). As revealed by Fig. 2E, the size distribution of the exosomes isolated using TiO2 is from 65 nm to 235 nm and centered at 133 nm, which is close to that before isolation (from 45 nm to 225 nm and centered at 122 nm, Fig. S1C†). Then, we verified the effect of directly lysing exosomes from the TiO2 surface. Proteins extracted from HeLa cells' exosomes, the supernatant after incubation, the washing solutions for three washes, and HeLa exosomes adsorbed on the surface of the TiO2 microspheres were separated by SDS-PAGE and subsequently silver-stained. As shown in Fig. S2,† no obvious proteins could be observed in the supernatant after incubation and washing solutions, and there are minimal differences between exosomal proteins before and after isolation by TiO2. To evaluate the possible interfering effect of glycoproteins and phosphoproteins on our exosome enrichment method, we prepared a mixture containing model phosphoproteins (α-casein and β-casein), model glycoproteins (horseradish peroxidase, HRP) (1 μg for each model protein) and model exosomes from UC (containing approximately 2 μg protein). As shown in Fig. S3,† though residual β-casein can still be visualized in the SDS-PAGE image (Fig. S3B†), majority of the inferring glycoproteins and phosphoproteins are efficiently removed after TiO2-based isolation and repeated washing, while maintaining a high recovery rate for exosomes. To evaluate the reproducibility of this TiO2-based isolation strategy, three technical replicates were conducted. Proteins extracted from the obtained exosomes were characterized by SDS-PAGE. As shown in Fig. S4,† almost identical protein band patterns are found in the three replicates (Lanes 1, 3 and 5 of Fig. S4†), demonstrating high reproducibility of the TiO2-based exosome isolation.
Kinetic study of the exosome adsorption process
After demonstrating the feasibility of using TiO2 microspheres for model exosome isolation, we further investigated the mechanism of the exosome adsorption process. Two possible kinetic models, namely, the pseudo-first order equation and the pseudo-second order equation, were proposed for the adsorption process. The pseudo-first order rate equation of Lagergren is expressed as follows:32| dqt/dt = k1(qe − qt) | (1) |
After integrating with the boundary conditions of t = 0 to t = t and qt = 0 to qt = qt, eqn (1) was rearranged to a linear form:
ln(qe − qt) = ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) qe − k1t qe − k1t | (2) |
The pseudo-second order rate equation is expressed as
| dqt/dt = k2(qe − qt)2 | (3) |
After integrating with the boundary conditions of t = 0 to t = t and qt = 0 to qt = qt, the integrated form of eqn (3) becomes
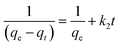 | (4) |
It can be rearranged to a linear form:
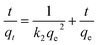 | (5) |
If the exosome adsorption on TiO2 fits the pseudo-first order rate equation, the plot of ln(qe − qt) against t of eqn (2) should give a linear relationship. If not, and the pseudo-second order kinetics are applicable, the plot of t/qt against t of eqn (5) should give a linear relationship. The theoretical value of the equilibrium adsorption capacity can be calculated by the Y-intercept and slope of the adsorption curves obtained by the pseudo-first order equation and the pseudo-second order equation, respectively. To determine the best-fitting kinetic model, the linear correlation coefficient (r2) and the theoretical qe values were used as evaluation indexes.
To fit eqn (2) and (5), various adsorption parameters, such as the amount of TiO2 and incubation time, were investigated. We first evaluated the effect of the amount of TiO2 microspheres on the exosome isolation efficiency determined by the greyscale value of the exosomal marker protein TSG101 extracted from the TiO2 isolated exosomes over that from the total exosomes in western blotting analysis. As the amount of the TiO2 microspheres increased from 0.5 mg to 10 mg, the isolation efficiency increased accordingly, and a maximum efficiency of 95.0% was reached at 2 mg TiO2 (Fig. 3A and S5A†). Further increase in the amount of TiO2 led to a decreased isolation efficiency, presumably due to the difficulty in sufficient mixing for a large amount of TiO2 microspheres. Therefore, 2 mg TiO2 was used for optimal incubation time evaluation. As shown in Fig. 3B and S5B,† the isolation efficiency increases from 53.1% to 93.4%, as the incubation time is extended from 30 s to 5 min. The isolation efficiency slightly increases to 95.5% when the incubation time is extended to 10 min. Based on the comprehensive consideration of the time requirements and isolation efficiency, 5 min was chosen as the optimal incubation time, with a maximum of 93.4% isolation efficiency achieved. The equilibrium adsorption capacity, qe, was determined using the adsorption capacity at 10 min (0.955 mg g−1).
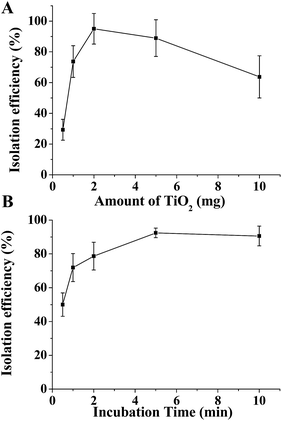 | ||
| Fig. 3 Optimization of the conditions for exosome isolation. Isolation efficiency of HeLa cells' exosomes as a function of the quantity of TiO2 (A) and incubation time (B). | ||
The plot of ln(qe − qt) against t for the pseudo-first order model and the plot of t/qt against t for the pseudo-second order model in the initial 10 min incubation of TiO2 and the HeLa cells' exosomes are shown in Fig. 4. A better linearity is achieved for the pseudo-second order model (r2 = 0.99974) than for the pseudo-first order model (r2 = 0.98591). Moreover, when the theoretical qe values are compared with the experimental qe values (0.955 mg g−1), very poor correlation is obtained for the pseudo-first order model (theoretical qe = 0.514 mg g−1) with a difference of up to 46%. In contrast, in the pseudo-second order model, the theoretical calculation (0.997 mg g−1) is quite close to the experimental data (0.955 mg g−1). These results suggest that the experimental data fit better with the pseudo-second order equation, indicating a chemical specific adsorption mechanism in the TiO2-based exosome enrichment. Presumably, the driving force of the chemical adsorption of exosomes on TiO2 is the coordinate binding between the phosphate groups of the phospholipid bilayer and TiO2. Therefore, specific and strong binding can be achieved because of the flexibility of the phospholipid bilayer, which may bend to facilitate multi-site interactions with the surface of TiO2. Then, five commercial titanium dioxide particles with different sizes, crystal forms and surface roughness were used to investigate their effect on exosome isolation. As shown in the SDS-PAGE image of the isolated exosome proteins in Fig. S6,† the protein band patterns are almost the same, indicating that titanium dioxide particles with different sizes, crystal forms and surface roughness have a similar effect on exosome isolation.
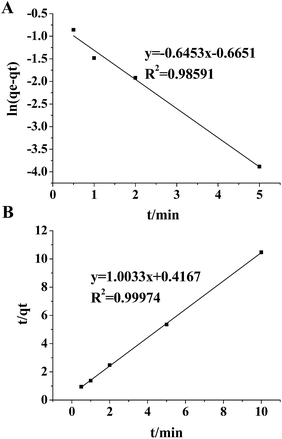 | ||
| Fig. 4 Pseudo first order sorption kinetics (A) and pseudo second order sorption kinetics (B) for adsorption of exosomes on TiO2. The average of three replicates was used to fit the linear equations. | ||
Serum exosome collection and proteomics analysis
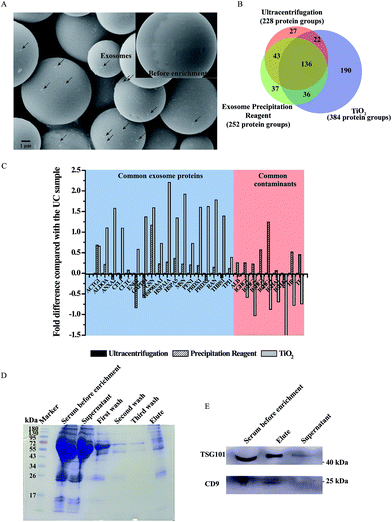
After demonstrating the feasibility of our TiO2-based exosome isolation strategy, we further applied it to human serum samples and compared it with the traditional methods (ultracentrifugation and coprecipitation kit) by proteome analysis (the process is shown in Fig. S7†). Serum exosomes are widely used in liquid biopsy for biomarker studies due to their high content of disease related DNA, RNA and proteins.12,33 To eliminate the interference of cell debris and large extracellular vesicles, the serum was pre-treated with filtration using 0.2 μm filters. Then, the filtered serum was subjected to the TiO2-based isolation process. As shown in Fig. 5A, particles with a size of approximately 100 nm were observed to adhere to the surface of TiO2 microspheres after incubation. Next, mass spectrometry was utilized to identify the proteins extracted from serum exosomes isolated using different strategies. In total, 384 protein groups were identified in the exosomes isolated by TiO2 from 100 μL serum samples in three biological replicates (Fig. 5B), which is obviously higher than that obtained using ultracentrifugation (228 protein groups) and a commercial coprecipitation kit (252 protein groups). Moreover, 73.4% of the protein groups could be identified in at least two of the three replicates by the TiO2-based method. Gene ontology analysis (DAVID Bioinformatics Resources34) of cell components of the identified proteins shows that the extracellular exosome is the most enriched item in the exosomes isolated by TiO2; however, only the third was enriched in exosomes isolated using ultracentrifugation and a commercial coprecipitation kit (Fig. S8†), indicating that higher isolation purity is achieved by the TiO2-based strategy. To acquire detailed information about the composition of the isolated exosomes, we further compared the exosomal protein groups identified in serum exosomes isolated by these three methods. Overall, 307, 171 and 187 exosomal protein groups were identified in serum exosomes isolated using TiO2, ultracentrifugation and a coprecipitation kit (ExoCarta database, http://www.exocarta.org/), respectively. Among the exosomal protein groups identified in serum exosomes isolated using ultracentrifugation and a coprecipitation kit, 79% and 76% were found to be overlapped in the TiO2-based method. Moreover, 144 out of the 190 protein groups uniquely obtained by our TiO2-based method are reported as exosome proteins in the ExoCarta exosome database.35 Gene ontology analysis (DAVID Bioinformatics Resources)34 shows that the cellular component of 127 out of the 190 protein groups is located in exosomes, which further indicates the reliability of our method. To further evaluate our method, we compared the quantity of the commonly found exosome proteins and contaminating proteins identified in the exosomes isolated by the three strategies using mass spectrometry analysis. In Fig. 5C, we listed 18 typical exosome markers of the top 100 proteins that are commonly identified in exosomes (ExoCarta). After normalization using the corresponding protein quantity obtained by ultracentrifugation as the reference, the log fold differences of the quantity of the 18 exosome markers obtained using TiO2 and a coprecipitation kit are shown in Fig. 5C. The quantity of most of the exosome markers is obviously higher (positive log value in the figure) in the TiO2 enriched sample compared with that isolated using ultracentrifugation and a coprecipitation kit. In contrast, the quantity of the common contaminants in serum (six highly abundant proteins, which represent approximately 85% of the total protein mass36), such as albumin (ALB), IgG (IGHG1–IGHG4), IgA (IGHA1 and IGHA2), haptoglobin (HP) and transferrin (TF), is obviously lower in the TiO2 enriched sample (negative log value in the figure) than that obtained using ultracentrifugation and a coprecipitation kit. In summary, the TiO2-based strategy shows improved exosome enrichment and reduced non-specific protein adsorption compared with the currently available methods for human serum exosome analysis.Then, in order to establish a facile, fast and individualized micro-serum exosome analysis strategy such as fingertip blood test, the volume of serum was further decreased to 1 μL. The serum before isolation and after isolation, washing buffer (first wash, second wash and third wash) and eluted exosomal proteins were separated by SDS-PAGE and stained with Coomassie Brilliant Blue. As shown in Fig. 5D, the band of human serum albumin (about 70 kDa) was visible in serum and the supernatant after isolation while it gradually faded with the increase of washings. The significant fading of human serum albumin in exosomal proteins indicated that our proposed TiO2-based exosome isolation method could reduce the interference of contaminating proteins after sufficient washing. The expression of exosomal proteins was also examined by western-blot analysis. TSG101 and CD9, the commonly used exosomal marker proteins, were enriched in serum and the eluent of isolated exosomes but almost undetectable in the supernatant after isolation (Fig. 5E). We also evaluated the recovery and assay time required for exosome isolation from serum samples using the TiO2-based method and other reported strategies, including ultracentrifugation, density gradient centrifugation, size exclusion chromatography, ultrafiltration, immuno-affinity and precipitation based on polymers. As shown in Table S1,† the exosome recovery reaches 80% in 5 minutes of incubation from 1 μL serum by the TiO2-based method (isolation efficiency is obtained by measuring the band intensity of TSG101 extracted from the isolated exosomes over total serum). Compared with published data of the other methods, our proposed method is the most efficient for serum exosome isolation. The number of identified proteins using different amounts of serum (1–200 μL) is listed in Table S2† (ESI†).
Serum exosomal proteins from healthy donors and pancreatic cancer patients
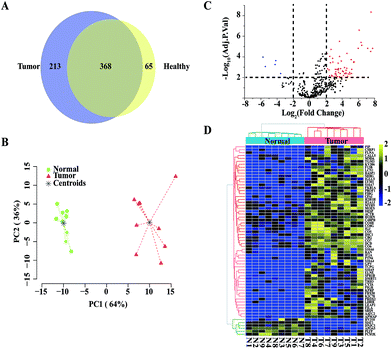
Next, we applied our TiO2-based exosome isolation strategy for proteome biomarker screening of pancreatic cancer. Pancreatic cancer is a highly lethal malignancy that is usually diagnosed at a later stage. Early diagnosis is essential to increase the chance of survival, so it is imperative to discover biomarkers for the detection of pancreatic cancer. Numerous studies have reported that exosomes carry proteins that provide valuable information for cancer diagnosis.4,12,25 Therefore, exosome protein profiles of healthy donors and pancreatic cancer patients were compared in our study. Using our TiO2-based strategy, exosomes were isolated from each serum sample of 9 healthy donors and 9 pancreatic cancer patients. In total, 433 and 581 serum exosomal protein groups were identified from the healthy group and the pancreatic cancer group, respectively. A total of 368 proteins were common in the healthy donors and the pancreatic cancer patients, while 65 proteins were unique in the healthy group and 213 proteins were unique in the pancreatic cancer group (Fig. 6A). Quantitative comparison of the two groups was conducted using label-free quantification intensity-based absolute quantification (iBAQ).37 Proteins identified more than three times in either group were used for relative quantification. Based on the results of exosomal protein quantification, we performed PCA. As shown in Fig. 6B, the two groups are well separated, indicating a significant difference between the healthy group and the pancreatic cancer group. Next, we performed a moderated t-statistic analysis to screen significantly regulated proteins in each group. Compared to the healthy group, 59 up-regulated proteins and 6 down-regulated proteins were identified in the tumour group (moderate t-statistic, Adj. P-Val < 0.01, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) FC > 2; Fig. 6C and Table S3†). The samples from the two groups were re-grouped by unsupervised hierarchical cluster analysis using expression data of the 65 differential proteins (Fig. 6D). As expected, the pancreatic cancer group and the healthy group could be well separated, demonstrating the potential of using exosomal protein profiling for biomarker screening for pancreatic cancer patients.
FC > 2; Fig. 6C and Table S3†). The samples from the two groups were re-grouped by unsupervised hierarchical cluster analysis using expression data of the 65 differential proteins (Fig. 6D). As expected, the pancreatic cancer group and the healthy group could be well separated, demonstrating the potential of using exosomal protein profiling for biomarker screening for pancreatic cancer patients.
Finally, the biological implications of the significantly regulated proteins were studied. Thirty up-regulated proteins in the serum exosomes of the pancreatic cancer patients were previously reported to be closely related to pancreatic cancer, including S100A4,38 S100A6,39,40 S100A9,40 Profilin-1,40–42 RAN,43 CALML5,44 TXN,45 PRDX2,46 and FGG.41,43 For other up-regulated proteins (HIST1H2BH, SPRR1B, and SBSN), though not reported in pancreatic cancer, evidence has demonstrated correlations with other cancers, such as neoplastic giant cell tumours,47 oral squamous cell cancer48 and esophageal squamous cell carcinoma.49 While the rest of the up-regulated proteins were not previously found to have direct association with cancers, these differential exosomal proteins may be considered as indicators of cancer progression.
Conclusions
In conclusion, we proposed a novel strategy for isolation of serum exosomes based on their specific interaction with TiO2. Pseudo-second order kinetics indicated a chemical specific adsorption mechanism in the TiO2-based exosome enrichment. Based on this principle, a high separation recovery (93.4%) is obtained within 5 min with good reproducibility for model exosomes. The captured exosomes can be either eluted to obtain intact exosomes or directly lysed for downstream proteome analysis. We further applied it to human serum samples and compared it with the traditional methods (ultracentrifugation and coprecipitation kit). The results demonstrated that the TiO2-based serum exosome isolation process is a simple and rapid method with high efficiency for serum exosome research compared with commonly used methods. Then, we compared the serum exosomal protein profile of pancreatic cancer patients with that of healthy donors. Proteomics analysis reveals that many differentially expressed proteins are closely associated with the development and progression of cancer, indicating the potential of this strategy for large-scale exosome-based biomarker screening for cancer diagnosis.Experimental
Collection of serum samples
Peking University Cancer Hospital and Peking University Third Hospital's Institutional Review Board approved the use of human serum samples. Blood samples from healthy donors and pancreatic cancer patients were obtained from consented donors. The normal control serum used in this investigation was obtained from one healthy volunteer. Twenty millilitres of peripheral blood were collected in tubes, allowed to sit at room temperature for 1 hour, and centrifuged at 2000 × g for 10 minutes. The separated serum was stored at −80 °C until use.Cell culture
HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco, UK) supplemented with 10% (v/v) fetal bovine serum (FBS, Gibco, UK), 100 U mL−1 penicillin and 100 mg mL−1 streptomycin (HyClone, USA) and incubated at 37 °C in a 5% CO2 humidified incubator. Once 80% confluency was achieved, the supernatant was carefully removed and the cells were washed twice with PBS. Next, the cells were cultured in exosome-free medium (DMEM with 10% exosome-depleted FBS) for 24 h and the culture medium was collected. Exosome-depleted FBS was prepared through ultracentrifugation at 110![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g overnight at 4 °C, followed by harvesting of the supernatant.
000 × g overnight at 4 °C, followed by harvesting of the supernatant.
Preparation of model samples of exosomes
Model samples of exosomes were obtained from HeLa cell culture medium by differential ultracentrifugation, according to the protocol described by Théry et al.50 with some modifications. In brief, the collected medium was centrifuged at 300 × g for 20 min, 2000 × g for 20 min and then 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g (19776 rotor, Sigma, USA) for 30 min to remove cells, dead cells and cell debris. Afterwards, the supernatants were ultracentrifuged at 110
000 × g (19776 rotor, Sigma, USA) for 30 min to remove cells, dead cells and cell debris. Afterwards, the supernatants were ultracentrifuged at 110![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g (SW 32 Ti rotor, Beckman Coulter, USA) for 70 min at 4 °C to pellet the crude exosomes. The pellets were washed twice with PBS, resuspended in a defined amount of PBS and stored at −80 °C.
000 × g (SW 32 Ti rotor, Beckman Coulter, USA) for 70 min at 4 °C to pellet the crude exosomes. The pellets were washed twice with PBS, resuspended in a defined amount of PBS and stored at −80 °C.
Characterization of model samples of exosomes
TiO2-based isolation of model samples and performance verification
Model samples derived from cultured HeLa cells were prepared by differential centrifugation as described above. Exosomes with a protein concentration of approximately 2 μg were diluted into DMEM containing no phenol red (Gibco, UK) to a final volume of 50 μL. The samples were mixed with 2 mg TiO2 microspheres and incubated for 5 min at 4 °C on a thermos-shaker to allow sufficient attachment. After simple centrifugation, the supernatant was discarded and the pellets were thoroughly rinsed thrice with PBS to remove non-specific molecules adsorbed on the microsphere surface.The exosomes attached to TiO2 were visualized by scanning electron microscopy (SEM), transmission electron microscopy (TEM) and immunofluorescence.
For SEM analysis, the TiO2 microspheres were first fixed with 1 mL of 2.5% glutaraldehyde in PBS for 2 h. After washing thrice with PBS, the fixed samples were dehydrated with a graded series of ethanol (30%, 50%, 75%, 90%, 95%, 100%, and 100%), each for 15 min. Then the samples were vacuum-dried overnight. The dried samples were spread on a double-sided conductive carbon tape and coated with gold using a sputter device for 60 s. Finally, images of TiO2 were generated using a ZEISS EVO LS10 SEM.
For TEM analysis, the TiO2 microspheres were first fixed with 1 mL of 2.5% glutaraldehyde in PBS for 2 h. After washing thrice with PBS, the particles were resuspended in PBS. Next, PBS solution containing TiO2 (20 μL) was added onto 200-mesh formvar carbon coated copper grids and allowed to adsorb to the formvar for 10 min. The excess solution was removed by blotting the edge of each grid with filter paper. Then, the sample was negatively stained with saturated uranyl acetate solution and incubated for 1 min at room temperature. The unevaporated solution was absorbed using filter paper and the grid was air-dried for TEM imaging. The samples were examined using a Hitachi h-7650 transmission electron microscope at 80 kV.
Two exosomal protein markers, CD81 and TSG101, were detected by immunofluorescence to verify the attachment of exosomes to the TiO2 surface. Three isotype control groups were used, including untreated TiO2 microspheres, TiO2 incubated with HeLa lysis solution and TiO2 incubated with exosomal proteins. These TiO2 microspheres were first fixed with 4% formaldehyde in PBS for 30 min. For detection of TSG101, which is an intracellular protein, an extra step of permeabilization was needed. The samples were permeabilized with 0.2% Triton X-100/PBS for 5 min and washed thrice with PBS. After blocking in 5% BSA/PBS for 1 h at room temperature, the samples were incubated with anti-CD81 rabbit polyclonal antibody (1:1000) (ab79559, Abcam, Cambridge, UK) or anti-TSG101 rabbit polyclonal antibody (1:500) (14497-1-AP, Proteintech Group, Chicago, USA) overnight at 4 °C. Subsequently, the microspheres were stained with secondary FITC-conjugated goat anti-rabbit antibodies (1:25) (CWBIO, China) or TRITC-conjugated goat anti-rabbit antibodies (1:25) (CWBIO, China) for 1 h at room temperature. Images were acquired with a Zeiss LSM 880 confocal microscope and processed using ZEN software. The same imaging parameters were used between the control group and the experimental group.
Recovery of the exosomes
The semiquantitative Western blot method was employed in order to eliminate the effect of protein contamination in model exosomes on quantification. The detailed process was described as follows: model exosomes were homogeneously re-suspended in PBS and divided into several replicates. Each model sample contained approximately 2 μg protein, as measured using a Micro BCA Protein Assay Kit (23235, Pierce, USA). As a control, one of the replicates of model samples subjected to the lysis process without isolation by TiO2. The rest of the replicates subjected to the incubation and wash process and directly lysed from the microspheres' surface for 20 min on ice with lysis buffer (4% SDS, 0.1 M Tris–HCl, pH 7.6) supplemented with protease inhibitor cocktails (Roche, Swiss). The lysates were loaded on a 12% SDS-polyacrylamide gel and subjected to Western blot using anti-Tsg101 antibody. For recovery calculations, the band intensity was measured using ImageJ software (band intensity of TSG101 extracted from the isolated exosomes over the control group). Triplicate standardization samples were used to evaluate each variable in the isolation of exosomes.Elution of captured exosomes
For the release of the captured exosomes, alkaline solution was used as the elution buffer.31 The microspheres were first incubated with 10% NH3·H2O for 10 min at 4 °C on a thermos-shaker to allow sufficient elution. After centrifuging at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 3 minutes, the supernatant was quickly substituted by PBS using an ultrafiltration system (Millipore, 30 kDa membrane) and subsequently washed several times with PBS. The solution inside the filter tubes was collected for further studies.
000 × g for 3 minutes, the supernatant was quickly substituted by PBS using an ultrafiltration system (Millipore, 30 kDa membrane) and subsequently washed several times with PBS. The solution inside the filter tubes was collected for further studies.
Isolation of exosomes from serum
To evaluate the efficiency of the proposed method, the results were compared with two conventional methods, ultracentrifugation and commercial exosome precipitation reagent, for exosome isolation.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 45 min to remove large debris and microvesicles. Then, the supernatant was ultracentrifuged at 110
000 × g for 45 min to remove large debris and microvesicles. Then, the supernatant was ultracentrifuged at 110![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 2 h to pellet crude exosomes. Next, the pellets were resuspended in PBS, filtered with a 0.2 μm filter, and washed twice with PBS. Finally, the exosome pellet aggregated at the bottom of the tube was directly lysed to extract the proteins.
000 × g for 2 h to pellet crude exosomes. Next, the pellets were resuspended in PBS, filtered with a 0.2 μm filter, and washed twice with PBS. Finally, the exosome pellet aggregated at the bottom of the tube was directly lysed to extract the proteins.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 15 min and the supernatant was removed. The tube was centrifuged at 10
000 × g for 15 min and the supernatant was removed. The tube was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 30 seconds for spinning down residual liquid. Finally, the exosome pellet aggregated was suspended in PBS for downstream experiments.
000 × g for 30 seconds for spinning down residual liquid. Finally, the exosome pellet aggregated was suspended in PBS for downstream experiments.
Proteomics analysis
The exosome loaded TiO2 microspheres were suspended in 4% SDS buffer containing 1% protease inhibitor cocktail. The suspension was ultrasonicated on ice for 20 min, followed by centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g at 4 °C for 3 min. The supernatant was collected and digested by the filter aided sample preparation (FASP) method. Briefly, the supernatant was transferred into a spin filter column (30 kDa) and the SDS was removed by washing the sample three times with 8 M UA. Afterwards the proteins were reduced by 10 mM DTT at 37 °C for 4 h and alkylated by 20 mM IAA at room temperature in the dark for 1 h. Then the buffer was exchanged with 50 mM NH4HCO3 by washing the membrane three times. Free trypsin was added into the protein solution at a ratio of 1
000 × g at 4 °C for 3 min. The supernatant was collected and digested by the filter aided sample preparation (FASP) method. Briefly, the supernatant was transferred into a spin filter column (30 kDa) and the SDS was removed by washing the sample three times with 8 M UA. Afterwards the proteins were reduced by 10 mM DTT at 37 °C for 4 h and alkylated by 20 mM IAA at room temperature in the dark for 1 h. Then the buffer was exchanged with 50 mM NH4HCO3 by washing the membrane three times. Free trypsin was added into the protein solution at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 and incubated at 37 °C overnight. The tryptic digests were recovered by centrifugation and additional wash with 50 mM NH4HCO3. The obtained solution was vacuum-dried and then adjusted to 10 μL with 0.1% formic acid prior to LC-MS/MS analysis.
50 and incubated at 37 °C overnight. The tryptic digests were recovered by centrifugation and additional wash with 50 mM NH4HCO3. The obtained solution was vacuum-dried and then adjusted to 10 μL with 0.1% formic acid prior to LC-MS/MS analysis.
LC-MS/MS analysis was carried out using an easy nLC-1000 system coupled with a Q Exactive HF mass spectrometer (Thermo Fisher Scientific, USA) using an ESI nanospray source. Mobile phase A was composed of 0.1% FA in water, and 0.1% FA in ACN was prepared as mobile phase B. The total flow rate was 600 nL min−1, and the gradient was performed as follows: 6% to 9% buffer B for 8 min; 9% to 14% buffer B for 16 min; 14% to 30% buffer B for 36 min; 30% to 40% buffer B for 15 min; and 40% to 95% buffer B for 3 min. After eluting with 95% buffer for 7 min, the separation system was equilibrated with 6% buffer B for 5 min. The spray voltage was set at 2.3 kV. The MS/MS spectra were acquired in data-dependent acquisition mode, and the full mass scan was acquired for m/z from 300 to 1400 with a resolution of 120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000.
000.
Label-free quantification analysis
Serum samples from healthy donors and pancreatic cancer patients were used for exosomal protein identification and relative quantification. The raw LC-MS/MS data files were analyzed using MaxQuant (version 1.5.2.8), with the spectra searched against the Uniprot human database (updated on July 21st, 2015). For identification of the peptides, the mass tolerances were 20 ppm for initial precursor ions and 0.5 Da for fragment ions. Two missed cleavages in tryptic digests were allowed. Cysteine residues were set as static modification. Oxidation of methionine was set as the variable modification. Filtering for the peptide identification was set at a 1% false discovery rate (FDR).Bioinformatics analysis was mainly performed using our in-house freely available software Perseus (www.perseus-framework.org). Analysis steps were performed in the statistical analysis environment R. For quantitative analysis of the proteomics data, the iBAQ intensities of exosome proteins from healthy donors and pancreatic cancer patients were extracted from the MaxQuant result files to represent the final expression of a particular protein across samples. The expression matrix was then normalized by using the quartile normalization method as previously described.51,52 To make the statistical analysis more precise, proteins identified more than three times in either group were used for relative quantification (the healthy group and the pancreatic cancer group). After filtering, a 534 × 18 protein expression matrix was generated for statistical analysis. Unsupervised principal component analysis (PCA) was used to determine whether the exosome proteins could effectively distinguish the two groups. Moderated t-statistics was implemented with R package ‘limma’ to select exosome proteins that were differentially expressed between the healthy group and the pancreatic cancer group.53 The P values were corrected for multiple testing using the Benjamini–Hochberg procedure and significant calls were made based on adjusted P values < 0.01 and the absolute value of log(FC) > 2. All statistical analyses were performed in the R statistical programming language (version 3.2.1).54
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported by the National Key Program for Basic Research of China (No. 2017YFA0505002, 2018YFC0910302, 2016YFA0501403, 2018YFF0212505, and 2017YFC0906703), the National Natural Science Foundation of China (No. 21675172, 21606181) and the China Postdoctoral Science Foundation (2017M613331).References
- R. M. Johnstone, M. Adam, J. R. Hammond, L. Orr and C. Turbide, J. Biol. Chem., 1987, 262, 9412–9420 CAS
.
- H. Shao, H. Im, C. M. Castro, X. Breakefield, R. Weissleder and H. Lee, Chem. Rev., 2018, 118, 1917–1950 CrossRef CAS PubMed
.
- Y. Sun, S. Liu, Z. Qiao, Z. Shang, Z. Xia, X. Niu, L. Qian, Y. Zhang, L. Fan and C.-X. Cao, Anal. Chim. Acta, 2017, 982, 84–95 CrossRef CAS PubMed
.
- S. A. Melo, L. B. Luecke, C. Kahlert, A. F. Fernandez, S. T. Gammon, J. Kaye, V. S. LeBleu, E. A. Mittendorf, J. Weitz and N. Rahbari, Nature, 2015, 523, 177–182 CrossRef CAS PubMed
.
- T. Pisitkun, R. F. Shen and M. A. Knepper, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 13368–13373 CrossRef CAS PubMed
.
- T. Chen, Q. Y. Xi, R. S. Ye, X. Cheng, Q. E. Qi, S. B. Wang, G. Shu, L. N. Wang, X. T. Zhu, Q. Y. Jiang and Y. L. Zhang, BMC Genomics, 2014, 15, 100 CrossRef PubMed
.
- B. B. Shenoda and S. K. Ajit, Clin. Med. Insights: Pathol., 2016, 9(Suppl. 1), 1–8 Search PubMed
.
- K. E. Richards, A. E. Zeleniak, M. L. Fishel, J. Wu, L. E. Littlepage and R. Hill, Oncogene, 2017, 36, 1770–1778 CrossRef CAS PubMed
.
- J. M. Pitt, G. Kroemer and L. Zitvogel, J. Clin. Invest., 2016, 126, 1139–1143 CrossRef PubMed
.
- B. Costa-Silva, N. M. Aiello, A. J. Ocean, S. Singh, H. Zhang, B. K. Thakur, A. Becker, A. Hoshino, M. T. Mark and H. Molina, Nat. Cell Biol., 2015, 17, 816–826 CrossRef CAS PubMed
.
- G. Chen, A. C. Huang, W. Zhang, G. Zhang, M. Wu, W. Xu, Z. Yu, J. Yang, B. Wang and H. Sun, Nature, 2018, 560, 382–386 CrossRef CAS PubMed
.
- I.-H. Chen, L. Xue, C.-C. Hsu, J. S. P. Paez, L. Pan, H. Andaluz, M. K. Wendt, A. B. Iliuk, J.-K. Zhu and W. A. Tao, Proc. Natl. Acad. Sci. U. S. A., 2017, 201618088 Search PubMed
.
- Y. Li, Q. Zheng, C. Bao, S. Li, W. Guo, J. Zhao, D. Chen, J. Gu, X. He and S. Huang, Cell Res., 2015, 25, 981–984 CrossRef CAS PubMed
.
- T. Matsumura, K. Sugimachi, H. Iinuma, Y. Takahashi, J. Kurashige, G. Sawada, M. Ueda, R. Uchi, H. Ueo and Y. Takano, Br. J. Cancer, 2015, 113, 275–281 CrossRef CAS PubMed
.
- Q. Li, Y. Shao, X. Zhang, T. Zheng, M. Miao, L. Qin, B. Wang, G. Ye, B. Xiao and J. Guo, Tumor Biol., 2015, 36, 2007–2012 CrossRef CAS PubMed
.
- B. K. Thakur, H. Zhang, A. Becker, I. Matei, Y. Huang, B. Costa-Silva, Y. Zheng, A. Hoshino, H. Brazier and J. Xiang, Cell Res., 2014, 24, 766–769 CrossRef CAS PubMed
.
- T. Tian, H.-X. Zhang, C.-P. He, S. Fan, Y.-L. Zhu, C. Qi, N.-P. Huang, Z.-D. Xiao, Z.-H. Lu and B. A. Tannous, Biomaterials, 2018, 150, 137–149 CrossRef CAS PubMed
.
- B. J. Tauro, D. W. Greening, R. A. Mathias, H. Ji, S. Mathivanan, A. M. Scott and R. J. Simpson, Methods, 2012, 56, 293–304 CrossRef CAS PubMed
.
- S. Mathivanan, J. W. Lim, B. J. Tauro, H. Ji, R. L. Moritz and R. J. Simpson, Mol. Cell. Proteomics, 2010, 9, 197–208 CrossRef CAS PubMed
.
- F. Yang, X. Liao, Y. Tian and G. Li, Biotechnol. J., 2017, 12, 1600699 CrossRef PubMed
.
- M. Wu, Y. Ouyang, Z. Wang, R. Zhang, P.-H. Huang, C. Chen, H. Li, P. Li, D. Quinn and M. Dao, Proc. Natl. Acad. Sci. U. S. A., 2017, 201709210 Search PubMed
.
- S. Shin, D. Han, M. C. Park, J. Y. Mun, J. Choi, H. Chun, S. Kim and J. W. Hong, Sci. Rep., 2017, 7, 9907 CrossRef PubMed
.
- C. Liu, J. Guo, F. Tian, N. Yang, F. Yan, Y. Ding, J. Wei, G. Hu, G. Nie and J. Sun, ACS Nano, 2017, 11, 6968–6976 CrossRef CAS PubMed
.
- Z. Zhao, Y. Yang, Y. Zeng and M. He, Lab Chip, 2016, 16, 489–496 RSC
.
- J. M. Lewis, A. D. Vyas, Y. Qiu, K. S. Messer, R. White and M. J. Heller, ACS Nano, 2018, 12, 3311–3320 CrossRef CAS PubMed
.
- P. Li, M. Kaslan, S. H. Lee, J. Yao and Z. Gao, Theranostics, 2017, 7, 789–804 CrossRef CAS PubMed
.
-
T. E. Thingholm and M. R. Larsen, in Phospho-Proteomics, Springer, 2016, pp. 135–146 Search PubMed
.
- A. Leitner, TrAC, Trends Anal. Chem., 2010, 29, 177–185 CrossRef CAS
.
- A. Sano and H. Nakamura, Anal. Sci., 2007, 23, 1285–1289 CrossRef CAS PubMed
.
- Y. Huang, Q. Zhou, J. Xiao and G. Xie, J. Sep. Sci., 2010, 33, 2184–2190 CrossRef CAS PubMed
.
- T. E. Thingholm, T. J. D. Jørgensen, O. N. Jensen and M. R. Larsen, Nat. Protoc., 2006, 1, 1929–1935 CrossRef CAS PubMed
.
-
S. K. Lagergren, Sven. Vetenskapsakad. Handingarl, 1898, vol. 24, pp. 1–39 Search PubMed
.
- E. Cocucci and J. Meldolesi, Trends Cell Biol., 2015, 25, 364–372 CrossRef CAS PubMed
.
- D. W. Huang, B. T. Sherman and R. A. Lempicki, Nat. Protoc., 2008, 4, 44–57 CrossRef PubMed
.
- S. Mathivanan and R. J. Simpson, Proteomics, 2009, 9, 4997–5000 CrossRef CAS PubMed
.
- N. Zolotarjova, J. Martosella, G. Nicol, J. Bailey, B. E. Boyes and W. C. Barrett, Proteomics, 2005, 5, 3304–3313 CrossRef CAS PubMed
.
- B. Schwanhäusser, D. Busse, N. Li, G. Dittmar, J. Schuchhardt, J. Wolf, W. Chen and M. Selbach, Nature, 2011, 473, 337–342 CrossRef PubMed
.
- K.-X. Ai, L.-Y. Lu, X.-Y. Huang, W. Chen and H.-Z. Zhang, World J. Gastroenterol., 2008, 14, 1931–1935 CrossRef PubMed
.
- D. Vimalachandran, W. Greenhalf, C. Thompson, J. Lüttges, W. Prime, F. Campbell, A. Dodson, R. Watson, T. Crnogorac-Jurcevic and N. Lemoine, Cancer Res., 2005, 65, 3218–3225 CrossRef CAS PubMed
.
- K.-K. Kuo, C.-J. Kuo, C.-Y. Chiu, S.-S. Liang, C.-H. Huang, S.-W. Chi, K.-B. Tsai, C.-Y. Chen, E. Hsi, K.-H. Cheng and S.-H. Chiou, Pancreas, 2016, 45, 71–83 CrossRef CAS PubMed
.
- R. Chen, C. Y. Eugene, S. Donohoe, S. Pan, J. Eng, K. Cooke, D. A. Crispin, Z. Lane, D. R. Goodlett and M. P. Bronner, Gastroenterology, 2005, 129, 1187–1197 CrossRef CAS PubMed
.
- M. Grønborg, J. Bunkenborg, T. Z. Kristiansen, O. N. Jensen, C. J. Yeo, R. H. Hruban, A. Maitra, M. G. Goggins and A. Pandey, J. Proteome Res., 2004, 3, 1042–1055 CrossRef PubMed
.
- Z. Lu, L. Hu, S. Evers, J. Chen and Y. Shen, Proteomics, 2004, 4, 3975–3988 CrossRef CAS PubMed
.
- J. Park, E. Lee, K.-J. Park, H.-D. Park, J.-W. Kim, H. I. Woo, K. H. Lee, K.-T. Lee, J. K. Lee and J.-O. Park, OncoTargets Ther., 2017, 8, 42761–42771 Search PubMed
.
- H. Nakamura, J. Bai, Y. Nishinaka, S. Ueda, T. Sasada, G. Ohshio, M. Imamura, A. Takabayashi, Y. Yamaoka and J. Yodoi, Cancer Detect. Prev., 2000, 24, 53–60 CAS
.
- S. Suenaga, Y. Kuramitsu, Y. Wang, B. Baron, T. Kitagawa, J. Akada, K. Tokuda, S. Kaino, S.-I. Maehara and Y. Maehara, Anticancer Res., 2013, 33, 4821–4826 CAS
.
- C. P. Lau, J. S. Kwok, J. C. Tsui, L. Huang, K. Y. Yang, S. K. Tsui and S. M. Kumta, J. Cell. Biochem., 2017, 118, 1349–1360 CrossRef CAS PubMed
.
- Y. Michifuri, Y. Hirohashi, T. Torigoe, A. Miyazaki, J. Fujino, Y. Tamura, T. Tsukahara, T. Kanaseki, J. Kobayashi and T. Sasaki, Biochem. Biophys. Res. Commun., 2013, 439, 96–102 CrossRef CAS PubMed
.
- J. Zhu, G. Wu, Q. Li, H. Gong, J. Song, L. Cao, S. Wu, L. Song and L. Jiang, Sci. Rep., 2016, 6, 21549 CrossRef CAS PubMed
.
-
C. Théry, S. Amigorena, G. Raposo and A. Clayton, in Current Protocols in Cell Biology, John Wiley & Sons, Inc., 2006, pp. 3.22.21–23.22.29 Search PubMed
.
- B. M. Bolstad, R. A. Irizarry, M. Åstrand and T. P. Speed, Bioinformatics, 2003, 19, 185–193 CrossRef CAS PubMed
.
- B. Zhang, J. Wang, X. Wang, J. Zhu, Q. Liu, Z. Shi, M. C. Chambers, L. J. Zimmerman, K. F. Shaddox and S. Kim, Nature, 2014, 513, 382–387 CrossRef CAS PubMed
.
- G. K. Smyth, Stat. Appl. Genet. Mol. Biol., 2004, 3, 1–25 Search PubMed
.
-
R Core Team, R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria, 2013 Search PubMed
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8sc04197k |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2019 |